Abstract
Background
Influenza viruses (IFVs) frequently achieve resistance to antiviral drugs, necessitating the development of compounds with novel mechanisms of action. DAS181 (Fludase®), a sialidase fusion protein, may have a reduced potential for generating drug resistance due to its novel host-targeting mechanism of action.
Methods
IFV strains B/Maryland/1/59 and A/Victoria/3/75 (H3N2) were subjected to >30 passages under increasing selective pressure with DAS181. The DAS181-selected IFV isolates were characterized in vitro and in mice.
Results
Despite extensive passaging, DAS181-selected viruses exhibited a very low level of resistance to DAS181, which ranged between 3- and 18-fold increase in EC50. DAS181-selected viruses displayed an attenuated phenotype in vitro, as exhibited by slower growth, smaller plaque size and increased particle to pfu ratios relative to wild-type virus. Further, the DAS181 resistance phenotype was unstable and was substantially reversed over time upon DAS181 withdrawal. In mice, the DAS181-selected viruses exhibited no greater virulence than their wild-type counterparts. Genotypic and phenotypic analysis of DAS181-selected viruses revealed mutations in the haemagglutinin (HA) and neuraminidase (NA) molecules and also changes in HA and NA function.
Conclusions
Results indicate that resistance to DAS181 is minimal and unstable. The DAS181-selected IFV isolates exhibit reduced fitness in vitro, likely due to altered HA and NA functions.
Keywords: resistance, antiviral, haemagglutinin, neuraminidase, sialic acid
Introduction
Influenza causes 250 000–500 000 deaths and 3–5 million cases of severe respiratory illness annually across the world.1 While useful, vaccination is an imperfect solution for preventing influenza virus (IFV) infection, due to difficulty in predicting future dominant strains, antigenic shift and drift, and reduced immunocompetence in certain populations. Furthermore, vaccines have limitations with regard to the time required for production and the need for annual reformulation, providing little utility if the predicted strains do not represent the majority of circulating infections. In contrast, antiviral compounds can be used in both prophylaxis and treatment regimens, remain effective without modification if resistance does not arise in the population and may be the only available therapy in immunocompromised patients.
Although antiviral drug development has resulted in several approved drugs to combat IFV, the development of drug resistance is a major concern for all antiviral agents due to the high error rate of the viral RNA polymerase and the ability of the virus to re-assort with other IFV strains. For instance, viral resistance to all M2 inhibitors (adamantanes) and the neuraminidase inhibitor (NAI) oseltamivir has become widespread in seasonal H3N2 and H1N1 IFV, respectively.2–8 The novel 2009 H1N1 IFV is also resistant to M2 inhibitors and has the potential to gain oseltamivir resistance.9–11 Only one other FDA-approved anti-IFV compound currently exists (zanamivir) and it is of the same class as oseltamivir (NAI).
DAS181 is a recombinant sialidase fusion protein in clinical development as a novel IFV prophylactic and therapeutic candidate. DAS181 is composed of the catalytic domain of Actinomyces viscosus sialidase and the epithelial anchoring domain of human amphiregulin and strongly inhibits infection when tested with numerous strains of IFV in vitro and in vivo.12–17 DAS181 acts by attaching to the respiratory epithelium and cleaving the IFV receptor, sialic acid, from cell surface glycans, thereby preventing virus binding.14,17 With this unique mechanism of action, DAS181 represents an entirely novel class of anti-IFV agents and is thus expected to be effective against strains resistant to the existing classes of antivirals. This has been demonstrated for the current oseltamivir-resistant H1N1 clinical isolates.15 Further, DAS181 is the first anti-influenza compound that acts on the host cell rather than the virus, and thus may be less likely to induce significant drug resistance compared with virus-targeting agents (e.g. M2 inhibitors, NAI inhibitors, viral RNA polymerase inhibitors, etc.). As DAS181 is currently in clinical trials, predictive information regarding resistance potential is important.
Here we provide characterization of two IFV strains, B/Maryland/1/59 and A/Victoria/3/75 (H3N2), passaged in Madin–Darby canine kidney (MDCK) cells under increasing DAS181 selective pressure. Specifically, we describe drug sensitivities and genotypes throughout passaging, the molecular characterization of mutant viral phenotypes and the relative in vivo fitness of the DAS181-selected (DS) IFV strains. These studies have revealed potential mechanisms by which IFV may adapt to DAS181 treatment.
Materials and methods
Cells and viruses
MDCK cells were obtained from ATCC (Manassas, VA, USA) and grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 1× Glutamax (Invitrogen, Carlsbad, CA, USA) and 1× antibiotic/antimycotic solution (Sigma, St Louis, MO, USA) at 37°C in a humidified atmosphere of 5% CO2. The IFVs B/Maryland/1/59 and A/Victoria/3/75 (H3N2) were obtained from ATCC. All viruses were amplified on MDCK cells, and viral titres were determined by plaque assay.
DAS181
Purified DAS181 was supplied in 1.7 mM acetate buffer, pH 5, and stored at −80°C until use, as previously described.14
Generation of DS mutant viruses
The limiting dilution method was used to isolate DS viruses, as previously described.18 Briefly, each virus was added to confluent MDCK cells in 24-well culture dishes for 1 h at 35°C at four different multiplicities of infection (moi) (10-fold serial dilutions ranging from 0.0001 to 0.1 moi), before washing once with PBS. Wild-type (wt) viruses were propagated in cells cultured in medium only [DMEM/F12, 0.2% insulin-transferrin-selenium (ITS) and 3 μg/mL acetylated trypsin], and the DS viruses were propagated in cells cultured in medium containing 2-fold increasing concentrations of DAS181, starting with the ∼EC50 for each particular virus (14 and G. B. Triana-Baltzer and F. Fang, unpublished observation). At 48–96 h post-infection virus was harvested from the wells that were inoculated with the lowest infectious dose of virus and the highest concentration of DAS181 that resulted in cell death compared with wt virus at the same infectious dose. The virus samples were cleared of cell debris by centrifugation, subdivided into aliquots and frozen at –80°C. DAS181 withdrawal (DW) virus was produced by taking passage 17 or 14 (B/Maryland/1/59 or A/Victoria/3/75, respectively) DS virus and switching to growth without drug. DS and wt viruses were minimally amplified from a single late passage (B/Maryland/1/59 p22, A/Victoria/3/75 passage 24) for use in the characterization studies.
DAS181 sensitivity in vitro
DAS181 sensitivity was determined using the plaque number reduction assay on MDCK cells. This protocol is modified from a previous publication.19 In short, ∼150 pfu of virus was applied to each well of confluent MDCK cells in six-well plate format. After binding for 1.5 h, unbound virus was washed off with PBS and the plates were overlaid with 1 : 1 Noble Agar and DMEM/F12 as in the plaque assay. Drug concentrations of 2× were included in the 2× DMEM/F12, so as to achieve 1× final concentration in the agar/medium mixture. After allowing the agar to cool, the plates were incubated, stained and counted as in the plaque assay. Data were graphed as plaque number per drug concentration, normalized to no drug and expressed as the percentage of plaques remaining. EC50 values were calculated as the concentration of drug reducing visible plaque number to 50% of the no-drug control. All viruses were tested in triplicate against each drug and values are represented as means ± SEM. All data were graphed with Prism 4.02 software. Drug sensitivity of all passages of wt and DS virus was tested simultaneously. DW virus sensitivity was compared with DS in parallel experiments.
Viral titre determination
Quantitative RT–PCR
Viral titres (total particle numbers) were determined by quantifying viral M-gene copy number at the University of Washington Medical Center Clinical Laboratory (Seattle, WA, USA) with specific primers and probes to influenza A and B.
Plaque assay
Infectious viral titre was determined by plaque assay on MDCK cells as previously described.15
Viral infection rate in MDCK cells
Confluent monolayers of MDCK cells were infected with wt and DS IFVs at equivalent infectious doses and allowed to incubate at 37°C. At selected timepoints the wells were fixed with Crystal Violet (0.5%) in 20% methanol and absorbance was quantified to determine the relative amount of live cells. The percentage of cells remaining was calculated by comparing with the uninfected control. Values represent means ± SD of triplicate samples.
