The crystal structure of phenolic acid decarboxylase from B. pumilus strain UI-670 has been determined and refined at 1.69 Å resolution. The enzyme is a dimer, with each subunit adopting a β-barrel structure belonging to the lipocalin fold.
Keywords: phenolic acid, decarboxylases, lipocalin, biocatalysis
Abstract
The decarboxylation of phenolic acids, including ferulic and p-coumaric acids, to their corresponding vinyl derivatives is of importance in the flavouring and polymer industries. Here, the crystal structure of phenolic acid decarboxylase (PAD) from Bacillus pumilus strain UI-670 is reported. The enzyme is a 161-residue polypeptide that forms dimers both in the crystal and in solution. The structure of PAD as determined by X-ray crystallography revealed a β-barrel structure and two α-helices, with a cleft formed at one edge of the barrel. The PAD structure resembles those of the lipocalin-fold proteins, which often bind hydrophobic ligands. Superposition of structurally related proteins bound to their cognate ligands shows that they and PAD bind their ligands in a conserved location within the β-barrel. Analysis of the residue-conservation pattern for PAD-related sequences mapped onto the PAD structure reveals that the conservation mainly includes residues found within the hydrophobic core of the protein, defining a common lipocalin-like fold for this enzyme family. A narrow cleft containing several conserved amino acids was observed as a structural feature and a potential ligand-binding site.
1. Introduction
Phenolic acids such as ferulic and p-coumaric acids are important components of plant cell-wall lignocellulosic materials, forming cross-links between hemicellulose and lignin. During the breakdown of lignocellulose, these phenolic acid components are released from their esterified form by ferulic acid (feruloyl) esterases (Benoit et al., 2006 ▶; Topakas et al., 2007 ▶; Wong, 2006 ▶). While the resulting free phenolic acids are toxic to some bacteria (Zaldivar & Ingram, 1999 ▶), other organisms, including some yeasts and fungi, have evolved metabolic pathways that are capable of transporting and biotransforming these compounds (Rosazza et al., 1995 ▶). The production of 4-vinylguaiacol (VG) from ferulic acid (FA) is the result of decarboxylation catalyzed by a cofactor-free ferulic acid decarboxylase, which was first purified from Bacillus pumilus (formerly Pseudomonas fluorescens) strain UI-670 (Huang et al., 1993 ▶, 1994 ▶; Rosazza et al., 1995 ▶). To date, several bacterial ferulic acid decarboxylases, also known as p-coumarate decarboxylases or by the more generic term phenolic acid decarboxylases (PAD), have been cloned, purified and characterized, including those from several bacteria, Bacillus ssp. (Barthelmebs et al., 2001 ▶; Cavin et al., 1997 ▶, 1998 ▶; Prim et al., 2003 ▶; Yang et al., 2009 ▶; Zago et al., 1995 ▶), Lactobacillus plantarum (Cavin et al., 1997 ▶; Rodríguez et al., 2008 ▶) and Pediococcus pentosaceus (Barthelmebs et al., 2000 ▶), and that from the yeast Brettanomyces bruxellensis (Godoy et al., 2008 ▶). These enzymes are generally small in size (161–178 amino acids) and normally function as dimers. They display broad substrate specificity, decarboxylating several related phenolic acids, including ferulic, p-coumaric and hydroxycinnamic acids. For the decarboxylation of FA to VG, Rosazza and coworkers carried out deuterium-exchange studies that provided evidence for a non-oxidative mechanism involving an initial enzymatic isomerization of FA to a quinoid intermediate that spontaneously decarboxylates to yield the vinyl derivative (Huang et al., 1993 ▶; Rosazza et al., 1995 ▶).
The crystallization of PAD from L. plantarum (Rodríguez et al., 2007 ▶) and its crystal structure (PDB code 2w2a; Rodríguez et al., 2010 ▶) have been described. This same structure has independently been determined by the Joint Center for Structural Genomics (PDB code 2gc9). Importantly, the study by Rodríguez et al. (2010 ▶) demonstrated that all of the critical active-site residues, specifically Tyr18, Tyr20, Arg48 and Glu71, are located in the N-terminal half of the protein, dismissing the notion of the variable C-terminal region as being important for substrate specificity or catalysis (Barthelmebs et al., 2001 ▶). The enzyme from L. plantarum shows 65–68% sequence identity to B. pumilus PAD. The crystal structure of B. subtilus PAD (PDB code 2p8g; Joint Center for Structural Genomics, unpublished work), which has 81% sequence identity to the enzyme from B. pumilus, has also been deposited in the Protein Data Bank (Berman et al., 2000 ▶). We carried out this study to structurally characterize the PAD from B. pumilus strain UI-670. Here, we report its crystal structure as well as a comparative structural analysis with other PADs and related proteins with the lipocalin fold.
