Abstract
Aims
We tested the hypothesis that the 9p21 risk locus promotes atherosclerosis by examining the association between rs10757278 and coronary artery disease (CAD) severity and progression determined by semi-quantitative angiographic scores.
Methods and results
The rs10757278 single nucleotide polymorphism (SNP) was genotyped as the marker for the 9p21 locus in 2334 Caucasian patients undergoing cardiac catheterization (mean age 63, male 67%). Angiographic CAD was assessed using two semi-quantitative scoring systems with one estimating severity (Gensini) and the other extent (Sullivan). A subset of 308 patients who underwent two or more coronary angiograms at least 6 months apart were examined for net change in Gensini and Sullivan scores over time to determine the rate of CAD progression by genotype and were further classified as ‘progressors’ or ‘non-progressors’ based on absolute change per year in angiographic severity score. We replicated the association between the rs10757278 SNP and myocardial infarction and binary (presence/absence) angiographic classifications of CAD. Furthermore, we observed a significant additive association with this SNP, and both severity and extent of CAD using angiographic scores, after adjustment for age, gender, body mass index, traditional cardiovascular risk factors, myocardial infarction, and statin use (Gensini P = 0.016, Sullivan P = 0.005). In addition, there was a significant linear association with CAD progression before and after adjustment for covariates (Gensini P = 0.023, Sullivan P = 0.003) with homozygotes for the risk variant having three-fold greater odds of CAD progression compared with the referent group.
Conclusion
The 9p21 risk locus is associated with angiographically defined severity, extent, and progression of CAD, suggesting a role for this locus in influencing atherosclerosis and its progression.
Keywords: Atherosclerosis, angiography, coronary disease, genetics, genomics, 9p21
See page 2977 for the editorial comment on this article (doi:10.1093/eurheartj/ehq333)
Introduction
Coronary artery disease (CAD) remains a significant health concern worldwide. While traditional risk factors account for much of this risk burden, heritable factors play a key role in the development of CAD.1 Unbiased genome-wide approaches have led to the identification of the 9p21.3 locus as a risk marker for myocardial infarction (MI) and prevalent CAD in predominantly Caucasian cohorts.2–5 This association has since been replicated in several studies and in non-Caucasian populations, and confirmed by two meta-analyses, making this one of the most robust genomic findings for coronary heart disease to date.6,7
A large prospective study demonstrated that 9p21 status is predictive of first revascularization in subjects with medically treated MI.8 In addition, recent functional studies have demonstrated enhanced expression of the non-coding RNA, ANRIL, in 9p21 carriers9 and this transcript has in turn been associated with greater atherosclerosis.10 More recently, deletion of the orthologous 70 kb non-coding interval on mouse chromosome 4 has provided direct evidence that the 9p21 CAD risk interval has a direct role in the regulation of CDKN2A/B expression and affects CAD progression by altering the dynamics of vascular cell proliferation.11 Despite these studies suggesting greater atherosclerotic activity as a potential mechanism, a positive and direct association between 9p21 carrier status and severity, extent, or progression of CAD is yet to be convincingly demonstrated in humans. This may in part be due to the inadequacy of phenotyping methods employed thus far to assess CAD severity, particularly as the effect size of common variants is often small.
Despite its many limitations, coronary angiography remains to be the gold standard for documenting the extent and severity of CAD. We sought to test the hypothesis that the 9p21 locus promotes atherosclerosis by examining its association with angiographically defined CAD severity and extent, as well as CAD progression by refining the phenotype using two validated semi-quantitative coronary scoring systems.
Methods
Study population
Study participants were recruited as part of the Emory Cardiology Biobank, consisting of 3492 consecutive patients enrolled prior to undergoing elective or emergent cardiac catheterization across three Emory Healthcare sites, between 2003 and 2008. Patients aged 20–90 years were interviewed to collect information on demographic characteristics, medical history, and behavioural (lifestyle) habits. Risk factor prevalence was determined by physician diagnosis and/or treatment for hypertension, hyperlipidaemia, and diabetes. Smoking was classified as non-smoker or ‘ever smoked’ if there was a lifetime history of smoking at least 100 cigarettes. Medical records were reviewed to confirm self-reported history of MI and other conditions as well as to document previous angiographic findings and prior coronary revascularization.
After excluding self-reported non-Caucasian ancestry, heart transplantation, missing or incomplete angiographic data and missing DNA/blood samples, 2334 subjects were deemed eligible for this analysis. The study was approved by the Institutional Review Board at Emory University, Atlanta, GA, USA. All subjects provided written informed consent at the time of enrolment.
Coronary angiography definitions and scoring
Two operators, evaluated all coronary angiograms by visual estimation of luminal narrowing in multiple segments based on a modified form of the AHA/ACC classification of the coronary tree.12 Using this data, coronary angiography phenotypes were estimated by the authors (R.S.P. and I.J.N.) including, any CAD > 50%, number of epicardial vessels with > 50% disease, left main and proximal vessel disease, and finally, quantitative angiographic scores using the Gensini and Sullivan extent systems.13,14 All coronary angiography evaluations were performed without the knowledge of genotype status.