Plaque size analysis
Confluent monolayers of MDCK in six-well plates were infected with IFVs, overlaid with agar/medium as above, incubated for ∼72 h and fixed/stained with Crystal Violet as above. Each wt, DS and DW virus set was fixed at the same time so plaque size could be compared directly. At least 24 representative plaques were measured for each virus and the mean ± SD was graphed with Prism 4.02 software.
Mouse infection studies
Female BALB/C mice (8–10 weeks old) were infected intranasally (50 μL/mouse) with the indicated infection levels of wt or DS IFV (normalized to equal M-gene copy number; also presented as pfu). For treatment, study mice received intranasal administration of PBS or DAS181 (50 μL/mouse) daily for 5 days beginning 6 h post-infection. Mice were anaesthetized with ketamine (200 mg/kg, intraperitoneally) prior to each intranasal administration. Survival and body weight were tracked daily, and viral titre in lung homogenates was determined at day 3 in a subset of the mice via plaque assay. All animal studies were conducted following the approval of the study protocols by the NexBio, Inc., Institutional Animal Care and Use Committee.
IFV genotyping
Most passages of wt, DS and DW virus were analysed to determine haemagglutinin (HA) and neuraminidase (NA) genotypes. Additionally, at passage 17 or 14 (B/Maryland/1/59 and A/Victoria/3/75, respectively) all eight viral gene segments were analysed. In brief, viral RNA was purified from the viral sample using either the QiaAMP Viral RNA Mini Kit (Qiagen, Valencia, CA, USA) or the MagMAX-96™ Viral RNA isolation kit (Applied Biosystems, Foster City, CA, USA). PCR was performed on reverse-transcribed RNA with primers to amplify the entire viral gene of interest. The primers were designed based on previously published universal primers20 and from published sequences of B/Maryland/1/59 and A/Victoria/3/75 genes. Upon confirming amplification of the correct size band, the PCR product was purified using a PCR purification kit (Qiagen) and sequenced using the same forward and reverse primers, as well as internal primers where necessary. Oligonucleotide sequences were aligned using ClustalW2 software (http://www.ebi.ac.uk/Tools/clustalw2/index.html) and sequence data noted with the H3 or N2 numbering scheme.21,22 Alignments are shown in Figures S1 to S4 (available as Supplementary data at JAC Online). Positions of mutated amino acids within HA or NA protein were determined by comparison with published crystal structures of A/duck/Ukraine/63 H3 trimer,22 B/HongKong/8/73 HA trimer23 or A/tern/Australia/G70C/75 N9 tetramer,24 using PyMOL Software (www.pymol.org).25 For B/Maryland/1/59, passages 1–18 were sequenced by reading two to four plaque-purified clones, while passages 19–31 were sequenced by reading a pool of virus sample. For A/Victoria/3/75, passages 1–14 were sequenced by reading two plaque-purified clones (mutation noted if observed in both clones), while passages 15–26 were sequenced by reading a pool of virus sample (mutation noted if observed as majority of population by chromatogram analysis).
Viral protein level quantification by ELISA
Viral stock solutions were equilibrated based on approximate NP protein levels (from pilot ELISA experiments), and 2-fold serial dilutions were coated on high-binding 96-well microplates (Corning, Corning, NY, USA) and incubated at 4°C overnight. The following day the plates were washed, blocked with 3% BSA + 0.1% Tween and HA, NA and NP viral antigens were detected with specific monoclonal antibodies [IFVA HA MAb (catalogue no. 10-I50F, clone no. 4090914; Fitzgerald, Concord, MA, USA), IFVA NA MAb (catalogue no. 11-227; Argene, North Massapequa, NY), IFVA NP MAb (catalogue no. 10-I50C, clone no. 322211; Fitzgerald), IFVB HA MAb (catalogue no. 66190; AbCAM, Cambridge, MA, USA), IFVB NA MAb (catalogue no. 19601; QED Biosciences, San Diego, CA, USA) and IFVB NP MAb (catalogue no. 10-I55B, clone no. 2110171; Fitzgerald)]. Medium was used in place of virus to determine background.
Viral NA activity assay
The viral NA activity assay was based on previously published versions.26–28 The wt and DS viruses were normalized on the basis of NA protein level (determined in protein level ELISA) and 2-fold serial dilutions of the equated viruses were pre-warmed to 37°C. Reaction buffer solution [2×, 400 μM 4-MUNANA (Sigma), 66 mM MES (Sigma) pH 6.5, 8 mM CaCl2, 0.2% NP40] was pre-warmed to 37°C. Virus dilution and 2× reaction buffer were mixed 1 : 1 in pre-warmed black 96-well microplates (BD Biosciences, San Jose, CA, USA) and incubated for 2 h at 37°C. Upon completion, stop solution was added (0.14 M NaOH/83% ethanol) and fluorescence intensity was determined immediately (excitation 365 nm, emission 455 nm, cut-off 435 nm). Medium was used in place of virus to determine background.
Viral binding dynamics on MDCK cells
The MDCK binding assay was performed as for the plaque size assay except that the amount of time the virus was allowed to bind to MDCK cells was varied before washing away unbound virus with PBS and overlaying with medium/agar. All binding was performed at room temperature, except for the maximal binding condition (2 h at 35°C). The number of resulting plaques with each binding condition was counted and normalized to the maximal binding condition.
Viral HA specificity determination
Glycan microarray analysis was performed by the Consortium for Functional Glycomics (CFG) (Core H, Emory University School of Medicine, Atlanta, GA, USA) under request number 1318. The Standard Tertiary Protocol was performed. In brief, virus, prepared as clarified tissue culture supernatant, was incubated for 1 h at 37°C on CFG glycan microarray v3.2 slides (containing 406 different glycans, each printed on six spots). After extensive washing, bound virus was detected with specific anti-IFVB HA MAb and Alexa488-labelled anti-mouse secondary antibody. Slides were washed extensively before detection of signal with a Perkin-Elmer Microarray XL4000 scanner. Data were analysed using Imagene (v6) image analysis software. Of the six replicate spots per glycan, the highest and lowest relative fluorescence unit (RFU) readings were discarded and the values reported are the means ± SEM of the four remaining spots. IFV A/Victoria/3/75 could not be tested on the glycan array due to lack of detectable binding with the wt virus.
Results
Generation of DS virus
To evaluate the potential for IFV to develop resistance to DAS181, we subjected two classical laboratory IFV strains, B/Maryland/1/59 and A/Victoria/3/75 (H3N2), to increasing selective pressure with DAS181 during multiple passages in MDCK cells. At each passage, a virus was cultured in parallel either in the presence of DAS181 (referred to as DAS181-selected or DS virus) or in the absence of DAS181 (referred to as wt virus) to enable comparison between the DS virus and the counterpart wt virus at each passage.
B/Maryland/1/59 was cultured for 102 days, comprising 34 passages, and A/Victoria/3/75 was cultured for 66 days, comprising 31 passages. DS and wt isolates of these two strains at selected passages were subjected to extensive phenotypic and genotypic analysis.
DAS181 sensitivity
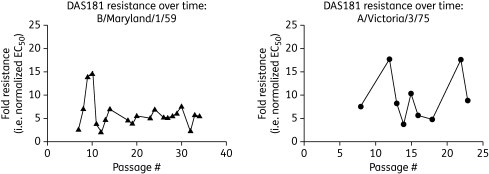
Evaluation of DAS181 sensitivity was performed on the wt and DS isolates from most passages using the plaque number reduction assay. The EC50 was calculated for each isolate, and fold resistance was determined as the ratio of DS EC50 to wt EC50. During DAS181 selection both B/Maryland/1/59 and A/Victoria/3/75 developed a low level (<20-fold) of resistance to DAS181 starting at passage 5–8; the fold resistance hovered around this level throughout the remaining passages (Figure 1 and Table 1). Due to the nature of the biological assay, EC50 results tend to vary (within 1 log); this was seen in that the extent of DAS181 resistance fluctuated between 3- and 15-fold for B/Maryland/1/59 and between 4- and 18-fold for A/Victoria/3/75 (Table 1). Importantly, the DS viruses maintained sensitivity to DAS181, albeit requiring slightly higher concentrations.
Figure 1.