2. Materials and methods
2.1. Cloning, expression and purification
Escherichia coli JM109 [pKFAD] carrying a cloned PAD-encoding gene (previously designated fdc) from B. pumilus strain UI-670 in the pKK223-3 vector was used as described previously (Yang et al., 2009 ▶). All purification procedures were performed at 277 K on an ÄKTA Explorer 100 Air chromatography system (GE Healthcare, Baie d’Urfe, Quebec, Canada). The crude extract was loaded onto a HiPrep DEAE Sepharose FF column (16/10) equilibrated with 20 mM sodium phosphate buffer pH 7.0 at a flow rate of 1.5 ml min−1. The column was washed with the same buffer containing 0.15 M NaCl until no protein could be detected (A 280nm) in the flowthrough and the enzyme was subsequently eluted with a linear gradient of 0.15–0.6 M NaCl. Active fractions were pooled and solid ammonium sulfate was slowly added to a final concentration of 30%(w/v). The enzyme-containing solution was then loaded onto a Butyl-S Sepharose 6 FF (16/10) column which had been previously equilibrated with 30%(w/v) ammonium sulfate in 20 mM sodium phosphate buffer pH 7.0. PAD was eluted using a linear gradient of 30–0%(w/v) ammonium sulfate. Active fractions were collected, pooled and concentrated by ultrafiltration with a membrane exclusion size of 10 kDa in a 50 ml Amicon stirred cell (Millipore, Billerica, Massachusetts, USA) and applied onto a HiLoad Superdex 200 prep-grade size-exclusion column (16/60) which was previously equilibrated with 20 mM sodium phosphate buffer pH 7.0 containing 150 mM NaCl. The protein was eluted with the same buffer (at a flow rate of 1.5 ml min−1) and collected in 2 ml fractions. During each chromatographic step the protein profile was monitored by its absorbance at 280 nm. The protein concentration was determined as described by Bradford (1976 ▶) and alternatively by a microbiuret method (Itzhaki & Gill, 1964 ▶). Bovine serum albumin was used as a protein standard.
2.2. Dynamic light scattering, ESI-MS and native PAGE
Dynamic light scattering (DLS) was performed at 295 K using a Wyatt plate reader (Wyatt Technologies, Santa Barbara, California, USA) at a protein concentration of 2 mg ml−1. Prior to DLS analysis, samples were centrifuged (11 600g, 10 min, 277 K) to remove dust and large aggregates. Analysis of a wild-type PAD sample by electrospray ionization time-of-flight (ESI-TOF) mass spectrometry was performed using a Waters Q-TOF mass spectrometer; the mass spectrum was analyzed using MassLynx v.4.0 software. Native PAGE analysis was performed on a 12.5%(w/v) polyacrylamide gel followed by staining with Coomassie Brilliant Blue.
2.3. Crystallization
Purified protein in buffer (20 mM Tris–HCl pH 7.2, 150 mM NaCl) was concentrated by ultrafiltration to 21.3 mg ml−1 as determined using the Bio-Rad protein assay (Bio-Rad Laboratories). Crystallization screening was performed by sitting-drop vapour diffusion using a Hydra-Plus-One crystallization robot (Thermo Scientific/Matrix Technologies, Hudson, New Hampshire, USA) by mixing 0.3 µl protein solution with 0.4 µl reservoir solution. Crystallization plates were then imaged using a CrystalFarm imaging system running Crystal Farm Navigator software (Bruker AXS, Madison, Wisconsin, USA). Initial crystallization conditions were obtained using the Classics I and Classics II screens (Qiagen, Mississauga, Ontario, Canada) as well as our own in-house sparse-matrix screens. Two crystal forms were obtained, one from 0.2 M sodium/potassium tartrate, 0.1 M trisodium citrate pH 5.6, 2.0 M ammonium sulfate (prisms) and another from 2.5–3.5 M sodium malonate pH 7 (plates). Both crystal forms diffracted to better than 2.0 Å resolution. The final crystals used in data collection were obtained by hanging-drop vapour diffusion and were formed by mixing 1 µl protein solution (21.3 mg ml−1) in buffer (20 mM Tris–HCl pH 7.2, 150 mM NaCl) with 1 µl reservoir solution (0.2 M sodium tartarate, 0.1 M trisodium citrate pH 5.6, 0.5 M ammonium sulfate). For cocrystallization with either FA (Fluka) or p-coumaric acid (Sigma), the respective ligands were first dissolved in neat dimethyl sulfoxide or 2-propanol at a concentration of 100 mM and diluted tenfold with the protein in buffer; the drops were set up as in the absence of ligands.
2.4. Data collection, structure determination and refinement
Crystals obtained using the sodium malonate conditions were mounted directly from the drop using nylon loops and flash-cooled in an N2 cold stream at 100 K. Crystals grown using ammonium sulfate as the precipitant were transferred to 1.8 M sodium malonate pH 7 prior to flash-cooling as above. X-ray diffraction data were collected on the LRL-CAT beamline at sector 31 of the Advanced Photon Source, Argonne National Laboratory. The crystals belonged to space group P21212, with unit-cell parameters a = 92.11, b = 109.96, c = 45.43 Å and Z = 8 (Table 1 ▶). There are two molecules (one dimer) in the asymmetric unit, giving a Matthews coefficient of 2.69 Å3 Da−1 (Matthews, 1968 ▶) and an estimated solvent content of 53.9%. X-ray diffraction data were integrated and scaled using HKL-2000 (Otwinowski & Minor, 1997 ▶). The structure was solved using the program MOLREP (Vagin & Teplyakov, 1997 ▶) from the CCP4 suite (Collaborative Computational Project, Number 4, 1994 ▶) using the coordinates of PAD from B. subtilis (PDB code 2p8g) as the search model (Table 2 ▶). The model was refined using REFMAC5 (Murshudov et al., 1999 ▶) and fitted to the electron density using the program Coot (Emsley & Cowtan, 2004 ▶). During restrained refinement, each chain within the asymmetric unit was defined as a separate TLS group. A sulfate ion and water molecules were added towards the end of the refinement. The quality of the final refined model was evaluated using the MolProbity server (http://kinemage.biochem.duke.edu/molprobity/; Chen et al., 2010 ▶). Final refinement statistics are given in Table 3 ▶. Coordinates and structure factors for the B. pumilus PAD structure have been deposited in the RCSB PDB (Berman et al., 2000 ▶) under accession code 3nad.