The Gensini score quantifies severity of CAD by a nonlinear points system for the degree of luminal narrowing along with a multiplier for specific coronary tree locations, thereby weighting each lesion score for prognostic significance. The total of the lesion scores is summed to give a final Gensini score. Thus, multiple severe proximal lesions gain the highest score.13
The Sullivan Extent score quantifies the percentage of the coronary intimal surface area affected by atheroma, without specific weighting for the degree of luminal narrowing. The percentage involvement of each vessel is estimated and multiplied by a factor representative of the surface area of that vessel in relation to the entire coronary tree. We used a modified version based on segments of each vessel with reported disease to derive percentage involvement. Four segments of right coronary artery (RCA) each contributing 25%; three segments of left anterior descending artery (LAD) each contributing 33% with the proximal segment further subdivided into two; left circumflex artery divided into three segments each contributing 33%.14
To determine the intra-class correlation coefficient, 25 patient angiograms were randomly chosen and examined independently by the authors (R.S.P. and I.J.N.). Lesions were visually estimated and recorded by coronary artery tree segments and then used to calculate Gensini and Sullivan Extent scores. The intra-class correlation coefficients were estimated at 0.88 (95% CI 0.74–0.95) and 0.90 (0.77–0.96) for Gensini and Sullivan Extent scores, respectively, which indicates good inter-observer agreement.
A subset of 308 patients who had undergone two or more coronary angiographies at least 6 months apart, were identified and the two angiograms furthest apart in time were quantified using the Gensini and Sullivan Extent scores described above. Given the variation in times between angiographies, the net change in angiographic score was divided by number of years between angiographies to give Gensini and Sullivan extent ‘rates’ as proxies for progression. Subjects were also arbitrarily categorized as ‘progressors’ and ‘non-progressors’ based on a Gensini rate of change of >1 or ≤0.5 points/year, respectively (as a guide, one point is equivalent to a 25% lesion in the RCA). Similarly for the Sullivan Extent score, progression and non-progression was defined simply as >1% and ≤0.5% change/year, respectively.
Genotyping
Genotyping for all samples was carried out at deCODE genetics in Reykjavik, Iceland, as part of the ongoing collaborative studies, with rs10757278 chosen as the representative single nucleotide polymorphism (SNP) for the 9p21 region based on our group's prior work.3 All single SNP (rs10757278) genotyping was carried out using the Centaurus (Nanogen) platform.15 The quality of each Centaurus SNP assay was evaluated by genotyping each assay on the Caucasian (CEU) samples and comparing the results with the HapMap data. All assays had a mismatch rate less than 0.5%.
Statistical analyses
Continuous variables are presented as means (SD) and categorical variables as proportions (%) with one-way analysis of variance and χ2 tests used to determine differences by genotype. Variables were tested for normality with Kolmogorov–Smirnov statistics and (+1 natural log) transformed for purposes of parametric analyses. Reverse log-transformation was applied to obtain clinically interpretable values. Haploview 4.0 software was used to compute Hardy–Weinberg equilibrium and minor allele frequency for rs10757278.
Logistic and linear regression models were constructed to test the additive effect of the SNPs on CAD phenotypes including severity and extent, with the SNP coded as 0, 1, or 2 based on the number of risk (G) alleles. Analyses were repeated after adjusting for age, gender, BMI, diabetes, hypertension, hyperlipidaemia, smoking, statin use, and history of MI. Analyses were also repeated after excluding subjects with normal coronary arteries (smooth or less than 10% luminal irregularities) to ensure any observed effect on graded severity was not being driven by those without any CAD in whom risk allele frequency is expected to be significantly lower. Interaction terms were tested for association between the 9p21 SNP and significant determinants of CAD severity, followed by stratified analysis to evaluate significant interactions.
CAD progression was tested as both a continuous variable (change in angiographic score/year) and as a categorical variable (progression vs. non-progression) with regression coefficients and odds ratios (ORs) calculated accordingly. Analyses were adjusted for age, gender, diabetes, statin use, smoking, and baseline angiographic score at first catheterization. A two-tailed P value < 0.05 was considered significant. All statistical analyses were performed using SPSS 17.0 (Chicago, IL, USA).
Results
A total of 2334 self-reported Caucasians were genotyped for the rs10757278 SNP and included in this study. The observed genotypic frequencies were consistent with Hardy–Weinberg equilibrium (P = 0.11) with a risk allele frequency of 0.50 (G allele). Patient characteristics at baseline by rs10757278 genotype are shown in Table 1. The mean age (SD) was 63.9 years (11.1) with a range of 24–90 years. No significant differences in patient characteristics were observed between rs10757278 genotypes for traditional risk factors, laboratory parameters, or medication usage.
Table 1.