DAS181 resistance level throughout passaging. wt and DS isolates from selected passages of IFV B/Maryland/1/59 (left-hand panel) were analysed in parallel for sensitivity to DAS181. Similarly, wt and DS isolates from selected passages of IFV A/Victoria/3/75 (right-hand panel) were analysed in parallel for sensitivity to DAS181. Fold resistances were quantified using derived EC50 values (DS EC50/wt EC50) at each passage.
Table 1.
Relative in vitro drug sensitivity and genotype of DS mutants
| DAS181 |
DS genotype |
|||||
|---|---|---|---|---|---|---|
| Virus | Passage no. | wt EC50 (µM) | DS EC50 (µM) | fold resistance | HA mutations | NA mutations |
| B/Maryland/1/59 | ||||||
| 7 | 0.07 | 0.22 | 3 | G137R | − | |
| 8 | 0.09 | 0.68 | 8 | G137R | − | |
| 10 | 0.05 | 0.73 | 15 | G137R | − | |
| 14 | 0.13 | 0.96 | 7 | G137R, S136T | − | |
| 18 | 0.20 | 0.94 | 5 | G137R, S136T | W438L | |
| 23 | 0.17 | 0.89 | 5 | G137R, S136T | W438L | |
| 34 | 0.14 | 0.81 | 6 | G137R, S136T | W438La | |
| A/Victoria/3/75 (H3N2) | ||||||
| 3 | NT | NT | NT | − | − | |
| 6 | NT | NT | NT | S186I | − | |
| 8 | 4.5 | 34.2 | 8 | NT | NT | |
| 12 | 1.7 | 30.8 | 18 | S186I | − | |
| 14 | 6.4 | 23.9 | 4 | S186I | K210R | |
| 15 | 1.6 | 16.1 | 10 | S186I | − | |
| 18 | 4.2 | 20.3 | 5 | NT | NT | |
| 22 | 1.7 | 30.4 | 18 | NT | NT | |
| 26 | NT | NT | NT | S186I | − | |
−, no mutations relative to wt virus; NT, not tested.
aHeterogeneity in viral pool.
IFV A/Victoria/3/75 and B/Maryland/1/59 were passaged with or without DAS181 at the concentrations indicated. Sensitivity to DAS181 in plaque reduction assay and HA/NA genotypes were subsequently determined for wt and DS samples from each passage and data for selected representative passages are shown. Values in bold indicate the maximum resistance level observed with simultaneous assessment of most available passages.
Viral growth and cytotoxicity in vitro
When MDCK cells were infected at an equal moi, the DS isolates grew at a slower rate than their wt counterparts, as indicated by the MDCK cell survival curves (Figure 2). While not a direct measure of viral replication, the cell survival curves do reflect the slower spread of the virus through the culture.
Figure 2.
DS IFV growth dynamics in vitro. Confluent monolayers of MDCK were infected with wt and DS IFVs at equivalent infectious doses. At selected timepoints the wells were fixed with Crystal Violet to determine the amount of live cells. The percentage of cells remaining was calculated by comparing with the uninfected control. Values represent means ± SD of triplicate samples.
In order to determine the particle to pfu ratio of the wt and DS viruses, virus quantification was measured by qRT–PCR as well as by plaque assay (Table 2). These two techniques allow the assessment of the viral M-gene copy number compared with infectious virions. These analyses demonstrated that the particle to pfu ratio was higher in both DS viruses relative to the wt viruses. In each case, the DS viruses had higher M-gene copy numbers by >1 log compared with the wt viruses. This result indicates that the DS viruses were impaired and produced a greater number of defective particles compared with the wt counterparts.
Table 2.
wt and DS viral titres as determined by distinct methods
| Virus | M-gene copy number/mL | pfu/mL | M gene:pfu ratio |
|---|---|---|---|
| B/Maryland/1/59 wt | 5.44 e10 | 1.3 e8 | 4.2 e2 : 1 |
| B/Maryland/1/59 DS | 5.72 e10 | 8.1 e6 | 7.1 e3 : 1 |
| A/Victoria/3/75 wt | 2.44 e10 | 1.2 e8 | 2.0 e2 : 1 |
| A/Victoria/3/75 DS | 6.98 e10 | 1.9 e7 | 3.7 e3 : 1 |
Undiluted solutions of IFV A/Victoria/3/75 wt and DS and IFV B/Maryland wt and DS, produced simultaneously in MDCK cells, were quantified for viral titre by two primary methods: qRT–PCR for viral M-gene copy number and plaque assay for pfu.
e = power of 10.
Plaque size
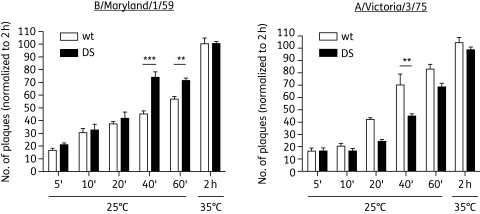
Consistent with reduced fitness, the DS viruses formed significantly smaller plaques than their wt counterparts in the absence of any drug. The plaques of the late-passage DS viruses were on average 0.5 mm in diameter whereas the wt plaques were 2–3 mm in diameter (Figure 3a and b). The representative pictures in Figure 3(a) also demonstrate that the DS virus is still sensitive to DAS181 at higher concentrations (see DAS181-treated DS plaque assay). Unlike the wt virus, the DS isolates also exhibited a DAS181-dependent increase in plaque size at low DAS181 concentrations. While plaque size of the wt virus was reduced in a dose-dependent manner in response to increasing DAS181 treatment, plaque size of the DS virus increased with increasing DAS181 concentrations up to 1.5 μM, at which point the DS virus plaques peaked at a size comparable to the untreated wt plaques. As DAS181 concentration increased further, the DS plaques decreased in size [Figure 3c and Figure S5 (available as Supplementary data at JAC Online)] and number (data not shown).
Figure 3.
Phenotypic characterization of DS IFV: plaque size. Confluent monolayers of MDCK cells were infected with wt and DS IFVs and overlaid with 1 : 1 agar/medium to allow plaque formation. Plaque sizes of wt (left-hand panel) and DS passage 22 IFV B/Maryland (right-hand panel) grown with PBS or DAS181 (a). Plaque sizes of wt (white bars) and DS (black bars) IFV B/Maryland/1/59 (passage 17) and A/Victoria/3/75 (passage 14) grown without drug (b). Plaque sizes of wt (broken line) or DS (continuous line) A/Victoria/3/75 virus (passage 13) in the presence of the indicated concentrations of DAS181 (c). Plaque sizes of wt, DS and DW viruses (normalized at each passage to wt) shown to indicate the return to near-wt plaque size in DW viruses after DAS181 removal (d and e). Values represent mean ± SD size of ≥24 plaques per condition.
Stability of DAS181 resistance phenotype
To examine the innate stability of the DS viruses, DS isolates from passage 14 (for A/Victoria/3/75) and passage 17 (for B/Maryland/1/59 strains) were subjected to further passaging in the absence of DAS181. These viruses, isolated after removal of DAS181 from culture, are referred to as DW viruses. Shortly after DAS181 withdrawal the DW isolates exhibited a partial restoration of DAS181 sensitivity. After six to nine passages, the B/Maryland/1/59 DW isolates returned to near wt sensitivity to DAS181 (92–99% reduction in DAS181 resistance) (data not shown). The A/Victoria/3/75 DW isolates also regained substantial sensitivity to DAS181 (90% reduction in DAS181 resistance) after three passages (data not shown). Consistent with the reversion to wt phenotype, the DW isolates exhibited a gradual increase in plaque size to near wt levels, coinciding with recovery of DAS181 sensitivity (Figure 3d and e). These results suggest that the DS phenotype was unstable and the innate tendency was for the virus to return to the wt phenotype in the absence of continuous selection by DAS181.
Virulence of the DS isolates in vivo
Consistent with the growth deficiency observed in cell culture, in mice the B/Maryland/1/59 DS isolates caused dramatically less lethality, weight loss and viral replication in mouse lungs compared with the wt counterparts at equivalent challenge doses measured in M-gene copy number (i.e. total viral particle number). When challenged with equivalent numbers of infectious viral particles (pfu), mice exhibited equivalent body weight loss (compare the highest DS dose group with the lowest wt dose group in Figure 4), indicating that the DS infectious virions cause equal virulence as measured by body weight loss. However, it should be noted that, at an equivalent pfu challenge dose, the mice treated with DS virus were exposed to roughly 17 times more viral particles than the mice challenged with the wt counterpart. Nevertheless, on a pfu-equated basis, the results indicate that the B/Maryland/1/59 DS isolate is no more virulent than the wt virus.