Table 1. Sample information.
| Macromolecule details | |
| Database code | PDB code 3nad |
| Component molecules | Phenolic acid decarboxylase (PAD; EC 4.1.1.) |
| Macromolecular assembly | PAD is a dimer contained within the asymmetric unit |
| Mass (Da) | 19082 (method: mass spectrometry) |
| Source organism | B. pumilus strain UI-670 |
| Macromolecule production | |
| Cloning | PCR-amplified from genomic DNA, cloned into the vector pKK223-3 and expressed in E. coli JM109 |
| Crystallization and crystal data | |
| Crystallization method | Vapour diffusion |
| Temperature (K) | 293 |
| Apparatus | Linbro crystallization plates |
| Atmosphere | Room air |
| Crystal-growth time (d) | 23 |
| Additional details | Crystals were grown by mixing 1l protein with 1l reservoir solution and equilibrating over 0.5ml reservoir solution |
| Crystallization solutions | |
| Macromolecule | 1l PAD (21.3mgml1), 20mM TrisHCl pH 7.2, 150mM sodium chloride pH 7.2 |
| Precipitant | 1.0l 0.2M disodium tartrate, 0.1M trisodium citrate pH 5.6, 0.5M ammonium sulfate pH 5.6 |
| Crystal data | |
| Crystal colour | Clear light |
| Crystal shape | Plates or prisms |
| Crystal dimensions (mm) | 0.2 0.05 0.1 |
| Matthews coefficient V M (3Da1) | 2.69 |
| Solvent content (%) | 53.9 |
| Unit-cell data | |
| Crystal system, space group | Orthorhombic, P21212 |
| Unit-cell parameters (, ) | a = 92.11, b = 109.96, c = 45.43, = = = 90 |
| No. of molecules in unit cell Z | 8 |
Table 2. Data-collection and structure-solution statistics.
Values for the outer shell are given in parentheses.
| Diffraction protocol | Single wavelength |
| Wavelength () | 0.9793 |
| Detector | Rayonix 225 HE |
| Temperature (K) | 100 |
| Resolution range () | 70.51.69 (1.751.69) |
| No. of unique reflections | 52226 (4990) |
| No. of observed reflections | 304116 |
| Completeness (%) | 98.7 (95.7) |
| Multiplicity | 5.9 (4.9) |
| I/(I) | 10.5 (2.1) |
| R merge | 0.095 (0.603) |
| Data-processing software | HKL-2000 |
| Phasing method | Molecular replacement |
| Starting model data set | 2p8g |
| Alterations to search model | None |
| Solution software | MOLREP from CCP4 |
| Phasing set | 0.9793 |
| Radiation source | APS ID31 |
| Temperature (K) | 100 |
| Phasing resolution range () | 1.6970.5 |
Table 3. Structure refinement and model validation.
Values for the outer shell are given in parentheses.
| Refinement software | REFMAC |
| Refinement on | F |
| cutoff | F > 0(F) |
| Resolution range () | 50.01.69 (1.7311.687) |
| No. of reflections used in refinement | 49179 |
| Final overall R factor | 0.185 (0.258) |
| Atomic displacement model | Isotropic |
| Overall average B factor (2) | 19.4 |
| No. of protein atoms | 2682 |
| No. of nucleic acid atoms | 0 |
| No. of ligand atoms | 5 |
| No. of solvent atoms | 578 |
| Total no. of atoms | 3265 |
| Noncrystallographic symmetry restraints | None |
| Final R work | 0.185 (0.258) |
| No. of reflections for R free | 2953 (162) |
| Final R free | 0.204 (0.286) |
| Ramachandran plot analysis | |
| Most favoured regions (%) | 98.4 |
| Additionally allowed regions (%) | 1.3 |
| Generously allowed regions (%) | 0 |
| Disallowed regions (%) | 0.003 |
3. Results and discussion
3.1. PAD expression and purification
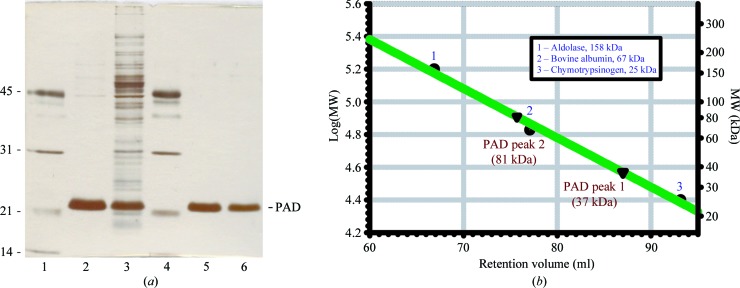
The PAD enzyme was overexpressed in E. coli JM109 and represented between 5 and 10% of the total soluble protein. A three-step purification procedure (Table 4 ▶) consisting of anion-exchange chromatography, hydrophobic interaction chromatography and gel filtration led to electrophoretically pure protein (Fig. 1 ▶ a). Interestingly, the second purification step using hydrophobic interaction chromatography on Butyl-S Sepharose revealed two distinct protein peaks which differed in the apparent quantity of protein based on the absorbance at 280 nm. Both elution peaks contained decarboxylase activity and showed only one protein band with a size of ∼22 kDa (calculated molecular mass of 19 kDa) when analyzed by SDS–PAGE. The mass of wild-type PAD was further confirmed using ESI-TOF MS analysis, giving a primary species with a mass of 19 082.5 Da, which is in good agreement with the calculated mass of 19 081 Da based on the sequence (NCBI gi:226348789). Size-exclusion chromatography showed that the native molecular weights of the two peaks following hydrophobic interaction chromatography were 37 and 81 kDa, respectively, suggesting the formation of homodimeric and homotetrameric species (Fig. 1 ▶ b). However, the homotetrameric form appeared to be a preparation artefact since it was unstable and dissociated to yield the homodimeric form (results not shown). Both DLS and SEC analysis of PAD are consistent with the enzyme behaving as a dimer in solution. Native PAGE showed that the protein runs as single discrete band, indicating that it is well behaved in solution.