Patient characteristics by rs10757278 genotype
| Patient characteristics (n = 2334) | rs10757278 genotype |
|||
|---|---|---|---|---|
| AA (557) | AG (1206) | GG (571) | P-value | |
| Age (years) | 64.4 (11) | 63.6 (11.1) | 64.2 (11.3) | 0.26 |
| Male (%) | 67.9 | 67.4 | 66.0 | 0.78 |
| BMI | 29.1 (6.1) | 29.5 (6.2) | 29.1 (5.6) | 0.24 |
| Diabetes (%) | 26.8 | 30.0 | 27.4 | 0.30 |
| Glucose (mg/dL) | 120.6 (41.9) | 122.6 (45.9) | 119.3 (38.5) | 0.34 |
| Hypertension (%) | 65.2 | 68.6 | 67.9 | 0.36 |
| Systolic BP (mmHg) | 142 (23.5) | 138 (22.6) | 141.4 (22.7) | 0.13 |
| Hyperlipidaemia (%) | 67.0 | 70.5 | 71.0 | 0.25 |
| Total cholesterol (mg/dL) | 168.5 (38.9) | 169.4 (43.4) | 166.1 (45.1) | 0.40 |
| LDL (mg/dL) | 96.6 (33.9) | 96.8 (35.0) | 92.7 (36.1) | 0.12 |
| HDL (mg/dL) | 41.1 (12.8) | 40.9 (12.4) | 40.7 (12.5) | 0.89 |
| Ever smoked (%) | 58.7 | 60.5 | 61.7 | 0.59 |
| Statin use (%) | 72.5 | 76.5 | 77.9 | 0.19 |
| Beta-blocker (%) | 61.5 | 64.1 | 66.3 | 0.28 |
| Serum creatinine (mg/dL) | 1.10 (0.56) | 1.08 (0.55) | 1.06 (0.53) | 0.42 |
| Prior MI (%) | 29.4 | 33.5 | 36.7 | 0.04 |
| Acute coronary syndrome (%) | 12.3 | 9.6 | 10.8 | 0.24 |
| Prior CABG (%) | 20.1 | 23.2 | 25.6 | 0.09 |
| Prior PCI (%) | 38.2 | 43.2 | 45.8 | 0.03 |
| Ejection fraction (%) | 54.3 (10.8) | 53.7 (11.2) | 53.4 (11.9) | 0.48 |
| Angiographic CAD (>50%) (%) | 72.4 | 79.0 | 80.1 | 0.004 |
| Coronary disease burden | 0.003 | |||
| Normal (%) | 27.6 | 21.0 | 19.9 | |
| Single vessel (%) | 22.9 | 21.5 | 19.5 | |
| Multi-vessel (%) | 49.5 | 57.5 | 60.5 | |
| Left main (%) | 5.6 | 8.5 | 10.2 | 0.017 |
| Proximal disease (%) | 60.8 | 72.1 | 72.7 | <0.001 |
| Gensini score, median (IQR) | 10 (0–49) | 17 (3–64) | 19 (3–76) | 0.001 |
| Sullivan extent, median (IQR) | 15 (3–32) | 19.9 (5–36) | 21.5 (7–37) | <0.001 |
Mean (SD) or % unless indicated. CAD, coronary artery disease; CABG, coronary artery bypass grafting; PCI, percutaneous coronary intervention; IQR, inter-quartile range; BMI, body mass index; MI, myocardial infarction; BP, blood pressure.
As described previously, we noted a significant association in the prevalence of prior MI, with increasing copies of the risk allele (P = 0.04, Table 1) equating to an allelic OR of 1.18 (95% CI 1.04–1.34).3 Similarly, there were significant associations with prior percutaneous coronary intervention [OR 1.17 (1.04–1.32)], CABG [OR 1.17(1.02–1.34)], and angiographically significant CAD defined as at least one vessel with 50% disease compared with normal coronary artery patients [OR 1.25 (1.08–1.45)]. When further classified as normal, single, and multi-vessel disease, there was a significant association with greater risk allele frequency with increasing CAD severity (P = 0.003). Angiographic traits considered to be especially heritable,16 were also more common in the carriers of the risk allele: Left main [OR = 1.36 (1.10–1.68)] and proximal disease [OR 1.32 (1.13–1.54)].
9p21 association with semi-quantitative coronary artery disease severity and extent scores
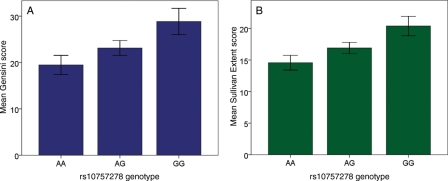
Table 1 shows the association between the rs10757278 genotype and severity/extent of CAD with respect to median Gensini and Sullivan Extent scores. There was a significant additive effect of the G allele on each measure of CAD. After adjusting for age, gender, BMI, traditional risk factors, statin use, and history of MI, the associations between rs10757278 and both scores remained significant (Gensini P = 0.016, Sullivan P = 0.005) (Table 2). Thus, possessing one copy of the risk variant equates to greater angiographic scores, which correspond to, for example, a 50% lesion in the proximal LAD, or 15% of the entire LAD intima area (Figure 1).
Table 2.