Figure 4.
Relative viral fitness in vivo. BALB/C mice were inoculated intranasally with wt and DS B/Maryland/1/59 (inocula shown in M-gene copy number and pfu). Survival (top) and body weight (middle) were tracked daily for 15 days post-infection; viral titre in lung homogenate was determined at 3 days post-infection for one dose group (bottom, 8.4–8.5 e8 copies). wt at 1.7 e8 copies had 100% survival; all DS groups had 100% survival. Animals with greatest body weight loss often did not survive infection, thus body weight recovery curves are in some cases escalated by the more modest weight loss of survivors. Values represent means ± SEM of eight mice per group in survival and body weight analyses, and three mice per group in viral titre analyses. ***P<0.001, significantly different from identical challenge dose of wt infection as determined by analysis of variance with the Bonferroni post-test.
At equivalent challenge doses (measured in pfu), infections by the A/Victoria/3/75 DS isolate were also associated with body weight loss comparable to that caused by wt infection and neither virus was lethal (data not shown). Finally, it should be noted that the mouse model is not an ideal setting to examine IFV with altered HA/NA properties as the pattern of sialylation in the mouse airway is not an accurate mimic of that in the human airway.
Genotypic characterization
The HA and NA gene segments of the wt, DS and DW isolates of B/Maryland/1/59 and A/Victoria/3/75 were sequenced at several selected passages (Tables S1 and S2, available as Supplementary data at JAC Online). In the B/Maryland/1/59 DS isolates, two stable mutations were observed in the HA gene, G137R appearing at passage 5 and S136T appearing at passage 13. In the A/Victoria/3/75 DS isolates, one stable mutation in the HA gene, S186I, was observed at passage 6 and beyond. Interestingly, in both IFV strains the appearance of the HA mutations around passage 5 or 6, G137R for B/Maryland/1/59 and S186I for A/Victoria/3/75, coincided with reduced sensitivity to DAS181 (Table 1).
In the IFV NA gene, stable mutations were only observed for the B/Maryland/1/59 DS isolates and these occurred late in passaging and with heterogeneity in the virus pool; W438L appeared at passage 18 and L38P appeared at passage 27. A transient NA mutation was observed in the A/Victoria/3/75 DS isolate, K210R at passage 14 only. Genotyping of all other viral genes (NP, NS, M1, PA, PB1, PB2) in the wt and DS B/Maryland/1/59 (passage 17) or A/Victoria/3/75 (passage 14) revealed no additional mutations.
Comparison with published HA and NA protein structures revealed that the HA mutations in the DS isolates are located near the sialic acid binding site (Figure 5).23,30 Thus, these HA mutations could potentially affect binding affinity and/or specificity to host cell sialic acid receptors. In contrast, the identified stable NA mutations are not close to the active site of the enzyme24,31 (Table 1 and Figure 6); thus, their significance remains unclear.
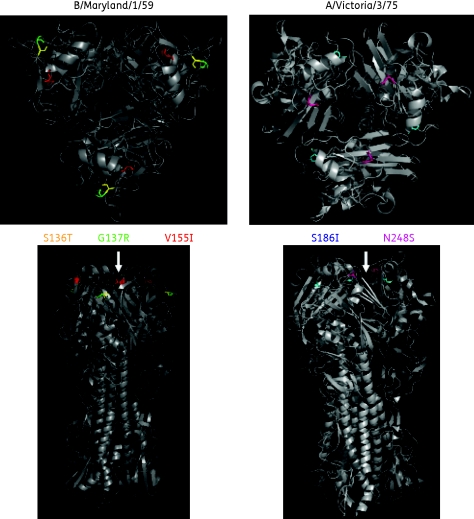
Figure 5.
Location of HA mutations identified in DS and DW IFV. The amino acid positions for the stable HA mutations (listed in Tables S1 and S2) were located on the published crystal structure of an IFV B or IFV A H3 HA homotrimer.22,23 In the IFV B/Maryland/1/59 model amino acid 136 is shown in yellow, amino acid 137 is shown in green and amino acid 155 is shown in red. In the IFV A/Victoria/3/75 model amino acid 186 is shown in blue and amino acid 248 is shown in pink; the T10I mutation in A/Victoria/3/75 wt HA lies in the transmembrane region and is not modelled. The top panels show a top-down view of the HA homotrimer while the bottom panels show a side view. The approximate site of sialic acid binding is indicated with arrows.23,30
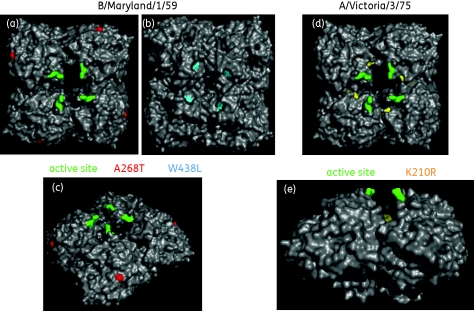
Figure 6.
Location of NA mutations identified in DS and DW IFV. The amino acid positions for the NA mutations (listed in Tables S1 and S2) were located on the published crystal structure of an IFV A N9 homotetramer.24 In the IFV B/Maryland/1/59 model (a–c) amino acid 268 is shown in red and amino acid 438 is shown in blue; amino acid 38 lies in the transmembrane region and is not modelled. In the IFV A/Victoria/3/75 model (d and e) amino acid 210 is shown in yellow. (a and d) Top-down view. (b) Bottom-up view. (c and e) Side view. The known active site is indicated with green residues.24,31
Interestingly, as the DW viruses regained near wt sensitivity to DAS181, they did not lose the DS mutations; instead, they acquired additional HA and NA mutations that coincided with the reversal to near wt phenotype (Tables S1 and S2). Of note, each DW virus exhibited one new stable HA mutation (V155I in B/Maryland/1/59 and N248S in A/Victoria/3/75), both of which are located near the sialic acid binding site (Figure 5). These results suggest that, after withdrawal from DAS181 selection, the acquisition of additional HA mutations could be responsible for the return of the DW IFVs to near wt DAS181 sensitivity.
Molecular characterization of the DS isolates
Viral protein levels
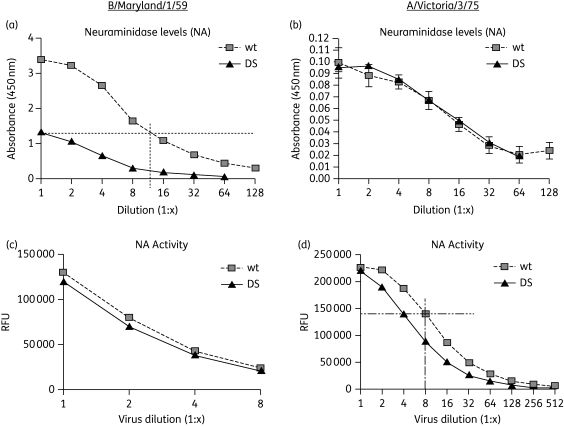
The relative HA, NA and NP protein levels in the DS and wt viruses were measured by an ELISA method with antibodies specific to the viral HA, NA and NP antigens. When applied to the ELISA at an approximately equal level of NP protein (which should remain unaffected during selection), equivalent amounts of HA protein were found in B/Maryland/1/59 wt and DS viruses (data not shown). However, the NA protein level was dramatically different. The amount of NA protein detected in the DS B/Maryland/1/59 was only about 10% of that in the wt B/Maryland/1/59 (Figure 7a). This result suggests that for B/Maryland/1/59 the DS isolate carries approximately 90% less NA protein compared with the wt virus when normalized by equal NP protein. Quantifying protein levels via dot blot assay revealed similar results (data not shown).
Figure 7.