Table 4. Purification of recombinant B. pumilus PAD expressed in E. coli .
n.a., not applicable.
| Purification step | Enzyme composition | Total protein (mg) | Total activity† (U) | Specific activity (Umg1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|---|
| Crude extract | n.a. | 945 | 6610 | 7 | 1 | 100 |
| DEAE Sepharose | n.a. | 144 | 8400 | 58 | 8 | 127 |
| Butyl-S Sepharose | 2 | 82 | 7950 | 97 | 14 | 120 |
| 4 | 3 | 582 | 194 | 28 | 9 |
The lower total units of activity in the crude extract fraction compared with subsequent purification steps is possibly the result of inhibition by some component of the culture medium.
Figure 1.
Purification of recombinant B. pumilus PAD expressed in E. coli. (a) SDS–PAGE of PAD. Lanes 1 and 4, molecular-mass standards as indicated on the left (kDa); lane 3, crude extract; lane 2, after DEAE-Sepharose chromatography; lane 5, peak 1 after Butyl-S chromatography; lane 6, peak 2 after Butyl-S chromatography. (b) Gel-filtration chromatography on Sephadex 200 pg. The two active enzyme fractions that show a single band on SDS–PAGE (lanes 5 and 6) differ in their native molecular weight (37 and 81 kDa, respectively). Molecular-mass markers are indicated as follows: (1) aldolase (158 kDa), (2) bovine albumin (67 kDa), (3) chymotrypsin (25 kDa).
3.2. Monomer structure
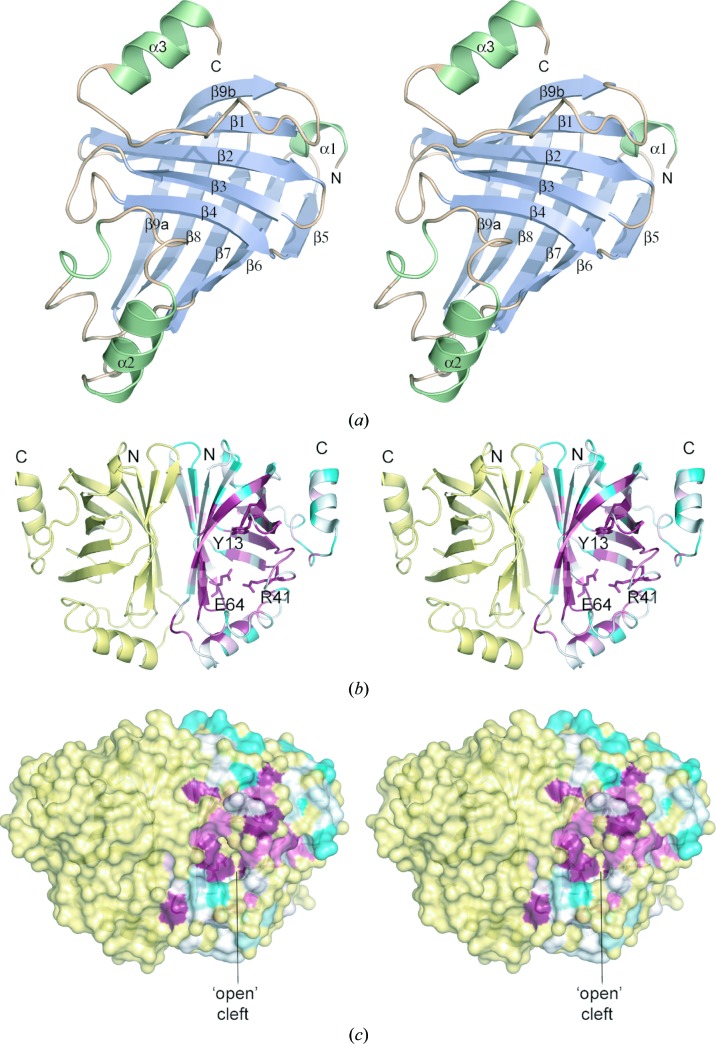
The crystal structure of B. pumilus PAD was determined using molecular replacement and refined at 1.69 Å resolution (Table 3 ▶). The model showed good geometry and included all residues (1–161) of chain A, residues 1–159 of chain B, 578 water molecules and one sulfate anion. The PAD structure consists almost entirely of β-sheets, with the exception of three surface-exposed short helices comprising residues Asp2–Val5 (α1), Ile106–Lys115 (α2) and, at the C-terminus, Gly151–Gly159 (α3). The basic core of the molecule is formed by two mutually perpendicular antiparallel five-stranded β-sheets, with the order of the β-strands being 9b-1-2-3-4 in one sheet and 5-6-7-8-9a in the other, with one of these β-strands (9a, 9b) being common to both β-sheets (Fig. 2 ▶ a). Together, these two β-sheets form a closed β-barrel structure. The structure belongs to the all-β class and adopts the lipocalin fold and superfamily as classified within the SCOP database (Murzin et al., 1995 ▶). Efforts to cocrystallize PAD with ferulic acid or p-coumaric acid were unsuccessful; this was a similar result to that obtained with L. plantarum PAD (Rodriguez et al., 2010 ▶).
Figure 2.
Crystal structure of B. pumilus PAD. (a) Structure of the PAD monomer with secondary-structure elements coloured pale blue (β-strands) or light green (α-helices). (b) Structure of the PAD dimer, with subunit A coloured by sequence conservation using the ConSurf server (http://consurf.tau.ac.il/; white represents unconserved and dark magenta represents highly conserved). The dimer is oriented to show the twofold axis relating the two monomers. Key active-site residues are shown in stick representation. (c) Molecular-surface representation of PAD coloured as in (b) showing the ‘open’ conformation of the active-site cleft in subunit A. These and subsequent figures were prepared using the program PyMOL (http://www.pymol.org).
