Cleavage of the serpin IRS-2 from the hard tick I. ricinus by contaminating proteolytic activity mimicked the specific processing of the serpin by its target protease and resulted in a more stable form of the serpin which produced crystals that diffracted to 1.8 Å resolution.
Keywords: serpins, ticks, proteolysis
Abstract
IRS-2 from the hard tick Ixodes ricinus belongs to the serpin family of protease inhibitors. It is produced in the salivary glands of the tick and its anti-inflammatory activity suggests that it plays a role in parasite–host interaction. Recombinant IRS-2 prepared by heterologous expression in a bacterial system was crystallized using the hanging-drop vapour-diffusion method. The crystals belonged to the primitive tetragonal space group P43 and diffracted to 1.8 Å resolution. Mass-spectrometric and electrophoretic analyses revealed that IRS-2 was cleaved by contaminating proteases during crystallization. This processing of IRS-2 mimicked the specific cleavage of the serpin by its target protease and resulted in a more stable form (the so-called relaxed conformation), which produced well diffracting crystals. Activity profiling with specific substrates and inhibitors demonstrated traces of serine and cysteine proteases in the protein stock solution.
1. Introduction
Serpins (serine protease inhibitors) are a broadly distributed family of protease inhibitors (Irving et al., 2000 ▶). The majority of serpins specifically inhibit serine proteases, but some serpins that inhibit papain-like cysteine proteases (Schick et al., 1998 ▶) and caspases (Ray et al., 1992 ▶) have also been identified. In rare cases, the serpins have lost their inhibitory function and act, for example, as hormone transporters (Pemberton et al., 1988 ▶), chaperones (Nagata, 1996 ▶) or tumour suppressors (Zou et al., 1994 ▶).
Serpins are relatively large molecules (comprising 330–500 amino-acid residues) which act as ‘single-use’ or ‘suicide’ inhibitors that undergo an extensive conformational change to inhibit proteases (Huntington et al., 2000 ▶). The serpin family is structurally well characterized; over 70 serpin structures have been determined (reviewed, for example, in Law et al., 2006 ▶). The remarkable structural change is induced by proteolysis of the reactive-centre loop (RCL), which in the native state (also called the S state) has an extended conformation that protrudes from the serpin domain. Upon cleavage, the amino-terminal part of the RCL inserts into the central β-sheet to form an additional β-strand. This structural rearrangement is crucial for protease inhibition and results in the so-called R state of serpin, which is more stable compared with the S state. In the final serpin–protease complex the protease remains covalently linked to the serpin. However, the protease can escape this trap and dissociate over time, leaving active protease and inactive serpin with its RCL cleaved in a substrate-like manner (Huntington et al., 2000 ▶).
Ticks are blood-feeding parasites of a variety of vertebrates, including domestic animals and humans. They are important (second only to mosquitoes) pathogen vectors worldwide (Sauer et al., 1995 ▶). The hard tick Ixodes ricinus is the most important European vector of Borrelia burgdorferi s.l., the agent of Lyme disease and tick-borne encephalitis virus.
In hard ticks, serpins have been identified in all tissues and have been proposed to be involved in both tick physiology and tick–host interaction. The salivary glands in particular produce a number of serpins, which may play roles in the modulation of immune response, coagulation, complement regulation and inflammation (Prevot et al., 2009 ▶). Tick serpins are thus considered to be promising antigens for the induction of host protective immunity against ticks (Andreotti et al., 2002 ▶; Imamura et al., 2005 ▶; Sugino et al., 2003 ▶).
The serpin IRS-2 has recently been identified in the salivary glands of the tick I. ricinus. Functional characterization has shown that its protease-inhibitor activity is directed against chymotrypsin-like proteases, resulting in an anti-inflammatory effect (Chmelar et al., manuscript in preparation).
No crystal structure of a serpin isolated from a parasitic organism is available to date. The closest homologues of IRS-2 among the serpin structures deposited in the Protein Data Bank are equine leukocyte elastase inhibitor (PDB code 1hle; Baumann et al., 1992 ▶) and human squamous cell carcinoma antigen 1 (PDB entry 2zv6; Zheng et al., 2009 ▶), with 35 and 32% sequence identity, respectively.