Multivariate predictors of CAD severity and extent
| log Gensini (CAD severity) |
log Sullivan (CAD extent) |
|||
|---|---|---|---|---|
| B (SE) | P-value | B (SE) | P-value | |
| Age (years) | 0.03 (<0.01) | <0.001 | 0.03 (<0.01) | <0.001 |
| Gender | 0.88 (0.07) | <0.001 | 0.70 (0.06) | <0.001 |
| BMI | −0.01 (0.01) | 0.028 | −0.01 (0.01) | 0.07 |
| Diabetes | 0.36 (0.08) | <0.001 | 0.34 (0.06) | <0.001 |
| Hyperlipidaemia | 0.30 (0.08) | <0.001 | 0.22 (0.06) | <0.001 |
| Hypertension | 0.08 (0.08) | 0.32 | 0.11 (0.06) | 0.06 |
| Smoking | 0.14 (0.07) | 0.05 | 0.19 (0.05) | <0.001 |
| Statin use | 0.83 (0.08) | <0.001 | 0.72 (0.06) | <0.001 |
| History of MI | 1.30 (0.07) | <0.001 | 0.89 (0.06) | <0.001 |
| rs10757278 | 0.12 (0.05) | 0.016 | 0.10 (0.04) | 0.005 |
Multivariate linear regression model includes age, gender, body mass index (BMI), diabetes, hypertension, hyperlipidaemia, smoking, statin use, history of myocardial infarction (MI), and rs10757278. Model R2 for log Gensini = 0.356 with R2 change for rs10757278 = 0.004; model R2 for log Sullivan = 0.343, R2 change for rs10757278 = 0.004. B (SE), unstandardized regression coefficient with standard error.
Figure 1.
Coronary artery disease (CAD) severity by rs10757278 genotype. CAD severity is illustrated here as (A) the mean Gensini score (P for trend < 0.001) and (B) the mean Sullivan Extent score (P for trend < 0.001), all adjusted for age, gender, body mass index, risk factors, statin use, myocardial infarction, and presented after reverse log-transformation. The risk allele is G.
Analyses were repeated after excluding subjects with normal coronary arteries in order to ensure that the observed effect was not being driven primarily by the absence of disease in one group. In this smaller group (n = 1849), the association with both Gensini and Sullivan Extent scores remained significant and independent of covariates (adjusted P = 0.03 for both).
Sensitivity analysis did not reveal any significant interactions with age, gender, or presence of diabetes, hypertension, hyperlipidaemia, statin use, or smoking (data not shown). However, we did observe an interaction with the history of MI (P = 0.03). Stratified analysis revealed no association between rs10757278 and CAD scores in subjects with MI (n = 751; Gensini P = 0.91, Sullivan P = 0.74), while those with no history of MI (n = 1583; Gensini and Sullivan P < 0.001) maintained a significant association with both scores.
9p21 association with coronary artery disease progression
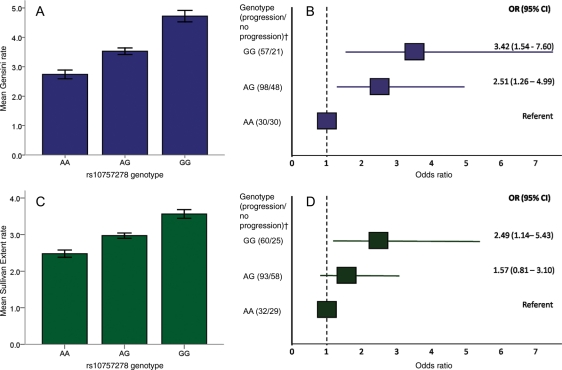
Of the 2334 patients, 308 were identified as having had repeat angiograms. These patients did not differ by genotype for basic characteristics and were similar to the main cohort (Table 3). The median length of time between angiographies was 4.5 years (IQR 2.5–7 years). There was a significant additive effect of the G allele on risk of progression when the net change in Gensini score per year was used to quantify progression (P = 0.023) with homozygotes for the risk allele progressing at a mean covariate adjusted rate of 5 Gensini points/year compared with the referent group progressing at under 3 points/year (Figure 2A). Furthermore when treated as a binary variable, heterozygotes for the risk allele were more than twice as likely to be progressors (Methods) [OR 2.51 (95% CI 1.26–4.99)] when compared with non-carriers, while homozygotes were greater than three times more likely [OR 3.42 (95% CI 1.54–7.6)] after adjustment for age, gender, diabetes, statin use, smoking, and baseline CAD Gensini score (Figure 2B). A similar trend was observed when the Sullivan Extent score was used to assess progression in this manner. After adjustment for the same covariates, we observed a significant association between rs10757278 and the net change in Sullivan Extent score/year (P = 0.003) as well as with a binary categorization of progression (Methods) [heterozygote OR 1.57 (95% CI 0.8–3.1); homozygote OR 2.49 (95% CI 1.1–5.4)] (Figure 2C and D).
Table 3.