Superposition of the two monomers revealed little difference in overall structure, with a root-mean-square deviation (r.m.s.d.) of 0.55 Å for 157 common Cα atoms. The exception is the loop between β1 and β2 (residues 13–18), in which the Cα atom of Asn15 is shifted by 3.8 Å. As a result of this conformational change, the active-site tunnels in the two subunits of the dimer do not have the same size and shape, with the tunnel being more ‘open’ in subunit A and more ‘closed’ in subunit B.
3.3. Dimer structure
Biochemical studies, including size-exclusion chromatography, of purified PAD enzymes from B. pumilus (this work; Huang et al., 1994 ▶; Zago et al., 1995 ▶), Br. bruxellensis (Godoy et al., 2008 ▶) and L. plantarum (Rodriguez et al., 2010 ▶) have shown these enzymes are homodimers, although the p-coumaric acid decarboxylase from L. plantarum was initially characterized as a homotetramer (Cavin et al., 1997 ▶). The solvent-accessible interface area consists of 1204 Å2 or 15.0% of the total subunit solvent-accessible surface as computed using the PISA server (Krissinel & Henrick, 2007 ▶). The PAD dimer is formed by interaction of the relatively flat antiparallel β-sheets, such that the strands of the two interacting β-sheets are parallel to one another (Fig. 2 ▶ b). Most of these contacts involve β6–β7–β8 of the two subunits and result in a total of 44 contacts (<3.5 Å) between the two monomers. This interface consists of both polar and apolar interactions, including a salt bridge between Lys121 NZ and Asp69 OD2 and hydrogen-bonding interactions (<3.2 Å) between Lys59 NZ and Ser71 O as well as between Thr54 OG1 and Glu78 OE2. Analysis of the contacting residues with respect to a multiple sequence alignment (Pfam; Finn et al., 2010 ▶) reveals that most of the residues are conserved, which is consistent with the dimeric arrangements of PADs from other organisms.
3.4. Comparison of PAD structures
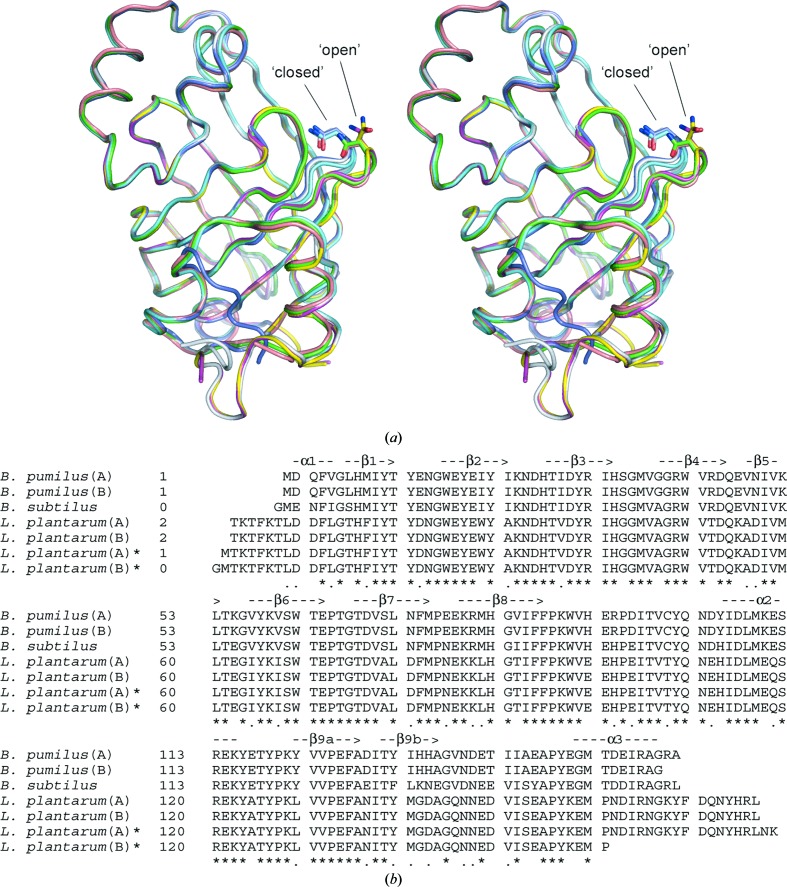
Among the currently available PAD structures, that of B. pumilus is the shortest enzyme (161 residues), with PAD from L. plantarum (PDB code 2w2a) being the longest (178 residues), although three residues are missing from the 2w2a model (Met1 and Asn177-Lys178). With respect to the B. pumilus PAD crystal structure, the L. plantarum structure contains extensions at both its N-terminus (Thr2–Asn8) and its C-terminus (Lys167–Leu176); the remainder of the structures are completely conserved in the two enzymes. Comparing chain A of B. pumilus PAD with either chain of L. plantarum PAD (PDB entry 2w2a) gives an r.m.s.d. of 0.46 Å for 159 common Cα atoms. With L. plantarum PAD (PDB entry 2gc9), the corresponding r.m.s.d.s are 0.59 Å for 156 Cα atoms and 0.61 Å for 143 Cα atoms for chains A and B, respectively. These small differences in structure between B. pumilus PAD and the two different structures of L. plantarum PAD are significant as they reflect differences in the β1–β2 loop conformation found in PDB entry 2w2a (both subunits ‘open’) versus PDB entry 2gc9 (both subunits ‘closed’) (Fig. 3 ▶ a). This contrasts with the two subunits of B. pumilus PAD, in which subunit A adopts the ‘open’ conformation and subunit B adopts the ‘closed’ conformation. The single subunit of B. subtilus PAD adopts the ‘open’ conformation. This change in the conformation of the β1–β2 loop, as well as the conserved Asn residue (Asn15 in B. pumilus PAD), in turn influences the shape and the size of the cavity which defines the active-site region. A structure-based sequence alignment of available PAD structures, showing for the most part a high residue identity in secondary-structure elements, is presented in Fig. 3 ▶(b).