In order to gain structural information on this newly discovered serpin, we initiated structural studies on this protein; here, we present the crystallization of recombinant IRS-2, analysis of its diffraction and an initial molecular-replacement solution.
2. Materials and methods
2.1. Protein expression and purification
The coding sequence of IRS-2 (UniProt entry Q06B74) was amplified from cDNA isolated from the salivary glands of an I. ricinus adult female and was cloned into pET-17b vector (Novagen).
Details of cloning and purification have been described elsewhere (Chmelar et al., manuscript in preparation). The 377 amino-acid residue IRS-2 protein was overexpressed in Escherichia coli BL21 (DE3) pLysS cells (Invitrogen) at 310 K upon induction by 0.5 mM IPTG. The expressed protein accumulated in inclusion bodies, which were separated. The inclusion bodies were dissolved in 6 M guanidine hydrochloride and the supernatant was diluted into a 150-fold volume of refolding buffer (20 mM Tris–HCl pH 8, 0.25 M l-arginine). The protein solution was then concentrated using a stirred-chamber concentrator (Millipore) and dialyzed against 20 mM Tris–HCl pH 8. Refolded and concentrated IRS-2 was purified on a Mono Q column with a 0–1 M gradient of NaCl using an ÄKTA FPLC system (Pharmacia) to a purity of >99% according to SDS–PAGE analysis. For crystallization, IRS-2 was dialyzed into 20 mM HEPES pH 7.2.
2.2. Protein crystallization
Screening for crystallization conditions was performed by sparse-matrix screening (Jancarik & Kim, 1991 ▶) using the commercially available Crystallization Basic and Extension Kits (Sigma–Aldrich) and the hanging-drop vapour-diffusion technique in 24-well Linbro plates at 293 K. Subsequent optimization of the initial crystallization conditions was performed in NeXtal plates (Qiagen). The reservoir contained 0.5 ml reservoir solution and the crystallization drop consisted of 2 µl IRS-2 protein solution (3.5 mg ml−1 in 20 mM HEPES pH 7.2) and 1 µl reservoir solution. Preliminary needle-shaped crystals grew in condition No. 22 of the Extension Kit: 0.1 M MES pH 6.5, 12%(w/v) PEG 20 000. Optimal crystals were obtained from the original kit solution diluted with water to a final composition of 75 mM MES pH 6.5, 9%(w/v) PEG 20 000.
2.3. Diffraction data collection
For data collection, crystals were soaked for 30 s in reservoir solution supplemented with 20%(v/v) PEG 400 and flash-cooled in liquid nitrogen. Diffraction data were collected at 100 K on the X12 EMBL beamline at DESY, Hamburg, Germany. A set of 107 images was recorded with a 0.5° oscillation angle, an exposure time of 10 s per image and a crystal-to-detector distance of 185 mm. Diffraction data were processed using the HKL-2000 suite of programs (Minor et al., 2006 ▶). The redundancy-independent merging R factor R r.i.m. as well as the precision-indicating merging R factor R p.i.m. were calculated using the program RMERGE (Weiss, 2001 ▶). Crystal parameters and data-collection statistics are summarized in Table 1 ▶.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| No. of crystals | 1 |
| Beamline | X12, DESY, Germany |
| Wavelength (Å) | 0.953 |
| Detector | MAR Mosaic 225 |
| Crystal-to-detector distance (mm) | 185 |
| Rotation range per image (°) | 0.5 |
| Total rotation range (°) | 54 |
| Exposure time per image (s) | 10 |
| Resolution range (Å) | 30.0–1.80 (1.86–1.80) |
| Space group | P43 |
| Unit-cell parameters (Å) | a = b = 84.6, c = 124.4 |
| Mosaicity (°) | 0.33 |
| Total no. of measured intensities† | 301874 |
| No. of unique reflections | 80249 (7790) |
| Multiplicity | 3.8 (3.3) |
| Average I/σ(I) | 32.8 (2.3) |
| Completeness (%) | 99.6 (97.3) |
| Rmerge‡ (%) | 4.6 (35.8) |
| Rr.i.m.§ (%) | 7.2 (53.7) |
| Rp.i.m.¶ (%) | 3.7 (29.2) |
| Overall B factor from Wilson plot (Å2) | 23.2 |
The criterion used for observed reflections was I/σ(I) > 0.