Relative NA protein and activity levels in DS IFV. Serial dilutions of wt and DS IFV B/Maryland/1/59 (a) or A/Victoria/3/75 (b) (pre-equilibrated by NP protein level) were probed for HA, NA or NP protein by ELISA with specific antibodies to each viral antigen. Only NA protein levels are shown. The horizontal and vertical broken lines in (a) indicate the relative amount of NA protein in DS IFV B/Maryland/1/59 (∼10% of wt). Serial dilutions of wt and DS IFV B/Maryland/1/59 (c) or A/Victoria/3/75 (d) (pre-equilibrated by NA protein level) were tested for NA activity level and RFU readout was plotted against viral dilution. The horizontal and vertical broken lines in (d) indicate the relative amount of NA activity in DS IFV A/Victoria/3/75 (∼50% of wt). Error bars indicate means ± SD of duplicate samples.
The DS and wt A/Victoria/3/75 had equivalent measurements of both HA and NA (data not shown and Figure 7b), indicating that the A/Victoria/3/75 DS virus has HA and NA protein compositions similar to those of the wt virus.
Viral NA activity
To determine the enzymatic activity of NA proteins in the DS isolates, relative NA activity levels of the wt and DS viruses were measured after equating the counterpart IFVs based on approximate NA protein levels (see Figure 7a and b). For B/Maryland/1/59, the DS and wt viruses had equivalent activity, while for A/Victoria/3/75 the DS isolate exhibited approximately 50% activity relative to the wt virus (Figure 7c and d). As no stable mutations were identified in the NA gene of the A/Victoria/3/75 DS isolate, it is not apparent why the NA function is diminished in this virus. Together with the results on NA protein level, these findings indicate that the DS isolates of both B/Maryland/1/59 and A/Victoria/3/75 have impaired NA function relative to that of the wt viruses, which is either due to a reduced NA level (for B/Maryland/1/59) or reduced NA enzymatic activity (for A/Victoria/3/75).
Viral binding dynamics
To assess HA/sialic acid binding affinity of the wt and DS isolates, we evaluated the relative dynamics of viral binding to MDCK cells by incubating an equivalent infectious dose (in pfu) of wt and DS virus with MDCK cells for varying durations, followed by washing of the unbound virus at the end of incubation and counting the resulting number of plaques. Graphing the percentage of maximal binding as a function of time revealed that the B/Maryland/1/59 DS virus bound significantly more efficiently to MDCK cells than did the wt counterpart virus (Figure 8). In contrast, the A/Victoria/3/75 DS virus bound to MDCK cells significantly less efficiently than did the wt counterpart virus (Figure 8). Comparable results were obtained when analysing viral binding to fetuin or synthetic sialylated glycans (data not shown). These results suggest that, compared with counterpart wt virus, B/Maryland/1/59 DS virus has an enhanced receptor binding ability, whereas A/Victoria/3/75 DS virus may be somewhat impaired in receptor binding. It should be noted, however, that only the infectious virions were assessed in these binding experiments. Since both of the DS viral populations contain a high level of defective virions, which may compete against the infectious virions, the real binding ability of the DS infectious virions could be significantly higher than the apparent binding by the whole viral population.
Figure 8.
Relative HA affinity of DS IFV: binding dynamics on MDCK cells. wt and DS IFV B/Maryland/1/59 (left-hand panel) and A/Victoria/3/75 (right-hand panel) were applied to MDCK cells at equal infection levels for the indicated incubation times and temperatures before washing away unbound virus and overlaying with agar. Three days later the number of plaques was counted for each binding condition and normalized to maximal binding. For B/Maryland/1/59, the DS virus bound significantly faster than wt. For A/Victoria/3/75, the DS virus bound significantly slower than wt. Values represent means ± SEM of triplicate samples, and the unpaired t-test was used to calculate statistical significance; **, *** = P < 0.01, P < 0.001, respectively, using ANOVA/Bonferroni post-test.
Viral HA specificity
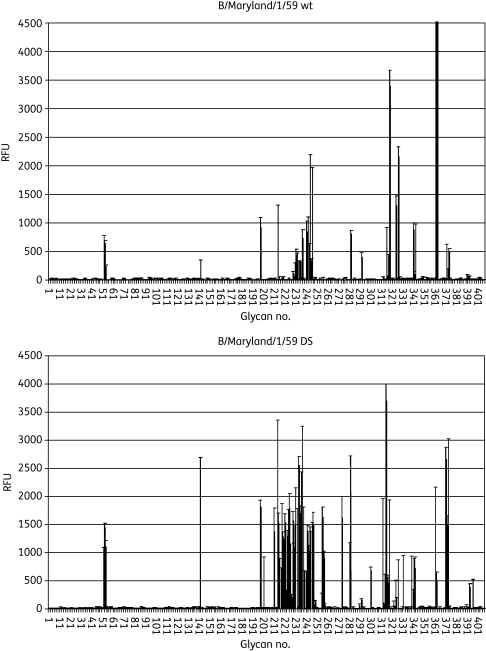
To assess the mechanism of increased receptor binding by B/Maryland/1/59 DS virus, we characterized receptor binding specificity of the B/Maryland/1/59 DS isolate using a glycan microarray that contained 406 discrete carbohydrates, including over 100 sialylated glycans.32–36 When binding intensity was compared across the panel of glycans, a pattern of differential glycan binding specificity between the wt and DS isolates was apparent (Figure 9). The wt B/Maryland/1/59 virus primarily recognized α2,6-linked sialic acids while the DS virus exhibited broader receptor specificity by acquiring significant recognition for α2,3-linked sialic acids in addition to the α2,6-linked sialic acids (Table S3, available as Supplementary data at JAC Online). Since MCDK cells express both α2,3- and α2,6-linked sialic acids,37,38 such broadened receptor binding specificity may represent an adaptive change by the B/Maryland/1/59 DS virus to have better overall receptor binding efficiency in MDCK cells.
Figure 9.
Relative HA specificity of DS IFV B/Maryland/1/59. Binding of IFV B/Maryland/1/59 to the CFG v3.2 glycan array was detected with specific antibodies to viral antigen; readout is described arbitrarily as RFU. wt and DS viruses were applied at approximately equivalent total fluorescence units (across the entire array) and the intensity of binding to each of the 406 glycans on the array was graphed. Note a relative increase in the number of glycans bound by the DS virus. Values represent means ± SEM of quadruplicate samples. Detailed information on the top 20 bound glycans in each graph is shown in Table S3.
The A/Victoria/3/75 viruses did not bind significantly on the glycan microarray for reasons that could not be identified despite repeated attempts, hence these analyses were not conducted for A/Victoria/3/75 wt and DS isolates.
Discussion
By subjecting two IFV laboratory strains to multiple passages in the presence of continuous selection with DAS181, DS viral isolates that are less sensitive to DAS181 than wt counterparts were obtained. These DS isolates are distinct from the well characterized NAI-resistant IFV strains. First, in contrast to the 1000- to 10 000-fold drug resistance commonly reported for NAI-resistant IFVs,18,39–53 the extent of observed DAS181 resistance was low (<20-fold). As a result, the DS viruses were still responsive to DAS181 treatment in vitro, albeit requiring a slightly higher dose of DAS181 for inhibition compared with wt virus. Second, NAI-resistant IFVs are phenotypically and genotypically stable once they are established46 and reversion to NAI sensitivity upon NAI withdrawal has never been reported. In fact, the H274Y oseltamivir-resistant IFV A is so stable and fit that H1N1 viruses carrying this mutation became the globally dominant seasonal strain in a period of 2 years, reaching nearly 100% prevalence.2,5–7,54,55 By contrast, resistance to DAS181 was unstable for both viruses in this study and the virus exhibited deficient growth characteristics in vitro. The DS isolates quickly returned to near wt sensitivity once they were passaged in the absence of DAS181 selective pressure. These findings suggest that the DS phenotype may be detrimental to the fitness of IFV, leading to its elimination by natural selection.
