The homodimeric nature of the ESAT-6 homologue GBS1074 and the potential for fibre-like assemblies are revealed by the 2 Å resolution crystal structure.
Keywords: ESAT-6, WXG100, GBS1074, four-helix bundle
Abstract
ESAT-6 is a well characterized secreted protein from Mycobacterium tuberculosis and represents the archetype of the WXG100 family of proteins. Genes encoding ESAT-6 homologues have been identified in the genome of the human pathogen Streptococcus agalactiae; one of these genes, esxA, has been cloned and the recombinant protein has been crystallized. In contrast to M. tuberculosis ESAT-6, the crystal structure of GBS1074 reveals a homodimeric structure similar to homologous structures from Staphylococcus aureus and Helicobacter pylori. Intriguingly, GBS1074 forms elongated fibre-like assemblies in the crystal structure.
1. Introduction
The secreted proteins ESAT-6 (early secreted antigen target 6) and CFP10 (culture filtrate protein 10) from Mycobacterium tuberculosis play a key role in virulence and elicit a strong antigenic response (forming the basis of a new diagnostic test), although their precise biological function has yet to be defined. NMR (Renshaw et al., 2005 ▶; PDB code 1wa8) and crystal (Poulsen et al., 2010 ▶; PDB code 3fav) structures have confirmed that the two proteins are homologous to one another and form a tight 1:1 heterodimeric complex composed of antiparallel helical hairpins arranged in a head-to-tail fashion and held together by a predominantly hydrophobic interface. Pallen (2002 ▶) used sequence analyses to show that ESAT-6 and CFP10 are archetypes of a large family of WXG100 proteins (so-called because of their central WXG amino-acid motif and a length commonly of ∼100 amino acids), members of which are widely distributed among Gram-positive bacteria. Drawing on these observations, Burts et al. (2005 ▶) showed that WXG100 proteins contribute to virulence in Staphylococcus aureus. Subsequently, crystal structures of WXG100 proteins from S. aureus (Sundaramoorthy et al., 2008 ▶; PDB codes 2vs0 and 2vrz) and Helicobacter pylori (Jang et al., 2009 ▶; PDB codes 3fx7 and 2gts) have shown these proteins to form homodimeric head-to-tail structures. Most recently, the M. tuberculosis Rv3019c–Rv3020c ESX complex has been shown to form both heterodimers and heterotetramers in solution in a relative ratio of approximately 15:1, but only the heterotetrameric complex could be crystallized (Arbing et al., 2010 ▶; PDB code 3h6p). The EsxR protein (the ESAT-6 homologue) folds as an antiparallel helical hairpin as expected, but strikingly the EsxS protein (the CFP10 homologue) folds as one long extended helix, with the C-terminal half of the helix domain-swapping with that of a second equivalent EsxRS complex to form a heterotetramer composed of two four-helix bundles (see Fig. 2 of Arbing et al., 2010 ▶).
Streptococcus agalactiae (group B streptococcus; GBS) is a leading cause of neonatal sepsis and infections in pregnant women (Larsen & Sever, 2008 ▶) and can also cause invasive infection in other settings (Sendi et al., 2008 ▶). Given the contribution of WXG100 proteins to virulence in M. tuberculosis and S. aureus, we searched for additional members of this protein family in the predicted proteome of S. agalactiae strain NEM316 (Herbert et al., 2005 ▶). We solved the crystal structure of one such protein, GBS1074, to 2.0 Å resolution. As with related WXG100 proteins from S. aureus and H. pylori, GBS1074 adopts a homodimeric structure, but intriguingly the crystal packing suggests the potential to form higher order polymers, with a second intermolecular interface being formed between the extended C-terminal helical regions.
2. Experimental
2.1. General methods
S. agalactiae strains were cultured in Todd–Hewitt broth and on Todd–Hewitt agar and Escherichia coli was cultured in LB medium. Genome, gene and protein analyses were performed with xBASE2 (http://xbase.bham.ac.uk/; Chaudhuri et al., 2008 ▶), PSI-BLAST and ClustalW (http://www.ebi.ac.uk/Tools). Genomic DNA was extracted with a DNeasy Blood and Tissue kit (Qiagen).