Patient characteristics by rs10757278 genotype at baseline of those with two or more angiograms
| Patient characteristics (n = 308) | rs10757278 genotypea |
|||
|---|---|---|---|---|
| AA (63) | AG (158) | GG (87) | P-value | |
| Age (years) | 64.4 (11.2) | 64.8 (10.3) | 66.0 (11.6) | 0.61 |
| Male (%) | 71.4 | 67.1 | 64.4 | 0.66 |
| BMI | 30.2 (7.2) | 29.8 (6.2) | 29.3 (5.9) | 0.66 |
| Diabetes (%) | 31.7 | 35.4 | 31.1 | 0.75 |
| Hypertension (%) | 74.6 | 77.0 | 64.4 | 0.09 |
| Hyperlipidaemia (%) | 76.2 | 75.2 | 74.3 | 0.92 |
| Ever smoked (%) | 52.5 | 62.1 | 64.0 | 0.32 |
| Statin use (%) | 77.6 | 72.6 | 81.9 | 0.27 |
| Baseline Gensini score, median (IQR) | 7.5 (1–14) | 10 (3–24) | 12 (6–24) | 0.07 |
| Baseline Sullivan Extent score, median (IQR) | 11.6 (5–23) | 15 (5–27) | 17 (8–27) | 0.11 |
| Gensini rate, median (IQR) | 0.8 (0–6) | 2.0 (0–5) | 2.7 (1–8) | 0.04 |
| Sullivan Extent rate, median (IQR) | 1.1 (0–4) | 1.8 (0–5) | 3.0 (0–5) | 0.08 |
Mean (SD) or % unless indicated. IQR, inter-quartile range; BMI, body mass index.
aHardy–Weinberg equilibrium test: P = 0.6.
Figure 2.
Coronary artery disease (CAD) progression by rs10757278 genotype. CAD progression by genotype using angiographic scores is illustrated here as a continuous parameter showing the mean Gensini (A) and Sullivan extent rate (C), defined as the net change in score/years between procedures, by genotype. Odds ratios and 95% confidence intervals for progression vs. non-progression are also illustrated for Gensini (B) and Sullivan Extent scores (D). All values are adjusted for age, gender, diabetes, statin use, smoking, and baseline CAD angiographic score and presented after reverse log-transformation. Dagger (†) denotes patients with intermediate rates of change who were omitted for Gensini (n = 24) and Sullivan extent (n = 11) definitions of progression/non-progression (Methods). The risk allele is G.
Discussion
Using detailed angiographic data and thereby refining the phenotype, we demonstrated a positive association between the rs10757278 SNP and the Gensini and Sullivan Extent scores that define the severity and extent of angiographic CAD. Furthermore, we demonstrated that each copy of the risk allele leads to a higher risk of CAD progression over time. Our findings add significantly to the existing clinical and functional studies linking the 9p21 risk locus to atherosclerosis, by demonstrating an independent association with a quantitative CAD phenotype, and importantly with CAD progression.
A quantitative measure of CAD is preferable to a binary phenotype as it (i) avoids misclassification bias owing to the time-sensitive nature of coronary disease, (ii) gives a better indication of lifelong cumulative burden of disease, and (iii) may be more sensitive to the small effect size of common variants. We therefore chose to use two validated semi-quantitative angiographic scores which can be easily applied as a means to estimate severity and extent of CAD. While moderately correlated with each other (r = 0.7, P < 0.01), each score represents a slightly different aspect of CAD.
Our results demonstrate a significant association with rs10757278 for both scores using an additive genetic model. As an example, each copy of the risk allele contributes approximately one 50% lesion in the LAD. Even after excluding subjects with normal coronary arteries, whose inclusion may potentially be driving the effect given, they were shown to have a lower frequency of the risk allele, the additive trend persisted. Interestingly, while we confirmed an absence of any significant interactions between 9p21 and common risk factors, we did observe an interaction with MI, with a non-significant relationship in those who had a previous MI. This likely represents a skewed distribution of disease, as MI patients tend to have a greater degree of CAD burden at the upper range of Gensini and Sullivan Extent scores, and thus a smaller range of disease in which to identify a trend.
The positive association between the 9p21 risk genotype and graded severity of CAD is a novel finding, not previously shown in angiographic cohorts. Initial studies demonstrated association with the presence/absence of CAD, either defined clinically or by 50% disease criteria on angiography.5,17 Early studies failed to demonstrate an association between 9p21 risk genotype and CAD severity by the number of vessels affected in Asian populations. This may have been owing to their low power to detect small effects along with a relatively insensitive estimate of severity.18,19 Another study, by Anderson and colleagues, demonstrated that presence of CAD was correlated with 9p21 in 2100 Caucasian subjects but not with the extent as assessed by a vessel score and the Duke CAD index.20 Both scores may be insensitive to the changes expected, given the complexity of the disease and the small effect size of this SNP. The Duke CAD index is a validated hierarchical prognostic score, including only vessels with >50% disease and is less suited to quantifying multiple lesions. For example, a left main lesion with 95% luminal stenosis would score a maximum of 100 for the Duke CAD Index with no room to quantify further disease that may exist in other vessels, unlike the Gensini score. Population stratification or differences in clinical selection criteria for coronary invasive investigation may also account for the divergent results.