R
merge = 100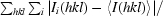
 , where I
i(hkl) is an individual intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the average intensity of reflection hkl with summation over all data.
, where I
i(hkl) is an individual intensity of the ith observation of reflection hkl and 〈I(hkl)〉 is the average intensity of reflection hkl with summation over all data.
R
r.i.m. = 100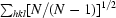
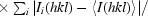
 , where N is the number of times the given reflection hkl was observed (Weiss, 2001 ▶).
, where N is the number of times the given reflection hkl was observed (Weiss, 2001 ▶).
R
p.i.m. = 100
 , where N is the number of times the given reflection hkl was observed (Weiss, 2001 ▶).
, where N is the number of times the given reflection hkl was observed (Weiss, 2001 ▶).
2.4. Mass spectrometry
Mass-spectrometric characterization of IRS-2 was performed by LC-MS/MS analysis and de novo sequencing of tryptic and chymotryptic digests. LC-MS/MS analysis was performed on a LTQ Orbitrap XL hybrid mass spectrometer (Thermo Scientific) coupled to a Rheos 2000 two-dimensional capillary LC system (Flux Instruments). The first-dimension column was a monolithic PS-DVB (200 µm × 10 mm; Dionex) and the second-dimension column was a C18 PepMap100 (75 µm × 150 mm × 3 µm; Dionex) with gradient elution in a 0.1% formic acid/acetonitrile system. The LC-MS/MS data were processed with SEQUEST and BioWorks (Thermo Scientific) and PEAKS (Bioinformatics Solutions) software.
2.5. Profiling of proteolytic activities
The residual proteolytic activities that were present in the IRS-2 protein were detected by hydrolysis of the following fluorogenic peptidic substrates containing a 7-amino-4-methylcoumarin group (AMC; Bachem): Z-Phe-Arg-AMC, MeoSuc-Ala-Ala-Pro-Val-AMC, Z-Gly-Gly-Leu-AMC, Suc-Ala-Ala-Pro-Phe-AMC and Suc-Leu-Tyr-AMC (Horn et al., 2009 ▶). The reaction mixture contained 15 µg ml−1 protein stock solution (final concentration), 40 µM substrate and 0.1 M MES buffer pH 6.5. A blank was prepared without addition of the IRS-2 protein sample. The reaction mixtures were incubated for 24 h at 310 K followed by reading the fluorescence of the liberated AMC using a GENios Plus fluorescence reader at 360 nm excitation and 465 nm emission wavelengths. For activity assays in the presence of protease inhibitors, the following inhibitors were added to the reaction mixture: 10 µM E-64 for cysteine proteases and 1 mM Pefabloc for serine proteases.
2.6. Serpin-cleavage assay
A 10 µl reaction mixture containing 0.4 µg protein from the IRS-2 stock solution in 0.1 M MES buffer pH 6.5 was incubated at 310 K for 6 d in the presence or absence of protease inhibitors (10 µM E-64 and 1 mM Pefabloc). The reaction was stopped by heating (at 343 K for 5 min) in reducing Laemmli sample buffer. Nonincubated IRS-2 was used as a control. The reaction mixture was separated by Laemmli SDS–PAGE (15% gels) and visualized by silver staining.
3. Results
IRS-2, a serpin from the hard tick I. ricinus, was refolded from inclusion bodies, producing an active inhibitor, and purified using ion-exchange chromatography. The recombinant IRS-2 contained 377 amino-acid residues with a molecular weight of 42.2 kDa. The purity of the sample was confirmed by the presence of a single band on silver-stained SDS–PAGE.