2.2. Cloning and expression of GBS1074
The GBS1074 coding sequence was amplified with primers GBS1074f_BamHI (5′-TAC GGA TCC ATG GCA CAA ATT AAA TTA ACA CC-3′) and GBS1074r_SalI (5′-GTC GAC GTA TCC ACT AAT TTG AGA AGC-3′) and cloned into pQE-80 (Qiagen) and recombinant GBS1074 protein with an N-terminal MRGS-HHHHHH-GS tag was expressed in E. coli XL1-Blue (Stratagene) at 293 K. Cells were induced with 1 mM IPTG at an OD600 of 0.6 and then cultured overnight at 293 K. Harvested cells were suspended in lysis buffer (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 10 mM imidazole) and lysed using a French press. The lysate was centrifuged at 18 000g for 1 h and the supernatant was loaded onto an Ni–NTA column, washed (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 20 mM imidazole) and eluted (50 mM NaH2PO4 pH 8.0, 300 mM NaCl, 250 mM imidazole). The protein was dialysed against 20 mM Tris pH 8.0 to remove imidazole. The histidine tag was not removed.
2.3. Crystallization, data collection and structure determination
Crystals of GBS1074 were prepared using the sitting-drop vapour-diffusion technique with a drop consisting of 2 µl protein stock solution (20 mg ml−1 GBS1074, 10 mM Tris buffer pH 8) mixed with 2 µl reservoir solution (0.75 M Li2SO4, 0.6 M ammonium sulfate, 0.1 M sodium citrate pH 5.1). Hexagonal-shaped crystals belonging to space group P6522 grew within a few days to a maximum dimension of approximately 0.5 mm. Crystals were transferred to a stabilization solution based on the mother liquor and were then cryoprotected by increasing the concentration of glycerol (in 5% steps to a final concentration of 25%) before flash-cooling and storage in liquid nitrogen. An iodine derivative was obtained by soaking a crystal as above but with a final solution supplemented with 1.9 mM I2 and 2.7 mM KI prepared by using a 250-fold dilution of the KI/I2 stock solution described in Evans & Bricogne (2002 ▶). The crystal was left in the derivative solution for 48 h, by which time the crystal was a strong yellow colour and the solution was completely colourless. Higher concentrations of KI/I2 cracked the crystals.
Both ‘Native1’ and ‘Iodine’ data sets were collected on our in-house X-ray facility (Rigaku MicroMax-007 HF with Saturn CCD and Kappa four-circle goniometer) with scans around multiple axes. To maximize the anomalous signal contribution from the iodine derivative, a highly redundant data set (∼60 h) was collected. A second native data set, ‘Native2’, was collected on ESRF beamline ID14-1. All data were indexed and integrated with XDS (Kabsch, 2010 ▶) and scaled using either SCALA (Native1 and Native2; Evans, 1993 ▶) or XSCALE (Iodine; Kabsch, 2010 ▶). Merging statistics were calculated in SCALA for all three data sets. Iodine positions were located and phases were determined using a SIRAS protocol (Iodine and Native1) with the program autoSHARP (Vonrhein et al., 2007 ▶), yielding a clear interpretable electron-density map. Six significant iodine sites were located and refined. All six sites showed single I atoms, with no evidence of I2 or I3 − binding (Evans & Bricogne, 2002 ▶); the shortest I—I distance was 6.05 Å. Three of the sites were buried in the hydrophobic interior of the protein, with two being noncrystallographically equivalent. Two further sites were in hydrophobic surface pockets with the potential to also form hydrogen bonds to solvent or protein and one site was located in a pocket formed at the interface of two protein molecules. RESOLVE (Terwilliger, 2002 ▶) was used to autotrace and build 145 amino acids (backbone and side chain) in four helical fragments, plus an additional 19 amino acids (backbone only) in a further four fragments, representing approximately 85% of the mature nontagged portion of the GBS1074 homodimer. The programs Coot (Emsley & Cowtan, 2004 ▶) and phenix.refine (Afonine et al., 2005 ▶) with MLHL targets were used to complete and refine the structure. The resulting refined Native1 structure was then refined against the 2.0 Å resolution Native2 data set. Data-collection and phasing statistics are presented in Table 1 ▶. Structural alignments were calculated using LSQKAB (Collaborative Computational Project, Number 4, 1994 ▶). Contact surfaces were analysed using the PISA server (Krissinel & Henrick, 2007 ▶) and AREAIMOL (Collaborative Computational Project, Number 4, 1994 ▶).