Importantly, our study adds additional information by demonstrating association with CAD progression over time. In subjects with repeat angiograms, we observed an additive effect of the risk allele on the rate of change of Gensini score per year. When classified as ‘progressors’ and ‘non-progressors’, homozygotes for the risk allele were three times more likely to be progressors compared with non-carriers, even when subjects without baseline coronary disease were excluded (data not shown). Similar findings were observed using the Sullivan Extent score to document progression. In contrast, one study based on a quantitative angiography analysis of subjects enrolled in a statin trial (treated 147, placebo 141) reported no evidence of progression over 2 years in relation to 9p21 genotype, despite post hoc calculations to suggest adequate power.21 This difference may be a consequence of strict patient selection or shorter follow-up time, compared with our study. On the other hand, supportive evidence comes from studies reporting progression of subclinical atherosclerosis with carotid intima media thickening22 and greater revascularization outcomes in carriers of the 9p21 risk allele.8
Our findings on the whole thus support the notion proposed by clinical and functional genomic studies that cell proliferation and atherosclerosis are mediated by this locus.9–11 In addition to studies associating this locus with coronary calcium scores23 and peripheral vascular disease,24 others have also reported association with intracranial aneurysms and arterial stiffness suggesting that the locus may also act outside of the traditional atherosclerotic pathway, perhaps by influencing vascular structure.25,26
Some strengths of our study include: (i) a large sample size, with a broad range of disease from normal to severe multi-vessel involvement enabling accurate assessment of severity; (ii) use of detailed coronary angiography phenotyping, moving beyond simple vessel scoring to carefully quantify disease burden and (iii) evaluation of progression of disease. There are also some important limitations to our study. First, the use of coronary angiography to visually quantify atherosclerosis is limited as remodelling may obscure substantial disease burden in arterial walls that can be detected by intravascular ultrasound,27,28 but relatively small and limited numbers of genomic ultrasound registries are available to date. Also, subjects undergoing first or repeat catheterization are a select group who are symptomatic or otherwise at high risk and thus may not be representative of the general population. Furthermore, variations in healthcare systems and referral patterns for angiography could also be a source of selection bias. Finally, we only ascertained the effect of one SNP in this region. However, this SNP was chosen as the marker of this region based on robust prior data and is in tight linkage disequilibrium with many other commonly used 9p21 markers (for example rs1333049, r2 = 1) and genotyping these would thus add little incremental value.
In conclusion, we have shown that the rs10757278 SNP at the 9p21 risk locus is associated with severity, extent, and progression of CAD in a population undergoing coronary angiography, suggesting a role for this locus in influencing atherosclerosis and its progression.
Funding
This work was supported by the American Heart Association (Postdoctoral Fellowship for R.S.P.), Robert W. Woodruff Health Sciences Center Fund, Emory Heart and Vascular Center Funds, and was supported in part by NIH grant UL1 RR025008 from the Clinical and Translational Science Award program, NIH grant R01HL089650-02, and from the Emory Neuroscience NINDS Core Facilities grant P30NS055077.
Conflict of interest: Authors whose affiliations are listed as deCODE Genetics are shareholders and/or employees of deCODE Genetics, Reykjavik, Iceland.
Acknowledgements
We would like to thank Salman Sher, Tanuj Kamineni, Felicia Warren, Erin Lyons, Imrana Qureshi, Hamid Syed, Shawn Arshad and Umair Janjua for help with patient recruitment and sample collection. We would also like to thank the consultants and fellows from the Divisions of Cardiology at Emory University and affiliated sites, as well as the staff of the cardiac catheterization laboratories for their continued support and assistance in recruiting patients for the Emory Cardiology Biobank.
References
- 1.Damani SB, Topol EJ. Future use of genomics in coronary artery disease. J Am Coll Cardiol. 2007;50:1933–1940. doi: 10.1016/j.jacc.2007.07.062. doi:10.1016/j.jacc.2007.07.062. [DOI] [PubMed] [Google Scholar]
- 2.Laurent S, Boutouyrie P. Arterial stiffness: a new surrogate end point for cardiovascular disease? J Nephrol. 2007;20(Suppl. 12):S45–S50. [PubMed] [Google Scholar]