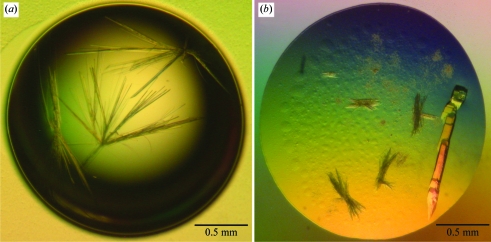
IRS-2 protein solution at 3.5 mg ml−1 in 20 mM HEPES pH 7.2 was used for crystallization at 293 K. Preliminary needle-shaped crystals were obtained in 5 d using reservoir solution consisting of 0.1 M MES pH 6.5, 12%(w/v) PEG 20 000 (Fig. 1 ▶ a). During optimization, larger crystals of final dimensions of about 0.3 × 0.1 × 0.1 mm appeared within 21 d using reservoir solution consisting of 75 mM MES pH 6.5, 9%(w/v) PEG 20 000 (Fig. 1 ▶ b).
Figure 1.
Crystals of IRS-2. (a) Initial crystals obtained from screening for crystallization conditions. (b) Crystals grown from optimized conditions. For data collection, part of the large crystal cluster on the right was used.
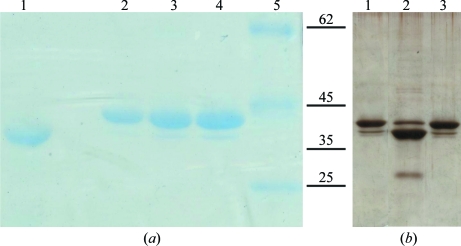
Analysis of dissolved crystals using SDS–PAGE indicated that the IRS-2 protein had been proteolyzed during the crystallization at a temperature of 293 K (Fig. 2 ▶ a, lane 1). In order to identify the cleavage site, IRS-2 was subjected to enzymatic digestion followed by LC-MS/MS analysis, which provided ∼85% peptide coverage. This revealed that the cleavage site was located between Tyr341 and Ser342, as the unexpected peptide 342–361 was identified in the tryptic digest. Cleavage at the identified site produced an N-terminal fragment of the serpin molecule with a molecular weight of 38 kDa corresponding to the band identified in the protein crystal (Fig. 2 ▶ a, lane 1) and a 4.2 kDa C-terminal fragment. The identified cleavage site is located within a region homologous to the reactive-centre loop (RCL) of other serpin inhibitors and is most likely to correspond to a natural cleavage site for target proteases.
Figure 2.
Analysis of IRS-2 proteolysis by SDS–PAGE. (a) 15% SDS–PAGE of an IRS-2 crystal (lane 1) and the protein sample used for crystallization experiments (lanes 2–4). Lane 1, dissolved protein crystal; lane 2, 0.75 µg IRS-2; lane 3, 1.1 µg IRS-2; lane 4, 1.5 µg IRS-2; lane 5, protein molecular-weight markers (Fermentas; labelled in kDa). (b) 15% SDS–PAGE of IRS-2 protein (0.4 µg) stored at 277 K (lane 1) and incubated for 6 d at 310 K without protease inhibitors (lane 2) and in the presence of 10 µM E-64 and 1 mM Pefabloc (lane 3).
As proteolysis was also obvious in protein sample stored at 277 K (Fig. 2 ▶ a, lanes 2–4), we concluded that the proteolysis was caused by contaminating proteases that were present in the protein sample and not in the solutions used for crystallization. In order to identify and characterize the contaminating proteases, we further investigated the proteolytic activity in the purified IRS-2 protein sample by testing a panel of specific peptidic substrates for serine and cysteine proteases (Table 2 ▶). The hydrolysis of these substrates was monitored using a continuous fluorimetric assay at the pH of the crystallization mixture. In addition, an activity assay was performed in the presence of selective protease inhibitors, namely Pefabloc and E64, which inhibit serine and cysteine proteases, respectively. Based on the activity profiling (Table 2 ▶), the detected activities were attributed to several protease types including the papain-like and calpain-like activities of the cysteine protease class and the trypsin-like and chymotrypsin-like activities of the serine protease class.