Indeed, the DS isolates appear to grow slower and are less infectious and less virulent compared with the wt counterpart viruses based on in vitro characterization. In mice, both DS isolates exhibited equivalent virulence to the respective parental wt virus when the mice were challenged at equivalent pfu, suggesting that DAS181 selection did not generate mutant strains with increased fitness. When normalized by equal M-gene copy number, the B/Maryland/1/59 DS virus exhibited reduced fitness in the mouse model. Molecular characterization of the B/Maryland/1/59 DS virus revealed an imbalance between the HA and NA functions; compared with the wt virus, the DS virus exhibited higher receptor binding affinity (likely due to a broadened spectrum of receptor recognition), but carried only 10% of the NA protein. Thus, for the B/Maryland/1/59 DS virus the functional ratio of HA/NA is skewed compared with the wt counterpart. Interestingly, this is functionally equivalent to the effect of NAI treatment and it may explain the attenuated features of the virus. For A/Victoria/3/75, the attenuated DS phenotype appears to be associated with reduced function of NA as well (even though a correlative amino acid mutation was not detected in this study), although the overall HA function is reduced rather than enhanced for this virus.
For B/Maryland/1/59, DAS181 resistance correlated with increased binding to α2,3-linked sialic acids, but this could not be tested for A/Victoria/3/75. While such a change may enable the virus to adapt to DAS181-treated MDCK cells, it is unlikely that such a change would confer a significant advantage for the virus in the human upper airway epithelium, which predominantly expresses α2,6-linked sialic acids. This notion is supported by the fact that IFV strains of avian origin, which predominantly recognize α2,3-linked sialic acids, do not effectively infect humans unless the virus becomes adapted by acquiring recognition of α2,6-linked sialic acid receptors,29,56,57 Finally, DAS181 efficiently cleaves all sialic acid structures tested, including numerous α2,3- and α2,6-linked sialic acid forms (F. Fang and G. B. Triana-Baltzer, unpublished observation); therefore, alteration of sialic acid binding specificity alone is unlikely to have significant impact on DAS181 effectiveness clinically.
As a segmented RNA virus, IFV is highly prone to errors during replication. However, in the normal host cell environment well-assembled fit virions have a selective advantage over deficient ones during amplification. By altering the cellular environment, DAS181 seems to eliminate this growth advantage for the fit virions. To this end, we have observed that the DS isolates contain significantly fewer virions capable of establishing infection; they not only exhibit altered receptor specificity but they are also deficient in NA activity. The emergence of impairment in NA of the DS isolates can potentially be attributed to DAS181, which is functionally homologous to IFV NA and thus makes the viral NA function superfluous. Being NA deficient, the DS virus may have impaired fitness in the absence of DAS181 (i.e. DAS181 dependence), as demonstrated by the increased plaque size of the DS virus when exposed to low levels of DAS181. In agreement with our findings, a previous study also reported that a sialidase-selected IFV carried large and unstable NA gene deletions and required the presence of exogenous sialidase to sustain its infection.58,59
Several HA mutations, located near the sialic acid binding site, have been identified in the DS isolates that correlate with a change in receptor specificity and reduced sensitivity to DAS181. The S136T and G137R mutations are found in the HA molecule of DS B/Maryland/1/5. Previously, a mutation corresponding to amino acid 137 in peramivir-selected IFV A/Singapore/1/57 (H2N2) was shown to reduce receptor binding affinity60 and this report supports the involvement of amino acid 137 in receptor binding. An S186I mutation was found in HA of the A/Victoria/3/75 DS virus. A similar mutation, S186F, was previously identified in IFV A/NWS/G70 (H1N9) grown in the presence of zanamivir.43 The S186F mutation conferred only 10-fold resistance to zanamivir, had little impact on receptor binding and was stably carried through additional passages.43 Notably, the DS S186I mutation in DS A/Victoria/3/75 confers no cross-resistance to zanamivir (data not shown).
Drug resistance associated with mutations in both the HA and the NA gene has been well documented with NAI-resistant IFVs.61 Of these two genes, HA is directly implicated in reduced DAS181 sensitivity, based on our findings. With the widespread occurrence of the H274Y mutation in NA, clinical NAI resistance surveillance is solely focused on detecting NA mutation and function, rather than HA changes. The lack of an accepted clinically relevant model of HA function is also an issue, due to the fact that any HA functional changes detected in MCDK cells, which have different sialic acid receptor expression patterns from the human respiratory epithelium, may not be relevant for IFV infection in the human airway and vice versa.62 This current report suggests that HA is also important in mediating reduced sensitivity to DAS181. Consequently, monitoring for DAS181 resistance in the clinical setting should be based on connecting a lack of clinical and virological response to DAS181 treatment in patients with specific HA, as well as NA, genotypic changes between the pre- and post-treatment clinical samples, rather than the MDCK cell-based phenotypic assays.
DAS181 is unique among all existing anti-influenza agents in regard to its host-targeting mechanism of action. Our studies have revealed that DAS181 may disrupt the foundation of robust IFV infection. In addition to preventing IFV from binding to sialic acid receptors, DAS181 may also encourage the growth of HA/NA imbalanced viruses by removing the selective growth advantage of normal virus production. Such fundamentally imbalanced viruses may be unlikely to circulate in the community due to a marked reduction in fitness.
Funding
The work at NexBio, Inc., was supported in part by NIH/National Institute of Allergy and Infectious Diseases (grant numbers U01AI070281 and R44AI056786, contract number HHSN266200600015C). The work at Consortium for Functional Glycomics was supported by the NIGMS (grant number GM62116).
Transparency declarations
G. B. T.-B., R. L. S., M. H., K. A. J., L. M. A., J. L. L. and F. F. are/were employees of NexBio, Inc., a developer of DAS181, and declare that they have competing financial interests including ownership of stock or options in the company.
Supplementary data
Supplementary Material
Acknowledgements
We thank the Consortium for Functional Glycomics and Core H at Emory University School of Medicine, Atlanta, GA, USA, for performing the glycan array analysis. We also thank the University of Washington Clinical Reference Laboratory for assistance with the qRT–PCR assays. We thank Eley K. Wong for assistance in cell culture and plaque reduction assay quantification and David Wurtman for critical evaluation of this manuscript.