Table 1. Data-collection and phasing statistics.
Values in parentheses are for the highest resolution shell.
| Native1 | Native2 | Iodine | |
|---|---|---|---|
| Crystal properties | |||
| Space group | P6522 | P6522 | P6522 |
| Unit-cell parameters (Å) | |||
| a = b | 77.2 | 76.2 | 77.1 |
| c | 152.8 | 151.3 | 153.7 |
| Data collection | |||
| Wavelength (Å) | 1.542 | 0.933 | 1.542 |
| No. of observations | 873191 (91459) | 211669 (30807) | 2259118 (303657) |
| No. of unique reflections | 16476 (2344) | 18255 (2591) | 5881 (818) |
| Completeness (%) | 99.9 (100) | 99.7 (100) | 99.6 (99.9) |
| Multiplicity | 53 (39) | 11.6 (11.9) | 384 (371) |
| Anomalous completeness (%) | 99.9 (100) | ||
| Anomalous multiplicity | 190 (191) | ||
| 〈I/σ(I)〉 | 58.6 (8.4) | 34 (6.7) | 174.7 (49.3) |
| Anomalous signal [|F+ − F−|/σ(ΔF)] | 4.7 (1.5) | ||
| Rmerge (%) | 5.3 (61.2) | 4.2 (39.6) | 8.4 (29.9) |
| Resolution range (Å) | 19.5–2.1 (2.21–2.10) | 40.1–2.0 (2.11–2.00) | 19.5–3.0 (3.16–3.00) |
| Wilson B (Å2) | 34.2 | 32.8 | 63.3 |
| Refinement | |||
| No. of non-H atoms | 1616 | ||
| No. of waters | 140 | ||
| R/Rfree† | 18.9/21.9 | ||
| R.m.s.d. | |||
| Bonds (Å) | 0.006 | ||
| Angles (°) | 0.742 | ||
| Ramachandran‡ | 172/4/0/0 | ||
| Average B (Å2) | 45 | ||
| PDB code | 3o9o | ||
R free was calculated for a random set of 5% of reflections that were not used in the refinement.
Number of nonglycine, nonproline amino acids in the core/allowed/generously allowed/disallowed regions of the Ramachandran plot.
3. Results
3.1. WXG100 proteins encoded in the S. agalactiae genome
We identified three genes encoding WXG100 proteins in S. agalactiae strain NEM316 (GBS0223, GBS1074 and GBS1979; data not shown). The gene encoding GBS1074 lies within a putative WXG100 secretion-system locus that encompasses homologues of the S. aureus essC, essB and esaB genes, which are essential for secretion of the WXG100 protein EsxA (Pallen, 2002 ▶; Burts et al., 2005 ▶). GBS1074 bears a canonical central WXG motif and shows 14.9% amino-acid identity to ESAT-6 from M. tuberculosis and 22.7% amino-acid identity to EsxA from S. aureus.
3.2. GBS1074 crystal structure
The crystal structure of recombinant GBS1074 was solved using the SIRAS technique with an iodine derivative (Evans & Bricogne, 2002 ▶; Fig. 1 ▶). The unbiased electron-density maps resulting from SIRAS phase calculations were of high quality and were easily interpretable, enabling automatic model building for approximately 85% of the Native1 structure, with the remainder being built manually. Owing to structural ambiguity of a few amino acids at the termini, the Native1 structure was refined against a second native data set, Native2, to produce a final model containing amino-acid residues 4–95 in subunit A and 0–95 in subunit B, where 0 is the last amino acid of the linker between the N-terminal His6 tag and the starting methionine (Table 1 ▶). The remainder of the linker and the His6 tags were not visible in the electron density. A portion of the final refined 2F o − F c electron-density map is shown in Fig. 2 ▶.
Figure 1.
GBS1074 crystal structure. Ribbon diagram of GBS1074 in two orthogonal views. Chain A is shown in green and blue and chain B in yellow and orange. The twofold NCS axis is indicated in grey.
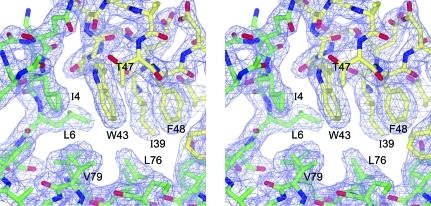
Figure 2.