Table 2. Profiling of proteolytic activities in IRS-2 protein preparation.
| Substrate | Protease type (catalytic class) | Proteolytic activity | Inhibitor sensitivity |
|---|---|---|---|
| Z-Phe-Arg-AMC | Papain-like (cysteine) | Yes | E64 |
| Trypsin-like (serine) | Pefabloc | ||
| MeoSuc-Ala-Ala-Pro-Val-AMC | Elastase-like (serine) | No | |
| Z-Gly-Gly-Leu-AMC | Chymotrypsin-like (serine) | Yes | Pefabloc |
| Suc-Ala-Ala-Pro-Phe-AMC | Chymotrypsin-like (serine) | No | |
| Suc-Leu-Tyr-AMC | Calpain-like (cysteine) | Yes | E64 |
| Chymotrypsin-like (serine) | Pefabloc |
To verify that the cleavage of IRS-2 protein was indeed caused by the residual protease activities detected in the protein sample, we incubated the protein sample for an extended period of time (6 d) at 310 K in the presence or absence of serine and cysteine protease inhibitors. SDS–PAGE analysis (Fig. 2 ▶ b) revealed specific cleavage of IRS-2 which is inhibited in the presence of the Pefabloc/E64 mixture. The cleavage produced an IRS-2 major fragment of ∼38 kDa corresponding to the fragment detected in the protein crystal. Further proteolysis to smaller fragments (∼30 kDa) was also observed.
Analysis of the crystals and protein sample led us to the conclusions that IRS-2 was cleaved by contaminating serine and cysteine proteases in the canonical serpin cleavage site and that the protein species which produced crystals is the cleaved and more stable R state.
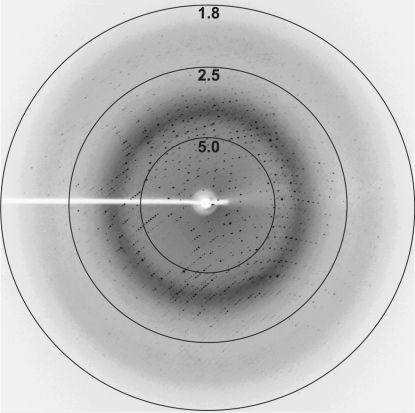
IRS-2 crystals were used to collect diffraction data after cryoprotection with 20% PEG 400. A complete data set was collected from a single crystal on the X13 beamline at BESSY (Hamburg) to 1.8 Å resolution (Fig. 3 ▶). The crystal exhibited the symmetry of space group P4, with systematic absences indicating the presence of a 41 or 43 screw axis. Crystal parameters and data-collection statistics are summarized in Table 1 ▶. Evaluation of the crystal-packing parameters indicated the presence of two molecules (containing 377 amino-acid residues each) in the asymmetric unit, with a solvent content of 53% and a Matthews coefficient of 2.62 Å3 Da−1 (Matthews, 1968 ▶).
Figure 3.
A diffraction image from an IRS-2 crystal. The numbers represent the resolution in Å.