References
- 1.WHO. Seasonal Influenza Factsheet. http://www.who.int/mediacentre/factsheets/fs211/en/index.html , 12 July 2010, date last accessed. [Google Scholar]
- 2.Dharan NJ, Gubareva LV, Meyer JJ, et al. Infections with oseltamivir-resistant influenza A(H1N1) virus in the United States. JAMA. 2009;301:1034–41. doi: 10.1001/jama.2009.294. doi:10.1001/jama.2009.294. [DOI] [PubMed] [Google Scholar]
- 3.Hatakeyama S, Sugaya N, Ito M, et al. Emergence of influenza B viruses with reduced sensitivity to neuraminidase inhibitors. JAMA. 2007;297:1435–42. doi: 10.1001/jama.297.13.1435. doi:10.1001/jama.297.13.1435. [DOI] [PubMed] [Google Scholar]
- 4.Hayden FG, Hay AJ. Emergence and transmission of influenza A viruses resistant to amantadine and rimantadine. Curr Top Microbiol Immunol. 1992;176:119–30. doi: 10.1007/978-3-642-77011-1_8. [DOI] [PubMed] [Google Scholar]
- 5.Moscona A. Oseltamivir resistance—disabling our influenza defenses. N Engl J Med. 2005;353:2633–6. doi: 10.1056/NEJMp058291. doi:10.1056/NEJMp058291. [DOI] [PubMed] [Google Scholar]
- 6.Moscona A. Global transmission of oseltamivir-resistant influenza. N Engl J Med. 2009;360:953–6. doi: 10.1056/NEJMp0900648. doi:10.1056/NEJMp0900648. [DOI] [PubMed] [Google Scholar]
- 7.Sheu TG, Deyde VM, Okomo-Adhiambo M, et al. Surveillance for neuraminidase inhibitor resistance among human influenza A and B viruses circulating worldwide from 2004 to 2008. Antimicrob Agents Chemother. 2008;52:3284–92. doi: 10.1128/AAC.00555-08. doi:10.1128/AAC.00555-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki H, Saito R, Masuda H, et al. Emergence of amantadine-resistant influenza A viruses: epidemiological study. J Infect Chemother. 2003;9:195–200. doi: 10.1007/s10156-003-0262-6. doi:10.1007/s10156-003-0262-6. [DOI] [PubMed] [Google Scholar]
- 9.CDC. Update: drug susceptibility of swine-origin influenza A (H1N1) viruses, April 2009. MMWR Morb Mortal Wkly Rep. 2009;58:433–5. [PubMed] [Google Scholar]
- 10.Couzin-Frankel J. Swine flu outbreak. What role for antiviral drugs? Science. 2009;324:705. doi: 10.1126/science.324_705. doi:10.1126/science.324_705. [DOI] [PubMed] [Google Scholar]
- 11.Scalera NM, Mossad SB. The first pandemic of the 21st century: a review of the 2009 pandemic variant influenza A (H1N1) virus. Postgrad Med. 2009;121:43–7. doi: 10.3810/pgm.2009.09.2051. doi:10.3810/pgm.2009.09.2051. [DOI] [PubMed] [Google Scholar]
- 12.Belser JA, Lu X, Szretter KJ, et al. DAS181, a novel sialidase fusion protein, protects mice from lethal avian influenza H5N1 virus infection. J Infect Dis. 2007;196:1493–9. doi: 10.1086/522609. doi:10.1086/522609. [DOI] [PubMed] [Google Scholar]
- 13.Chan RW, Chan MC, Wong AC, et al. DAS181 inhibits H5N1 influenza virus infection of human lung tissues. Antimicrob Agents Chemother. 2009;53:3935–41. doi: 10.1128/AAC.00389-09. doi:10.1128/AAC.00389-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Malakhov MP, Aschenbrenner LM, Smee DF, et al. Sialidase fusion protein as a novel broad-spectrum inhibitor of influenza virus infection. Antimicrob Agents Chemother. 2006;50:1470–9. doi: 10.1128/AAC.50.4.1470-1479.2006. doi:10.1128/AAC.50.4.1470-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Triana-Baltzer GB, Gubareva LV, Klimov AI, et al. Inhibition of neuraminidase inhibitor-resistant influenza virus by DAS181, a novel sialidase fusion protein. PLoS ONE. 2009;4:e7838. doi: 10.1371/journal.pone.0007838. doi:10.1371/journal.pone.0007838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Triana-Baltzer GB, Gubareva LV, Nicholls JM, et al. Novel pandemic influenza A(H1N1) viruses are potently inhibited by DAS181, a sialidase fusion protein. PLoS ONE. 2009;4:e7788. doi: 10.1371/journal.pone.0007788. doi:10.1371/journal.pone.0007788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Triana-Baltzer GB, Babizki M, Chan MC, et al. DAS181, a sialidase fusion protein, protects human airway epithelium against influenza virus infection: an in vitro pharmacodynamic analysis. J Antimicrob Chemother. 2010;65:275–84. doi: 10.1093/jac/dkp421. doi:10.1093/jac/dkp421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKimm-Breschkin JL, Blick TJ, Sahasrabudhe A, et al. Generation and characterization of variants of NWS/G70C influenza virus after in vitro passage in 4-amino-Neu5Ac2en and 4-guanidino-Neu5Ac2en. Antimicrob Agents Chemother. 1996;40:40–6. doi: 10.1128/aac.40.1.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hayden FG, Cote KM, Douglas RG., Jr Plaque inhibition assay for drug susceptibility testing of influenza viruses. Antimicrob Agents Chemother. 1980;17:865–70. doi: 10.1128/aac.17.5.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann E, Stech J, Guan Y, et al. Universal primer set for the full-length amplification of all influenza A viruses. Arch Virol. 2001;146:2275–89. doi: 10.1007/s007050170002. doi:10.1007/s007050170002. [DOI] [PubMed] [Google Scholar]
- 21.Colman PM, Hoyne PA, Lawrence MC. Sequence and structure alignment of paramyxovirus hemagglutinin-neuraminidase with influenza virus neuraminidase. J Virol. 1993;67:2972–80. doi: 10.1128/jvi.67.6.2972-2980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha Y, Stevens DJ, Skehel JJ, et al. X-ray structure of the hemagglutinin of a potential H3 avian progenitor of the 1968 Hong Kong pandemic influenza virus. Virology. 2003;309:209–18. doi: 10.1016/s0042-6822(03)00068-0. doi:10.1016/S0042-6822(03)00068-0. [DOI] [PubMed] [Google Scholar]
- 23.Wang Q, Tian X, Chen X, et al. Structural basis for receptor specificity of influenza B virus hemagglutinin. Proc Natl Acad Sci USA. 2007;104:16874–9. doi: 10.1073/pnas.0708363104. doi:10.1073/pnas.0708363104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Varghese JN, Epa VC, Colman PM. Three-dimensional structure of the complex of 4-guanidino-Neu5Ac2en and influenza virus neuraminidase. Protein Sci. 1995;4:1081–7. doi: 10.1002/pro.5560040606. doi:10.1002/pro.5560040606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; 2002. http://www.pymol.org . [Google Scholar]
- 26.Gubareva LV, Nedyalkova MS, Novikov DV, et al. A release-competent influenza A virus mutant lacking the coding capacity for the neuraminidase active site. J Gen Virol. 2002;83:2683–92. doi: 10.1099/0022-1317-83-11-2683. [DOI] [PubMed] [Google Scholar]
- 27.Kobasa D, Kodihalli S, Luo M, et al. Amino acid residues contributing to the substrate specificity of the influenza A virus neuraminidase. J Virol. 1999;73:6743–51. doi: 10.1128/jvi.73.8.6743-6751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKimm-Breschkin J, Trivedi T, Hampson A, et al. Neuraminidase sequence analysis and susceptibilities of influenza virus clinical isolates to zanamivir and oseltamivir. Antimicrob Agents Chemother. 2003;47:2264–72. doi: 10.1128/AAC.47.7.2264-2272.2003. doi:10.1128/AAC.47.7.2264-2272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matrosovich M, Tuzikov A, Bovin N, et al. Early alterations of the receptor-binding properties of H1, H2, and H3 avian influenza virus hemagglutinins after their introduction into mammals. J Virol. 2000;74:8502–12. doi: 10.1128/jvi.74.18.8502-8512.2000. doi:10.1128/JVI.74.18.8502-8512.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem. 2000;69:531–69. doi: 10.1146/annurev.biochem.69.1.531. doi:10.1146/annurev.biochem.69.1.531. [DOI] [PubMed] [Google Scholar]
- 31.Air GM, Laver WG. The neuraminidase of influenza virus. Proteins. 1989;6:341–56. doi: 10.1002/prot.340060402. doi:10.1002/prot.340060402. [DOI] [PubMed] [Google Scholar]
- 32.Belser JA, Blixt O, Chen LM, et al. Contemporary North American influenza H7 viruses possess human receptor specificity: implications for virus transmissibility. Proc Natl Acad Sci USA. 2008;105:7558–63. doi: 10.1073/pnas.0801259105. doi:10.1073/pnas.0801259105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumari K, Gulati S, Smith DF, et al. Receptor binding specificity of recent human H3N2 influenza viruses. Virol J. 2007;4:42. doi: 10.1186/1743-422X-4-42. doi:10.1186/1743-422X-4-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens J, Blixt O, Glaser L, et al. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol. 2006;355:1143–55. doi: 10.1016/j.jmb.2005.11.002. doi:10.1016/j.jmb.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 35.Stevens J, Blixt O, Tumpey TM, et al. Structure and receptor specificity of the hemagglutinin from an H5N1 influenza virus. Science. 2006;312:404–10. doi: 10.1126/science.1124513. doi:10.1126/science.1124513. [DOI] [PubMed] [Google Scholar]