Figure 3.
(a) Structural superposition of PAD subunits from B. pumilus (PDB code 3nad; this study; chain A in green, chain B in cyan), L. plantarum [PDB codes 2w2a (chain A in magenta, chain B in yellow) and 2gc9 (chain A in light blue, chain B in dark blue)] and B. subtilus (PDB code 2p8g; pink). The differences in structure of the β1–β2 loop, which contains a conserved Asn residue and adopts either a ‘closed’ or ‘open’ conformation, are shown. (b) Structure-based sequence alignment of PAD subunits based on the structure superposition shown in (a). Identical residues (*) and residues with similar properties (.) are indicated below the alignment. Secondary-structure elements based on the B. pumilus crystal structure are shown above the alignment. The structural superposition and structure-based alignment were generated using Swiss-PdbViewer (Guex & Peitsch, 1997 ▶).
At the level of the dimers, the two PADs show an r.m.s.d. of 0.55 Å for 315 common Cα atoms. When the B. pumilus PAD dimer is superposed with the alternative L. plantarum PAD dimer (PDB entry 2gc9), the r.m.s.d. is 0.61 Å for 301 common Cα atoms. The sequences of the B. subtilus and B. pumilus enzymes contain the same number of residues (161), although the B. subtilus structure starts with a Gly residue which presumably originates from the vector used to express the protein. Unlike other PAD structures, only one subunit is found within the asymmetric unit for B. subtilus PAD (PDB entry 2p8g), giving an r.m.s.d. of 0.41 Å for 159 common Cα atoms. Based on a structure-based alignment of the available PAD structures (Fig. 3 ▶ b), residues forming inter-subunit hydrogen bonds appear to be mainly conserved among the different structures, with the exception of Asn73 (B. pumilus/B. subtilus), which is replaced by an Asp in the L. plantarum structures.
3.5. PADs share a common lipocalin/calycin fold
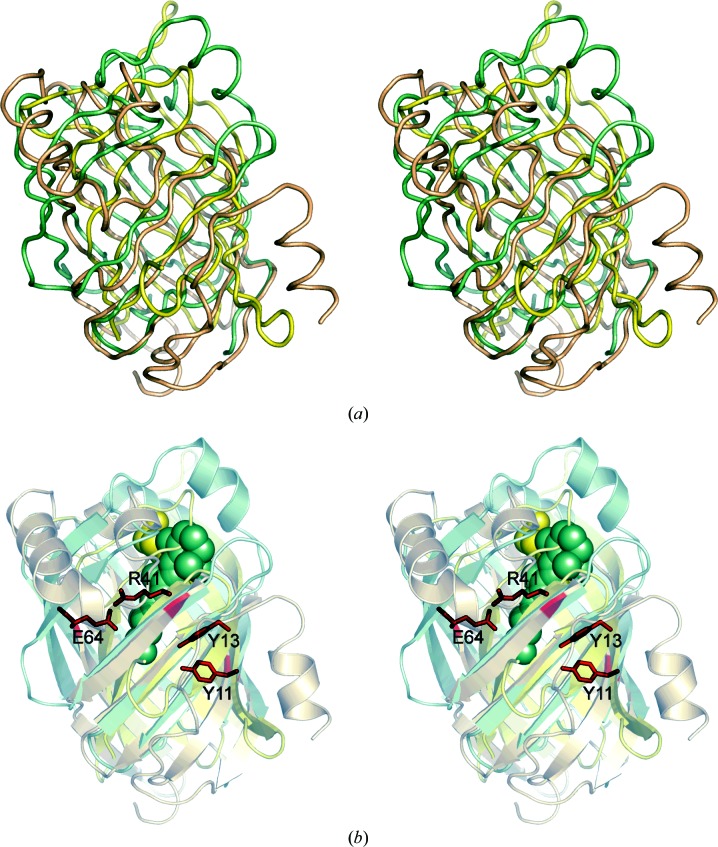
Analysis of the distribution of conserved residues from a multiple sequence alignment mapped onto the PAD molecular surface using the ConSurf server (http://consurf.tau.ac.il/; Glaser et al., 2003 ▶) revealed that many of the most highly conserved residues are hydrophobic residues that are found within the core of the β-barrel, thereby defining a common fold for this protein family (Figs. 2 ▶ b and 2 ▶ c). A search for structurally related proteins in the PDB was performed using SSM v.2.34 (Krissinel & Henrick, 2004 ▶; http://www.ebi.ac.uk/msd-srv/ssm/) and DaliLite v.3 (Holm et al., 2008 ▶; http://ekhidna.biocenter.helsinki.fi/dali_server/start). The most closely structurally related proteins include rhizavidin/avidin/streptavidin from various sources, which share r.m.s.d.s of 2.7–3.3 Å for ∼100 common Cα atoms, as well as several retinoic acid and fatty-acid-binding proteins, which share a level of structural similarity that is comparable to that of the streptavidins. The structural relatedness between lipocalins and streptavidins has been noted previously (Flower, 1993 ▶). The most structurally similar regions of these proteins are the eight to ten β-strands that form the two antiparallel β-sheets (Fig. 4 ▶ a). The pairwise sequence identity and similarity between these proteins are ∼10% and ∼30%, respectively. Mapping of the identical residues common to PAD and the fatty-acid-binding and retinol-binding proteins show that these residues lie primarily within the core of the β-sheets and probably contribute to defining the common β-barrel fold shared amongst these proteins.
Figure 4.