Wall-eyed stereoview of the refined 2F o − F c electron-density map (contour level 1.2σ) showing part of the hydrophobic core around Trp43. Atoms are coloured as follows: oxygen, red; nitrogen, blue; carbon, yellow (subunit A) or green (subunit B).
3.3. Homodimeric structure
There are two molecules per asymmetric unit, forming a homodimer, with each subunit contributing 1850 Å2 of solvent-accessible surface area to the interface. Owing to the head-to-tail symmetrical nature of the dimer, inter-subunit contacts are preserved in the twofold symmetry; therefore, amino acids in one polypeptide chain are distinguished from those in the other by a prime (′). The r.m.s.d. between subunits is 1.32 Å when comparing the maximum 92 pairs of Cα atoms, but this improves to 0.62 Å for 85 pairs of Cα atoms when the last seven amino acids of each chain are excluded. Although a continuation of the second helix, the helix axes over the last two helical turns at the C-termini diverge, probably as a result of crystal packing (see below). The homodimer interface is almost exclusively hydrophobic and is further stabilized by peripheral hydrogen bonding and salt bridges in a typical heptad amino-acid repeat.
The GBS1074 protein folds as an antiparallel coiled-coil pair to form a four-helix bundle (Fig. 1 ▶) typified by the structure of the protein ROP. Each subunit forms two long helices, Pro8–Glu41 and Asp49–Ile94, with a 30° kink in the second helix at Leu68. Amino acids Trp43-Asp44-Gly45, the conserved WXG motif, form the apex of the loop connecting the antiparallel helices. The side chain of Trp43 is part of an extensive hydrophobic core and Gly45 is the only amino acid in the refined model to be in the lower right-hand corner of the Ramachandran plot, a region that is disallowed for nonglycine amino acids.
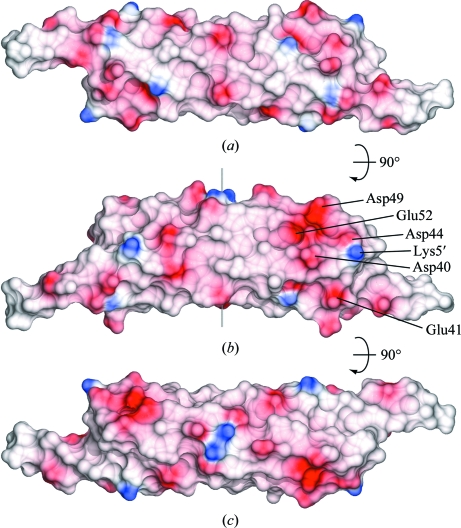
The N-terminus of one polypeptide chain interacts with the WXG motif of the other (Figs. 1 ▶, 2 ▶ and 3 ▶). Ile4 forms van der Waals interactions with Trp43′ and together with Leu6 and Met1 (in the B chain only) forms an extension of the hydrophobic core that holds the two pairs of antiparallel helices together into a four-helix bundle. Lys5 and Asp44′, two amino acids that are conserved in many WXG100-family members, form a salt bridge and in the only intersubunit main-chain–main-chain hydrogen bonding in the whole molecule the backbone O atom of Gln3 forms a hydrogen bond to Asp44′ N and Lys5 N hydrogen bonds to Asn42′ O, so that Ile4 forms an intersubunit β-bridge with Trp43′. Overall, the homodimer is approximately cylindrical, with a diameter of 25 Å and a length of 95 Å, and has a distinctive negative patch on the surface formed by amino acids Asp40, Asp44, Asp49, Glu52 and, slightly further away, Glu41 (Fig. 4 ▶).
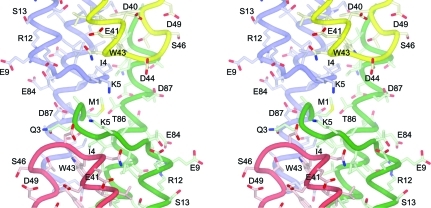
Figure 3.
Wall-eyed stereoview of the crystallographic symmetry interactions between one molecule and the next along the chain. Chain B is coloured green and chain A is coloured red. In the symmetry mate (mostly shown in the upper half of the figure) chain A is coloured blue and chain B is coloured yellow (see also Fig. 5 ▶).