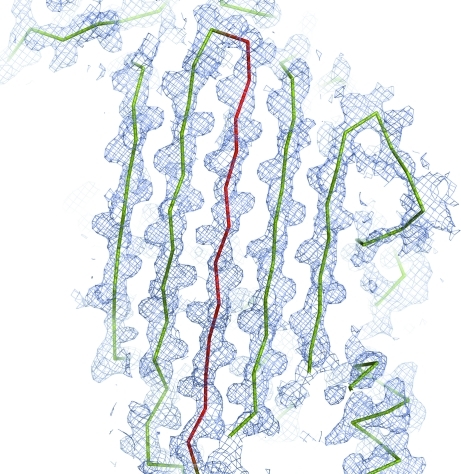
Determination of the structure by molecular replacement with MOLREP (Vagin & Teplyakov, 2000 ▶) was attempted using the structures of the two serpins that share the highest sequence identity with IRS-2: human squamous cell carcinoma antigen 1 (PDB entry 2zv6; Zheng et al., 2009 ▶) and equine leukocyte elastase inhibitor (PDB entry 1hle; Baumann et al., 1992 ▶). The solution in space group P43 using a polyalanine model derived from the structure of the cleaved (R-state) equine leukocyte elastase inhibitor gave the best results (R factor 0.55, score 0.252). The two molecules pack sensibly in the unit cell with no clashes and the electron densities calculated from the molecular-replacement solution appeared to be suitable for model building. Examination of the electron-density maps calculated from the initial phases permitted us to identify that the structure indeed represents an R form of serpin with the RCL inserted into the central β-sheet to form as an additional β-strand (Fig. 4 ▶). Model building and refinement of the model is currently in progress and the structure analysis will be described in a subsequent manuscript (Chmelar et al., manuscript in preparation).
Figure 4.
Electron-density map (2F obs − F calc, contoured at 1σ) in the region of the central β-sheet of IRS-2 calculated from the initial phases after molecular replacement. The protein model (a polyalanine model derived from the structure of the cleaved equine leukocyte elastase inhibitor) is shown as a ribbon, with the region of the reactive-centre loop (RCL; residues 362–377) coloured red. The β-strand conformation of the RCL is well supported by the electron-density map. This figure was generated using the program PyMOL (DeLano, 2002 ▶).
4. Discussion
Partial proteolysis has been shown to be an important event that is required for the successful crystallization of many protein samples. Successful crystallization of proteins after proteolysis by a contaminating protease originating from the protein sample or crystallization solution has been reported (Mandel et al., 2006 ▶) and the inclusion of a protease in the crystallization experiment (in situ proteolysis) is becoming a widely used technique (Dong et al., 2007 ▶). For most proteins, the positive effect of limited proteolysis on crystallization stems from the removal of flexible parts or highly hydrophobic segments from the protein surface.
In our case, we observed a specific cleavage of the serpin IRS-2 from the tick I. ricinus by contaminating proteases originating from the bacterial expression system. Interestingly, the cleavage was located in the RCL, which is the region of the serpin that is responsible for interaction with the target protease. This proteolytic treatment of IRS-2 mimicked specific cleavage by a target protease and caused a conformational transition of the serpin molecule from the S form into a more stable form (the R form) which produced crystals that were suitable for diffraction analysis.
The cleavage of serpins during crystallization has been reported previously. The first crystal structure of a cleaved serpin, that of human α1-proteinase inhibitor, was the result of an unsuccessful attempt to crystallize its complex with the protease zymogen (Loebermann et al., 1984 ▶). During long-term crystallization, the serpin reacted with traces of active proteinase to produce the covalent complex, which then dissociated to give the cleaved serpin, which crystallized. We show here that serpins can be efficiently cleaved by proteases originating from the expression host and that these contaminating activities can be identified using specific substrates and inhibitors as diagnostic tools.
Acknowledgments
This work was supported by the Grant Agency of the Czech Republic (grant No. P207/10/2183) and in part by research centre No. LC06009 from the Ministry of Education and projects Z40550506 and Z50520514 awarded by the Academy of Sciences of the Czech Republic. Diffraction data were collected on beamline X12 at the EMBL Hamburg Outstation at DESY in Hamburg, Germany; beamline MX14.2 at BESSY, Berlin, Germany was used for preliminary diffraction analysis.
References
- Andreotti, R., Gomes, A., Malavazi-Piza, K. C., Sasaki, S. D., Sampaio, C. A. & Tanaka, A. S. (2002). Int. Immunopharmacol.2, 557–563. [DOI] [PubMed]
- Baumann, U., Bode, W., Huber, R., Travis, J. & Potempa, J. (1992). J. Mol. Biol.226, 1207–1218. [DOI] [PubMed]
- DeLano, W. L. (2002). PyMOL. DeLano Scientific, Palo Alto, California, USA.