PAD adopts the lipocalin/calycin fold. (a) Structure superposition of B. pumilus PAD monomer (wheat), rhizavidin-biotin complex (PDB entry 3ew2; yellow; Meir et al., 2009 ▶) and human holo cellular retinol-binding protein II bound to retinol (PDB entry 2rct; pale green; Tarter et al., 2007 ▶). (b) The same orientation as in (a) with key PAD residues shown in stick representation and biotin and retinol ligands shown in stick and space-filling representation.
Comparing the topology of PAD with known structures reveals that it adopts the lipocalin/calcyin fold, with the same topology and connectivity of secondary-structure elements and an extension of two additional β-strands (β9a and β9b), resulting in two five-stranded β-sheets instead of four-stranded sheets (Skerra, 2008 ▶). However, PADs do not share the three defining conserved sequence motifs characteristic of lipocalins or the long Ω loop (L1) between β1 and β2 (Flower et al., 1993 ▶, 2000 ▶). Lipocalins are a family of small β-barrel proteins that are characterized as binding and often transporting small hydrophobic ligands; in a few cases they have been ascribed an enzymatic function, as exemplified by prostaglandin D synthase in the isomerization of prostaglandin H2 to prostaglandin D (Åkerstrom et al., 2000 ▶; Flower et al., 2000 ▶; Grzyb et al., 2006 ▶). The relationship in structure between molecules such as retinoic acid and the short-chain phenolic acids is intriguing and could suggest that this fold, which was originally intended for binding small mainly hydrophobic ligands, was later adopted to not only bind similar molecules but also to act on them enzymatically. The conjugated nature of the substrates appears to be a common feature among the enzymatic lipocalins, including those catalyzing epoxidation reactions (Grzyb et al., 2006 ▶). Lipocalins are thus far more abundant in eukaryotes and have been less frequently identified in bacteria (Bishop, 2000 ▶), although microbial genome-sequencing projects have increased the number of known proteins to more than 90 (Bishop et al., 2006 ▶).
Superposition and analysis of the molecular surfaces of PADs and related lipocalin/calycin structures revealed a common cavity or cleft located between the two β-sheets of each subunit (Fig. 4 ▶ b). In PAD this cavity is larger near the surface of the molecule and becomes progressively narrower as the enzyme is entered. Residues near the surface of the cavity include Asn15, Tyr19, Phe87 and His92. Both Arg41 and Glu64 sit near the bottom of this tunnel, distal from the solvent. Other residues lining the tunnel include Tyr13, Tyr19, Ile21, Ile29, Ile33, Val38, Trp62, Thr66, Thr68, Val70, Leu72, Gly83, Ile85, Val91 and Thr98. Most of these residues are highly conserved among PADs, suggesting that the tunnel is a common structural feature of these enzymes. The closed nature of the tunnel implies that the substrate enters and the product leaves the enzyme through the same opening. In the B. pumilus PAD dimer, chain A is more open than chain B owing to the difference in the conformation of residues 13–18 in the β1–β2 loop. Interestingly, the flexible β1–β2 loop in PAD structures corresponds topologically to the larger Ω loop of lipocalins, a flexible element that often folds back to restrict access to the ligand-binding pocket (Flower et al., 1993 ▶). The residues that have been implicated as important for the activity of PAD from L. plantarum (Tyr18, Tyr20, Arg48 and Glu71; Rodriguez et al., 2010 ▶), corresponding to Tyr11, Tyr13, Arg41 and Glu64 in B. pumilus PAD, are structurally conserved between the two enzymes.
4. Summary
The three-dimensional structure of PAD from B. pumilus strain UI-670 shows very high overall similarity to the PADs from L. plantarum and B. subtilus, although the latter structure has not been described apart from the deposited coordinates (PDB entry 2p8g). Nonetheless, a significant conformational change is observed in the β1–β2 loop between the B. pumilus and L. plantarum structures, a region that influences the shape and size of the cavity that defines the active site. As a new enzyme exhibiting the lipocalin/calcyin fold, the structure of PAD provides a basis for rational design with alternative ligand specificity (Skerra, 2008 ▶) or other improved properties.
Supplementary Material
PDB reference: phenolic acid decarboxylase, 3nad
Acknowledgments
We wish to thank Rong Shi for assistance with structure refinement, Christine Munger for assistance with crystallization, Jack Rosazza for the B. pumilus UI-670 strain and Mirek Cygler for advice and encouragement. Use of the Advanced Photon Source at Argonne National Laboratory is supported by the US Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-06CH11357. Use of the Lilly Research Laboratory Collaborative Access Team (LRL-CAT) beamline at sector 31 of the Advanced Photon Source was provided by Eli Lilly & Company, who operate the facility. The Canadian Light Source is supported by NSERC, NRC, CIHR and the University of Saskatchewan. This research was supported by funding from the National Research Council Canada (PCKL). This is NRC publication No. 53127.
References
- Åkerstrom, B., Flower, D. R. & Salier, J.-P. (2000). Biochim. Biophys. Acta, 1482, 1–8. [DOI] [PubMed]
- Barthelmebs, L., Diviès, C. & Cavin, J.-F. (2001). Appl. Environ. Microbiol. 67, 1063–1069. [DOI] [PMC free article] [PubMed]
- Barthelmebs, L., Lecomte, B., Diviès, C. & Cavin, J.-F. (2000). J. Bacteriol. 182, 6724–6731. [DOI] [PMC free article] [PubMed]
- Benoit, I., Navarro, D., Marnet, N., Rakotomanomana, N., Leasage-Messen, L., Sigoilot, J.-C., Asther, M. & Asther, M. (2006). Carbohydrate Res. 341, 1820–1827. [DOI] [PubMed]
- Berman, H., Westbrook, J., Feng, Z., Gilliland, G., Bhat, T. N., Weissig, H., Shindyalov, I. N. & Bourne, P. E. (2000). Nucleic Acids Res. 28, 235–242. [DOI] [PMC free article] [PubMed]
- Bishop, R. E. (2000). Biochim. Biophys. Acta, 1482, 73–83. [DOI] [PubMed]
- Bishop, R. E., Cambillau, C., Privé, G. G., Hsi, D., Tillo, D. & Tillier, E. R. M. (2006). Lipocalins, edited by B. Åkerström, N. Borregaard, D. R. Flower & J.-P. Salier, pp. 28–40. Austin: Landes Bioscience.