- 3.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T, Jonasdottir A, Jonasdottir A, Sigurdsson A, Baker A, Palsson A, Masson G, Gudbjartsson DF, Magnusson KP, Andersen K, Levey AI, Backman VM, Matthiasdottir S, Jonsdottir T, Palsson S, Einarsdottir H, Gunnarsdottir S, Gylfason A, Vaccarino V, Hooper WC, Reilly MP, Granger CB, Austin H, Rader DJ, Shah SH, Quyyumi AA, Gulcher JR, Thorgeirsson G, Thorsteinsdottir U, Kong A, Stefansson K. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316:1491–1493. doi: 10.1126/science.1142842. doi:10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 4.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, Hinds DA, Pennacchio LA, Tybjaerg-Hansen A, Folsom AR, Boerwinkle E, Hobbs HH, Cohen JC. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007;316:1488–1491. doi: 10.1126/science.1142447. doi:10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, Dixon RJ, Meitinger T, Braund P, Wichmann HE, Barrett JH, Konig IR, Stevens SE, Szymczak S, Tregouet DA, Iles MM, Pahlke F, Pollard H, Lieb W, Cambien F, Fischer M, Ouwehand W, Blankenberg S, Balmforth AJ, Baessler A, Ball SG, Strom TM, Braenne I, Gieger C, Deloukas P, Tobin MD, Ziegler A, Thompson JR, Schunkert H. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. doi:10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schunkert H, Gotz A, Braund P, McGinnis R, Tregouet D-A, Mangino M, Linsel-Nitschke P, Cambien F, Hengstenberg C, Stark K, Blankenberg S, Tiret L, Ducimetiere P, Keniry A, Ghori MJR, Schreiber S, El Mokhtari NE, Hall AS, Dixon RJ, Goodall AH, Liptau H, Pollard H, Schwarz DF, Hothorn LA, Wichmann HE, Konig IR, Fischer M, Meisinger C, Ouwehand W, Deloukas P, Thompson JR, Erdmann J, Ziegler A, Samani NJ for the Cardiogenics Consortium. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008;117:1675–1684. doi: 10.1161/CIRCULATIONAHA.107.730614. doi:10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palomaki GE, Melillo S, Bradley LA. Association between 9p21 genomic markers and heart disease: a meta-analysis. JAMA. 2010;303:648–656. doi: 10.1001/jama.2010.118. doi:10.1001/jama.2010.118. [DOI] [PubMed] [Google Scholar]
- 8.Lina D, Notarangelo MF, Merlini PA, Berzuini C, Mannucci PM, Peyvandi F, Tubaro M, Foco L, Bernardinelli L, Ardissino D. Abstract 4011. Influence of rs1333040, a newly discovered 9p21.3 genetic variant, on clinical outcomes in early-onset myocardial infarction. Circulation. 2008;118:S_818. [Google Scholar]
- 9.Jarinova O, Stewart AF, Roberts R, Wells G, Lau P, Naing T, Buerki C, McLean BW, Cook RC, Parker JS, McPherson R. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler Thromb Vasc Biol. 2009;29:1671–1677. doi: 10.1161/ATVBAHA.109.189522. doi:10.1161/ATVBAHA.109.189522. [DOI] [PubMed] [Google Scholar]
- 10.Holdt LM, Beutner F, Scholz M, Gielen S, Gabel G, Bergert H, Schuler G, Thiery J, Teupser D. ANRIL expression is associated with atherosclerosis risk at chromosome 9p21. Arterioscler Thromb Vasc Biol. 2010;30:620–627. doi: 10.1161/ATVBAHA.109.196832. doi:10.1161/ATVBAHA.109.196832. [DOI] [PubMed] [Google Scholar]
- 11.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412. doi: 10.1038/nature08801. doi:10.1038/nature08801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Austen WG, Edwards JE, Frye RL, Gensini GG, Gott VL, Griffith LS, McGoon DC, Murphy ML, Roe BB. A reporting system on patients evaluated for coronary artery disease. Report of the Ad Hoc Committee for Grading of Coronary Artery Disease, Council on Cardiovascular Surgery, American Heart Association. Circulation. 1975;51(suppl):5–40. doi: 10.1161/01.cir.51.4.5. [DOI] [PubMed] [Google Scholar]
- 13.Gensini G. Coronary arteriography. New York: NY: Futura Publishing Co.; 1975. [Google Scholar]
- 14.Sullivan DR, Marwick TH, Freedman SB. A new method of scoring coronary angiograms to reflect extent of coronary atherosclerosis and improve correlation with major risk factors. Am Heart J. 1990;119:1262–1267. doi: 10.1016/s0002-8703(05)80173-5. doi:10.1016/S0002-8703(05)80173-5. [DOI] [PubMed] [Google Scholar]
- 15.Kutyavin IV, Milesi D, Belousov Y, Podyminogin M, Vorobiev A, Gorn V, Lukhtanov EA, Vermeulen NM, Mahoney W. A novel endonuclease IV post-PCR genotyping system. Nucleic Acids Res. 2006;34:e128. doi: 10.1093/nar/gkl679. doi:10.1093/nar/gkl679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fischer M, Broeckel U, Holmer S, Baessler A, Hengstenberg C, Mayer B, Erdmann J, Klein G, Riegger G, Jacob HJ, Schunkert H. Distinct heritable patterns of angiographic coronary artery disease in families with myocardial infarction. Circulation. 2005;111:855–862. doi: 10.1161/01.CIR.0000155611.41961.BB. doi:10.1161/01.CIR.0000155611.41961.BB. [DOI] [PubMed] [Google Scholar]