- Dong, A. et al. (2007). Nature Methods, 4, 1019–1021. [DOI] [PMC free article] [PubMed]
- Horn, M., Nussbaumerova, M., Sanda, M., Kovarova, Z., Srba, J., Franta, Z., Sojka, D., Bogyo, M., Caffrey, C. R., Kopacek, P. & Mares, M. (2009). Chem. Biol.16, 1053–1063. [DOI] [PMC free article] [PubMed]
- Huntington, J. A., Read, R. J. & Carrell, R. W. (2000). Nature (London), 407, 923–926. [DOI] [PubMed]
- Imamura, S., da Silva Vaz Junior, I., Sugino, M., Ohashi, K. & Onuma, M. (2005). Vaccine, 23, 1301–1311. [DOI] [PubMed]
- Irving, J. A., Pike, R. N., Lesk, A. M. & Whisstock, J. C. (2000). Genome Res.10, 1845–1864. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411.
- Law, R. H., Zhang, Q., McGowan, S., Buckle, A. M., Silverman, G. A., Wong, W., Rosado, C. J., Langendorf, C. G., Pike, R. N., Bird, P. I. & Whisstock, J. C. (2006). Genome Biol.7, 216. [DOI] [PMC free article] [PubMed]
- Loebermann, H., Tokuoka, R., Deisenhofer, J. & Huber, R. (1984). J. Mol. Biol.177, 531–557. [PubMed]
- Mandel, C. R., Gebauer, D., Zhang, H. & Tong, L. (2006). Acta Cryst. F62, 1041–1045. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Minor, W., Cymborowski, M., Otwinowski, Z. & Chruszcz, M. (2006). Acta Cryst. D62, 859–866. [DOI] [PubMed]
- Nagata, K. (1996). Trends Biochem. Sci.21, 22–26. [DOI] [PubMed]
- Pemberton, P. A., Stein, P. E., Pepys, M. B., Potter, J. M. & Carrell, R. W. (1988). Nature (London), 336, 257–258. [DOI] [PubMed]
- Prevot, P. P., Beschin, A., Lins, L., Beaufays, J., Grosjean, A., Bruys, L., Adam, B., Brossard, M., Brasseur, R., Zouaoui Boudjeltia, K., Vanhamme, L. & Godfroid, E. (2009). FEBS J.276, 3235–3246. [DOI] [PubMed]
- Ray, C. A., Black, R. A., Kronheim, S. R., Greenstreet, T. A., Sleath, P. R., Salvesen, G. S. & Pickup, D. J. (1992). Cell, 69, 597–604. [DOI] [PubMed]
- Sauer, J. R., McSwain, J. L., Bowman, A. S. & Essenberg, R. C. (1995). Annu. Rev. Entomol.40, 245–267. [DOI] [PubMed]
- Schick, C., Pemberton, P. A., Shi, G. P., Kamachi, Y., Cataltepe, S., Bartuski, A. J., Gornstein, E. R., Bromme, D., Chapman, H. A. & Silverman, G. A. (1998). Biochemistry, 37, 5258–5266. [DOI] [PubMed]
- Sugino, M., Imamura, S., Mulenga, A., Nakajima, M., Tsuda, A., Ohashi, K. & Onuma, M. (2003). Vaccine, 21, 2844–2851. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2000). Acta Cryst. D56, 1622–1624. [DOI] [PubMed]
- Weiss, M. S. (2001). J. Appl. Cryst.34, 130–135.
- Zheng, B., Matoba, Y., Kumagai, T., Katagiri, C., Hibino, T. & Sugiyama, M. (2009). Biochem. Biophys. Res. Commun.380, 143–147. [DOI] [PubMed]
- Zou, Z., Anisowicz, A., Hendrix, M. J., Thor, A., Neveu, M., Sheng, S., Rafidi, K., Seftor, E. & Sager, R. (1994). Science, 263, 526–529. [DOI] [PubMed]