- Bradford, M. M. (1976). Anal. Biochem. 72, 248–254. [DOI] [PubMed]
- Cavin, J.-F., Barthelmebs, L. & Diviès, C. (1997). Appl. Environ. Microbiol. 63, 1939–1944. [DOI] [PMC free article] [PubMed]
- Cavin, J.-F., Dartois, V. & Diviès, C. (1998). Appl. Environ. Microbiol. 64, 1466–1471. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Finn, R. D., Mistry, J., Tate, J., Coggill, P., Heger, A., Pollington, J. E., Gavin, O. L., Gunesekaran, P., Ceric, G., Forslund, K., Holm, L., Sonnhammer, E. L., Eddy, S. R. & Bateman, A. (2010). Nucleic Acids Res. 38, D211–D222. [DOI] [PMC free article] [PubMed]
- Flower, D. R. (1993). FEBS Lett. 333, 99–102. [DOI] [PubMed]
- Flower, D. R., North, A. C. T. & Attwood, T. K. (1993). Protein Sci. 2, 753–761. [DOI] [PMC free article] [PubMed]
- Flower, D. R., North, A. C. T. & Sansom, C. E. (2000). Biochim. Biophys. Acta, 1482, 9–24. [DOI] [PubMed]
- Glaser, F., Pupko, T., Paz, I., Bell, R. E., Bechor-Shental, D., Martz, E. & Ben-Tal, N. (2003). Bioinformatics, 19, 163–164. [DOI] [PubMed]
- Godoy, L., Martinez, C., Carrasco, N. & Ganga, M. A. (2008). Int. J. Food Microbiol. 127, 6–11. [DOI] [PubMed]
- Grzyb, J., Latowski, D. & Strzalka, K. (2006). J. Plant Physiol. 163, 895–915. [DOI] [PubMed]
- Guex, N. & Peitsch, M. C. (1997). Electrophoresis, 18, 2714–2723. [DOI] [PubMed]
- Holm, L., Kääriäinen, S., Rosenström, P. & Schenkel, A. (2008). Bioinformatics, 24, 2780–2781. [DOI] [PMC free article] [PubMed]
- Huang, Z., Dostal, L. & Rosazza, J. P. N. (1993). Appl. Environ. Microbiol. 59, 2244–2250. [DOI] [PMC free article] [PubMed]
- Huang, Z., Dostal, L. & Rosazza, J. P. N. (1994). J. Bacteriol. 176, 5912–5918. [DOI] [PMC free article] [PubMed]
- Itzhaki, R. F. & Gill, D. M. (1964). Anal. Biochem. 9, 401–410. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2004). Acta Cryst. D60, 2256–2268. [DOI] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Meir, A., Helppolainen, S. H., Podoly, E., Nordlund, H. R., Hytönen, V. P., Määttä, J. A., Wilchek, M., Bayer, E. A., Kulomaa, M. S. & Livnah, O. (2009). J. Mol. Biol. 386, 379–390. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A., Lebedev, A., Wilson, K. S. & Dodson, E. J. (1999). Acta Cryst. D55, 247–255. [DOI] [PubMed]
- Murzin, A. G., Brenner, S. E., Hubbard, T. & Chothia, C. (1995). J. Mol. Biol. 247, 536–540. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Prim, N., Pastor, F. I. J. & Diaz, P. (2003). Appl. Microbiol. Biotechnol. 63, 51–56. [DOI] [PubMed]
- Rodríguez, H., Angulo, I., de las Rivas, B., Campillo, N., Páez, J. A., Muñoz, R. & Mancheño, J. M. (2010). Proteins, 78, 1662–1676. [DOI] [PubMed]
- Rodríguez, H., de las Rivas, B., Muñoz, R. & Mancheño, J. M. (2007). Acta Cryst. F63, 300–303. [DOI] [PMC free article] [PubMed]
- Rodríguez, H., Landete, J. M., Curiel, A., de las Rivas, B., Mancheño, M. & Muñoz, R. (2008). J. Agric. Food Chem. 56, 3068–3072. [DOI] [PubMed]
- Rosazza, J. P. N., Huang, Z., Dostal, L., Volm, T. & Rousseau, B. (1995). J. Ind. Microbiol. 15, 457–471. [DOI] [PubMed]
- Skerra, A. (2008). FEBS J. 275, 2677–2683. [DOI] [PubMed]
- Tarter, M., Capaldi, S., Carrizo, M. E., Ambrosi, E., Perduca, M. & Monaco, H. L. (2007). Proteins, 70, 1626–1630. [DOI] [PubMed]
- Topakas, E., Vafiadi, C. & Christakopoulos, P. (2007). Process Biochem. 42, 497–509.
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst. 30, 1022–1025.
- Wong, D. W. S. (2006). Appl. Biochem. Biotechnol. 133, 87–112. [DOI] [PubMed]
- Yang, J., Wang, S., Lorrain, M.-J., Rho, D., Abokitse, K. & Lau, P. C. K. (2009). Appl. Microbiol. Biotechnol. 84, 867–876. [DOI] [PubMed]
- Zago, A., Degrassi, G. & Bruschi, C. V. (1995). Appl. Environ. Microbiol. 61, 4484–4486. [DOI] [PMC free article] [PubMed]
- Zaldivar, J. & Ingram, L. O. (1999). Biotechnol. Bioeng. 66, 203–210. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: phenolic acid decarboxylase, 3nad