- 36.Stevens J, Blixt O, Chen LM, et al. Recent avian H5N1 viruses exhibit increased propensity for acquiring human receptor specificity. J Mol Biol. 2008;381:1382–94. doi: 10.1016/j.jmb.2008.04.016. doi:10.1016/j.jmb.2008.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hatakeyama S, Sakai-Tagawa Y, Kiso M, et al. Enhanced expression of an alpha2,6-linked sialic acid on MDCK cells improves isolation of human influenza viruses and evaluation of their sensitivity to a neuraminidase inhibitor. J Clin Microbiol. 2005;43:4139–46. doi: 10.1128/JCM.43.8.4139-4146.2005. doi:10.1128/JCM.43.8.4139-4146.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matrosovich M, Matrosovich T, Carr J, et al. Overexpression of the alpha-2,6-sialyltransferase in MDCK cells increases influenza virus sensitivity to neuraminidase inhibitors. J Virol. 2003;77:8418–25. doi: 10.1128/JVI.77.15.8418-8425.2003. doi:10.1128/JVI.77.15.8418-8425.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Barnett JM, Cadman A, Burrell FM, et al. In vitro selection and characterisation of influenza B/Beijing/1/87 isolates with altered susceptibility to zanamivir. Virology. 1999;265:286–95. doi: 10.1006/viro.1999.0058. doi:10.1006/viro.1999.0058. [DOI] [PubMed] [Google Scholar]
- 40.Baum EZ, Wagaman PC, Ly L, et al. A point mutation in influenza B neuraminidase confers resistance to peramivir and loss of slow binding. Antiviral Res. 2003;59:13–22. doi: 10.1016/s0166-3542(03)00011-1. doi:10.1016/S0166-3542(03)00011-1. [DOI] [PubMed] [Google Scholar]
- 41.Baz M, Abed Y, Boivin G. Characterization of drug-resistant recombinant influenza A/H1N1 viruses selected in vitro with peramivir and zanamivir. Antiviral Res. 2007;74:159–62. doi: 10.1016/j.antiviral.2006.10.012. doi:10.1016/j.antiviral.2006.10.012. [DOI] [PubMed] [Google Scholar]
- 42.Blick TJ, Tiong T, Sahasrabudhe A, et al. Generation and characterization of an influenza virus neuraminidase variant with decreased sensitivity to the neuraminidase-specific inhibitor 4-guanidino-Neu5Ac2en. Virology. 1995;214:475–84. doi: 10.1006/viro.1995.0058. doi:10.1006/viro.1995.0058. [DOI] [PubMed] [Google Scholar]
- 43.Blick TJ, Sahasrabudhe A, McDonald M, et al. The interaction of neuraminidase and hemagglutinin mutations in influenza virus in resistance to 4-guanidino-Neu5Ac2en. Virology. 1998;246:95–103. doi: 10.1006/viro.1998.9194. doi:10.1006/viro.1998.9194. [DOI] [PubMed] [Google Scholar]
- 44.Cheam AL, Barr IG, Hampson AW, et al. In vitro generation and characterisation of an influenza B variant with reduced sensitivity to neuraminidase inhibitors. Antiviral Res. 2004;63:177–81. doi: 10.1016/j.antiviral.2004.04.004. doi:10.1016/j.antiviral.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 45.Giannecchini S, Campitelli L, Bandini G, et al. Characterization of human H1N1 influenza virus variants selected in vitro with zanamivir in the presence of sialic acid-containing molecules. Virus Res. 2007;129:241–5. doi: 10.1016/j.virusres.2007.07.010. doi:10.1016/j.virusres.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 46.Gubareva LV, Bethell R, Hart GJ, et al. Characterization of mutants of influenza A virus selected with the neuraminidase inhibitor 4-guanidino-Neu5Ac2en. J Virol. 1996;70:1818–27. doi: 10.1128/jvi.70.3.1818-1827.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gubareva LV, Robinson MJ, Bethell RC, et al. Catalytic and framework mutations in the neuraminidase active site of influenza viruses that are resistant to 4-guanidino-Neu5Ac2en. J Virol. 1997;71:3385–90. doi: 10.1128/jvi.71.5.3385-3390.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McKimm-Breschkin JL, Sahasrabudhe A, Blick TJ, et al. Mutations in a conserved residue in the influenza virus neuraminidase active site decreases sensitivity to Neu5Ac2en-derived inhibitors. J Virol. 1998;72:2456–62. doi: 10.1128/jvi.72.3.2456-2462.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molla A, Kati W, Carrick R, et al. In vitro selection and characterization of influenza A (A/N9) virus variants resistant to a novel neuraminidase inhibitor, A-315675. J Virol. 2002;76:5380–6. doi: 10.1128/JVI.76.11.5380-5386.2002. doi:10.1128/JVI.76.11.5380-5386.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nedyalkova MS, Hayden FG, Webster RG, et al. Accumulation of defective neuraminidase (NA) genes by influenza A viruses in the presence of NA inhibitors as a marker of reduced dependence on NA. J Infect Dis. 2002;185:591–8. doi: 10.1086/339358. doi:10.1086/339358. [DOI] [PubMed] [Google Scholar]
- 51.Smee DF, Sidwell RW, Morrison AC, et al. Characterization of an influenza A (H3N2) virus resistant to the cyclopentane neuraminidase inhibitor RWJ-270201. Antiviral Res. 2001;52:251–9. doi: 10.1016/s0166-3542(01)00168-1. doi:10.1016/S0166-3542(01)00168-1. [DOI] [PubMed] [Google Scholar]
- 52.Tai CY, Escarpe PA, Sidwell RW, et al. Characterization of human influenza virus variants selected in vitro in the presence of the neuraminidase inhibitor GS 4071. Antimicrob Agents Chemother. 1998;42:3234–41. doi: 10.1128/aac.42.12.3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang MZ, Tai CY, Mendel DB. Mechanism by which mutations at his274 alter sensitivity of influenza a virus n1 neuraminidase to oseltamivir carboxylate and zanamivir. Antimicrob Agents Chemother. 2002;46:3809–16. doi: 10.1128/AAC.46.12.3809-3816.2002. doi:10.1128/AAC.46.12.3809-3816.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gooskens J, Jonges M, Claas EC, et al. Morbidity and mortality associated with nosocomial transmission of oseltamivir-resistant influenza A(H1N1) virus. JAMA. 2009;301:1042–6. doi: 10.1001/jama.2009.297. doi:10.1001/jama.2009.297. [DOI] [PubMed] [Google Scholar]
- 55.Hurt AC, Holien JK, Parker MW, et al. Oseltamivir resistance and the H274Y neuraminidase mutation in seasonal, pandemic and highly pathogenic influenza viruses. Drugs. 2009;69:2523–31. doi: 10.2165/11531450-000000000-00000. doi:10.2165/11531450-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 56.Chandrasekaran A, Srinivasan A, Raman R, et al. Glycan topology determines human adaptation of avian H5N1 virus hemagglutinin. Nat Biotechnol. 2008;26:107–13. doi: 10.1038/nbt1375. doi:10.1038/nbt1375. [DOI] [PubMed] [Google Scholar]
- 57.Yamada S, Suzuki Y, Suzuki T, et al. Haemagglutinin mutations responsible for the binding of H5N1 influenza A viruses to human-type receptors. Nature. 2006;444:378–82. doi: 10.1038/nature05264. doi:10.1038/nature05264. [DOI] [PubMed] [Google Scholar]
- 58.Liu C, Air GM. Selection and characterization of a neuraminidase-minus mutant of influenza virus and its rescue by cloned neuraminidase genes. Virology. 1993;194:403–7. doi: 10.1006/viro.1993.1276. doi:10.1006/viro.1993.1276. [DOI] [PubMed] [Google Scholar]
- 59.Yang P, Bansal A, Liu C, et al. Hemagglutinin specificity and neuraminidase coding capacity of neuraminidase-deficient influenza viruses. Virology. 1997;229:155–65. doi: 10.1006/viro.1996.8421. doi:10.1006/viro.1996.8421. [DOI] [PubMed] [Google Scholar]
- 60.Bantia S, Ghate AA, Ananth SL, et al. Generation and characterization of a mutant of influenza A virus selected with the neuraminidase inhibitor BCX-140. Antimicrob Agents Chemother. 1998;42:801–7. doi: 10.1128/aac.42.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zambon M, Hayden FG. Position statement: global neuraminidase inhibitor susceptibility network. Antiviral Res. 2001;49:147–56. doi: 10.1016/s0166-3542(01)00124-3. doi:10.1016/S0166-3542(01)00124-3. [DOI] [PubMed] [Google Scholar]
- 62.Tisdale M. Monitoring of viral susceptibility: new challenges with the development of influenza NA inhibitors. Rev Med Virol. 2000;10:45–55. doi: 10.1002/(sici)1099-1654(200001/02)10:1<45::aid-rmv265>3.0.co;2-r. doi:10.1002/(SICI)1099-1654(200001/02)10:1<45::AID-RMV265>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.