- 17.Muendlein A, Saely CH, Rhomberg S, Sonderegger G, Loacker S, Rein P, Beer S, Vonbank A, Winder T, Drexel H. Evaluation of the association of genetic variants on the chromosomal loci 9p21.3, 6q25.1, and 2q36.3 with angiographically characterized coronary artery disease. Atherosclerosis. 2009;205:174–180. doi: 10.1016/j.atherosclerosis.2008.10.035. doi:10.1016/j.atherosclerosis.2008.10.035. [DOI] [PubMed] [Google Scholar]
- 18.Chen Z, Qian Q, Ma G, Wang J, Zhang X, Feng Y, Shen C, Yao Y. A common variant on chromosome 9p21 affects the risk of early-onset coronary artery disease. Mol Biol Rep. 2009;36:889–893. doi: 10.1007/s11033-008-9259-7. [DOI] [PubMed] [Google Scholar]
- 19.Hinohara K, Nakajima T, Takahashi M, Hohda S, Sasaoka T, Nakahara K, Chida K, Sawabe M, Arimura T, Sato A, Lee BS, Ban JM, Yasunami M, Park JE, Izumi T, Kimura A. Replication of the association between a chromosome 9p21 polymorphism and coronary artery disease in Japanese and Korean populations. J Hum Genet. 2008;53:357–359. doi: 10.1007/s10038-008-0248-4. doi:10.1007/s10038-008-0248-4. [DOI] [PubMed] [Google Scholar]
- 20.Anderson JL, Horne D, Kolek MJ, Muhlestein JB, Mower CP, Park JJ, May HT, Camp NJ, JF C. Genetic variation at the 9p21 locus predicts angiographic coronary artery disease prevalence but not extent and has clinical utility. Am Heart J. 2008;156:1155–1162. doi: 10.1016/j.ahj.2008.07.006. e1152 doi:10.1016/j.ahj.2008.07.006. [DOI] [PubMed] [Google Scholar]
- 21.Chen SN, Ballantyne CM, Gotto AM, Jr., Marian AJ. The 9p21 susceptibility locus for coronary artery disease and the severity of coronary atherosclerosis. BMC Cardiovasc Disord. 2009;9:3. doi: 10.1186/1471-2261-9-3. doi:10.1186/1471-2261-9-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ye S, Willeit J, Kronenberg F, Xu Q, Kiechl S. Association of genetic variation on chromosome 9p21 with susceptibility and progression of atherosclerosis: a population-based, prospective study. J Am Coll Cardiol. 2008;52:378–384. doi: 10.1016/j.jacc.2007.11.087. doi:10.1016/j.jacc.2007.11.087. [DOI] [PubMed] [Google Scholar]
- 23.Assimes TL, Knowles JW, Basu A, Iribarren C, Southwick A, Tang H, Absher D, Li J, Fair JM, Rubin GD, Sidney S, Fortmann SP, Go AS, Hlatky MA, Myers RM, Risch N, Quertermous T. Susceptibility locus for clinical and subclinical coronary artery disease at chromosome 9p21 in the multi-ethnic ADVANCE Study. Hum Mol Genet. 2008;17:2320–2328. doi: 10.1093/hmg/ddn132. doi:10.1093/hmg/ddn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, Jones GT, Rinkel GJ, Blankensteijn JD, Ronkainen A, Jaaskelainen JE, Kyo Y, Lenk GM, Sakalihasan N, Kostulas K, Gottsater A, Flex A, Stefansson H, Hansen T, Andersen G, Weinsheimer S, Borch-Johnsen K, Jorgensen T, Shah SH, Quyyumi AA, Granger CB, Reilly MP, Austin H, Levey AI, Vaccarino V, Palsdottir E, Walters GB, Jonsdottir T, Snorradottir S, Magnusdottir D, Gudmundsson G, Ferrell RE, Sveinbjornsdottir S, Hernesniemi J, Niemela M, Limet R, Andersen K, Sigurdsson G, Benediktsson R, Verhoeven EL, Teijink JA, Grobbee DE, Rader DJ, Collier DA, Pedersen O, Pola R, Hillert J, Lindblad B, Valdimarsson EM, Magnadottir HB, Wijmenga C, Tromp G, Baas AF, Ruigrok YM, van Rij AM, Kuivaniemi H, Powell JT, Matthiasson SE, Gulcher JR, Thorgeirsson G, Kong A, Thorsteinsdottir U, Stefansson K. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224. doi: 10.1038/ng.72. doi:10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 25.Bilguvar K, Yasuno K, Niemelä M, Ruigrok YM, von Und Zu Fraunberg M, van Duijn CM, van den Berg LH, Mane S, Mason CE, Choi M, Gaál E, Bayri Y, Kolb L, Arlier Z, Ravuri S, Ronkainen A, Tajima A, Laakso A, Hata A, Kasuya H, Koivisto T, Rinne J, Ohman J, Breteler MM, Wijmenga C, State MW, Rinkel GJ, Hernesniemi J, Jääskeläinen JE, Palotie A, Inoue I, Lifton RP, Günel M. Susceptibility loci for intracranial aneurysm in European and Japanese populations. Nat Genet. 2008;40:1472–1477. doi: 10.1038/ng.240. doi:10.1038/ng.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel RS, Al Mheid I, Jorgensen JP, Uphoff I, Qureshi I, Ahmed Y, Aznaouridis K, Zhao J, Clements SD, Helgadottir A, Gulcher J, Zafari AM, Vaccarino V, Quyyumi AA. Abstract 3075. The chromosome 9p21 risk variant rs10757278 is associated with increased arterial stiffness. Circulation. 2008;118:S810. [Google Scholar]
- 27.Glagov S, Weisenberg E, Zarins CK, Stankunavicius R, Kolettis GJ. Compensatory enlargement of human atherosclerotic coronary arteries. N Engl J Med. 1987;316:1371–1375. doi: 10.1056/NEJM198705283162204. [DOI] [PubMed] [Google Scholar]
- 28.Nissen S. Coronary angiography and intravascular ultrasound. Am J Cardiol. 2001;87(suppl.):15A–20A. doi: 10.1016/s0002-9149(01)01420-5. doi:10.1016/S0002-9149(01)01420-5. [DOI] [PubMed] [Google Scholar]