Figure 4.
Molecular surfaces coloured according to electrostatic potential using the program CCP4MG with default colour settings (Potterton et al., 2004 ▶). (b) has the same orientation as Fig. 1 ▶(a).
4. Discussion
4.1. Structural homology
The available structures of WXG100 proteins are highly similar. While the archetypal pair ESAT-6 and CFP10 form a heterodimer and a minor component of the related M. tuberculosis EsxRS complex forms a domain-swapped heterotetramer, other WXG100 proteins of known structure, now including GBS1074, form homodimers. Although the NMR structure of ESAT-6 shows a relatively unstructured C-terminus, with the deviation from regular helicity starting at residue Ala79, this C-terminal region has been described as having a high helical propensity (Renshaw et al., 2005 ▶). In the crystal structure of GBS1074, as in the X-ray structure of the ESAT-6–CFP10 complex and in similar structures of WXG100 proteins from S. aureus and H. pylori, the second helix is continuous until the end of the polypeptide chain. Interestingly, in GBS1074 the C-terminal helical region forms extensive intermolecular contacts with neighbouring molecules (see below).
4.2. Crystal packing and fibre-like structures
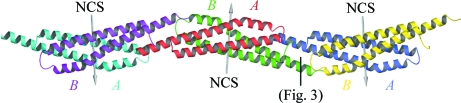
A packing analysis of the GBS1074 crystal structure offers the intriguing possibility that GBS1074 can form fibre-like structures (Fig. 5 ▶) or some higher order assembly. In the crystal, GBS1074 molecules are aligned end-to-end in a chain fashion, with alternating up–down links. Each homodimer contributes 850 Å2 of solvent-accessible surface area to the interface with the next homodimer along the chain, the second largest interface in the crystal structure (Fig. 5 ▶). The protruding C-terminal helices continue the antiparallel coiled-coil interactions along the chain. In addition, the backbone amide of Met1 hydrogen bonds to the conserved Asp87 OD1 (2.6 Å) of a chain-forming symmetry mate, an interaction that would be a salt bridge if the N-terminal tag were not present (Fig. 3 ▶). The conserved hydrophobic positions 90 and 94 (both Ile) of one homodimer form van der Waals interactions with Trp43 of another, each contributing approximately 100 Å2 of solvent-accessible surface to the potential fibre-forming interface. Such end-to-end chain interactions were not observed in the crystal structures of EsxA from S. aureus, although Sundaramoorthy et al. (2008 ▶) describe a potential peptide-binding groove on the surface of the protein. Despite positions 90 and 94 in EsxA being hydrophobic (Leu and Phe, respectively), only subunit A of PDB entry 2vrz has a structurally ordered C-terminus. The last 10–15 amino acids are disordered in chain B of PDB entry 2vrz and in both subunits of PDB entry 2vs0. End-to-end chain interactions are not seen in the crystal structure of HP0062 either, but here the sequence is shorter. The intermolecular interactions observed in the GBS1074 crystal structure are reminiscent of the crystal packing and oligomer assembly in the structure of the secreted mature FadA (PDB entry 3etw), a bacterial adhesion protein from the opportunistic human pathogen Fusobacterium nucleatum that has been shown to be important for host-cell attachment and invasion (Nithianantham et al., 2009 ▶). FadA also forms a helical hairpin structure, but does not form dimers. Instead, the molecules form an elongated structure in what Nithianantham and coworkers describe as a leucine chain, in which the extended C-terminal helix forms a leucine-rich hydrophobic interaction with the head of the next molecule in a coiled-coil-type assembly.
Figure 5.
Crystal packing showing potential chain-forming interactions. NCS, noncrystallographic symmetry. Three dimers in the chain are shown. The asymmetric unit (x, y, z) is the dimer in the centre coloured red and green. The dimer to the left of the figure, coloured cyan and magenta (x, x − y, −z + 5/6), and the dimer to the right, coloured blue and yellow (x, x − y, −z + 5/6), are related by unit-cell translations (2a + b). The A and B monomers are labelled in matching colours. The interface shown in Fig. 3 ▶ is indicated.
5. Concluding remarks
Sundaramoorthy et al. (2008 ▶) make a strong case for EsxA (and, by extension, other WXG100 proteins) acting as chaperones that aid the delivery of effector proteins across the bacterial cell envelope. However, it is hard to reconcile the oligomerization we observe in the GBS1074 crystal structure with a role as chaperone. Instead, parallels with the structure of the secreted mature FadA protein hint at a possible role for GBS1074, and perhaps other WXG100 proteins, in mediating interactions with host cells. Clearly, determining whether WXG100 oligomerization occurs under physiological conditions remains a key target for research on this protein family.
Supplementary Material
PDB reference: homodimeric GBS1074, 3o9o
Acknowledgments
We thank the University of Birmingham genomics laboratory for core sequencing and the ESRF for travel and access to synchrotron X-ray facilities. This research was supported by grants from Birmingham Women’s NHS Foundation Trust and The Birmingham Children’s Hospital Research Fund.
References
- Afonine, P. V., Grosse-Kunstleve, R. W. & Adams, P. D. (2005). CCP4 Newsl.42, contribution 8.
- Arbing, M. A., Kaufmann, M., Phan, T., Chan, S., Cascio, D. & Eisenberg, D. (2010). Protein Sci.19, 1692–1703. [DOI] [PMC free article] [PubMed]
- Burts, M. L., Williams, W. A., DeBord, K. & Missiakas, D. M. (2005). Proc. Natl Acad. Sci. USA, 102, 1169–1174. [DOI] [PMC free article] [PubMed]
- Chaudhuri, R. R., Loman, N. J., Snyder, L. A., Bailey, C. M., Stekel, D. J. & Pallen, M. J. (2008). Nucleic Acids Res.36, D543–D546. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, G. & Bricogne, G. (2002). Acta Cryst. D58, 976–991. [DOI] [PubMed]
- Evans, P. R. (1993). Proceedings of the CCP4 Study Weekend. Data Collection and Processing, edited by L. Sawyer, N. Isaacs & S. Bailey, pp. 114–122. Warrington: Daresbury Laboratory.
- Herbert, M. A., Beveridge, C. J., McCormick, D., Aten, E., Jones, N., Snyder, L. A. & Saunders, N. J. (2005). BMC Microbiol.5, 31. [DOI] [PMC free article] [PubMed]
- Jang, S.-B., Kwon, A.-R., Son, W.-S., Park, S. J. & Lee, B.-J. (2009). J. Biochem.146, 535–540. [DOI] [PubMed]
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol.372, 774–797. [DOI] [PubMed]
- Larsen, J. W. & Sever, J. L. (2008). Am. J. Obstet. Gynecol.198, 440–448. [DOI] [PubMed]
- Nithianantham, S., Xu, M., Yamada, M., Ikegami, A., Shoham, M. & Han, Y. W. (2009). J. Biol. Chem.284, 3865–3872. [DOI] [PMC free article] [PubMed]
- Pallen, M. J. (2002). Trends Microbiol.10, 209–212. [DOI] [PubMed]
- Potterton, L., McNicholas, S., Krissinel, E., Gruber, J., Cowtan, K., Emsley, P., Murshudov, G. N., Cohen, S., Perrakis, A. & Noble, M. (2004). Acta Cryst. D60, 2288–2294. [DOI] [PubMed]
- Poulsen, C., Holton, S., Geerlof, A., Wilmanns, M. & Song, Y.-H. (2010). FEBS Lett.584, 669–674. [DOI] [PubMed]
- Renshaw, P. S., Lightbody, K. L., Veverka, V., Muskett, F. W., Kelly, G., Frenkiel, T. A., Gordon, S. V., Hewinson, R. G., Burke, B., Norman, J., Williamson, R. A. & Carr, M. D. (2005). EMBO J.24, 2491–2498. [DOI] [PMC free article] [PubMed]
- Sendi, P., Johansson, L. & Norrby-Teglund, A. (2008). Infection, 36, 100–111. [DOI] [PubMed]
- Sundaramoorthy, R., Fyfe, P. K. & Hunter, W. N. (2008). J. Mol. Biol.383, 603–614. [DOI] [PMC free article] [PubMed]
- Terwilliger, T. C. (2002). Acta Cryst. D58, 1937–1940. [DOI] [PubMed]
- Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. (2007). Methods Mol. Biol.364, 215–230. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: homodimeric GBS1074, 3o9o