Abstract
AIM: To investigate the prognostic value of KRAS mutation, and phosphatase and tensin (PTEN) expression in Chinese metastatic colorectal cancer metastatic colorectal cancer (mCRC) patients treated with cetuximab.
METHODS: Ninety Chinese mCRC patients treated with cetuximab were evaluated for KRAS mutation and PTEN protein expression by DNA sequencing of codons 12 and 13 and immunohistochemistry, respectively. We then selected 61 patients treated with cetuximab, either in combination with chemotherapy, or alone as a second-line or third-line regimen to assess whether KRAS mutation or PTEN protein expression is associated with the response and the survival time of mCRC patients treated with cetuximab.
RESULTS: KRAS mutation was found in 30 (33.3%) tumor samples from the 90 patients, and positive PTEN expression was detected in 58 (64.4%) of the 90 patients. Among the 61 patients who were treated with cetuximab as a second-line or third-line regimen, the resistance to cetuximab was found in 22 patients with KRAS mutation and in 39 patients without KRAS mutation, with a response rate of 4.5% and 46.1% respectively (P = 0.001), a shorter median progression-free survival (PFS) time of 14 ± 1.3 wk and 32 ± 2.5 wk respectively (P < 0.001), a median overall survival (OS) time of 11 ± 1.2 mo and 19 ± 1.8 mo respectively (P < 0.001), as well as in 24 patients with negative PTEN expression and in 37 patients with positive PTEN expression respectively (P < 0.001), with a responsive rate of 4.2% and 48.6% respectively, a shorter median PFS survival time of 17 ± 2.0 wk and 28 ± 1.9 wk respectively (P = 0.07), and a median OS time of 11 ± 1.3 mo and 18 ± 1.9 mo respectively (P = 0.004). Combined KRAS mutation and PTEN expression analysis showed that the PFS and OS time of patients with two favorable prognostic factors were longer than those of patients with one favorable prognostic factor or no favorable prognostic factor (P < 0.001).
CONCLUSION: KRAS mutation and PTEN protein expression are significantly correlated with the response rate and survival time of Chinese mCRC patients treated with cetuximab.
Keywords: Cetuximab, Metastatic colorectal cancer, KRAS mutation, Phosphatase and tensin protein expression
INTRODUCTION
The incidence of colorectal cancer (CRC) has been increasing in the past decades and CRC is the third-leading cause of cancer-related deaths in China. During the past few years, several new biological agents have been evaluated in metastatic colorectal cancer (mCRC) with a remarkable anti-mCRC activity. Epidermal growth factor receptor (EGFR), one of the most promising targets, can activate the proliferation and prolong the survival time of cancer cells through the Ras/Raf/mitogen-activated protein kinase (MEK)/EPH receptor B2 (ERK) pathway or the phosphoinositide-3-kinase (PI3K)/PTEN/AKT pathway[1].
Cetuximab (Erbitux®, Merck KgaA, Darmstadt, Germany), a chimeric mouse/human antibody against the extracellular domain of EGFR, has a single-agent activity in mCRC refractory to irinotecan, oxaliplatin and fluoropyrimidines, and restores chemosensitivity in irinotecan-refractory mCRC patients[2-4]. However, only a small number of patients can benefit from cetuximab. The response rate to the combined cetuximab and irinotecan is about 23%[2]. Immunohistochemical studies showed that EGFR protein expression in CRC patients is not a useful predictor for the response to cetuximab[5,6].
Recent reports are available on the EGFR pathways, such as KRAS/BRAF/MAPKs, and on their potential correlation with cetuximab activity. KRAS somatic mutation occurs in approximately 40% of CRC patients. The negative predictive value of KRAS mutation has been confirmed in CRYSTAL study of first-line fluorouracil, leucovorin, and irinotecan (FOLFIRI) with or without cetuximab, demonstrating that only the patients with KRAS wild-type mutations benefit from cetuximab treatment[7-9].
Increasing interest in anti-EGFR therapy has been focused on another EGFR pathway, and PI3K/AKT/PTEN. PTEN encodes phosphatase with phosphatidylinositol-3, 4, 5-triphosphate (PIP-3) produced by the activity of PI3K as its major substrate. Loss of PTEN function increases PIP-3 concentration, and subsequent AKT hyperphosphorylation stimulates the proliferation of cancer cells[10].
It was reported that PTEN protein expression and KRAS mutation can predict the outcome of mCRC patients treated with cetuximab plus irinotecan, and negative PTEN expression in mCRC patients can predict the resistance to cetuximab plus irinotecan. Combined PTEN expression and KRAS mutational analysis can help to identify a subgroup of mCRC patients who have a greater chance of benefiting from EGFR inhibition[11].
KRAS and PTEN are the important molecular determinants of the EGFR downstream signal pathway and play an important role in anti-EGFR therapy in Western countries. However, little is known about the correlation between KRAS mutation and PTEN protein expression with the activity of anti-EGFR mono-antibody in Asian populations. This retrospective study was to evaluate the prognostic value of EGFR downstream cascade members, KRAS and PTEN, in Chinese mCRC patients treated with cetuximab plus chemotherapy.
MATERIALS AND METHODS
Patients
We retrospectively assessed 90 mCRC patients (59 males and 31 females with a median age of 53.0 ± 13.9 years) treated with cetuximab in Sun Yat-Sen University Cancer Center and Beijing Cancer Hospital from June 2000 to August 2008. The patients had histologically proven colorectal adenocarcinoma and the tumor response to cetuximab treatment was evaluable. Tissue samples of primary colorectal tumor were taken. KRAS mutation and PTEN protein expression in the patients were analyzed. Of the 90 patients, 3 received cetuximab monotherapy, 58 received cetuximab in combination with irinotecan-based chemotherapy, and 29 received cetuximab in combination with oxaliplatin-based chemotherapy. Cetuximab was administered as the first- fourth lines of treatment in 29, 23, 28, and 10 patients, respectively (Table 1). Paraffin-embedded tumor tissue samples from 100 mCRC patients (69 males and 31 females with a mean age of 50.5 ± 12.1 years) not treated with cetuximab were used for gene analysis. Furthermore, KRAS mutation in these patients was analyzed to confirm the mutation frequency of KRAS.
Table 1.
Data about patients enrolled in this study
| Characteristics | Patients, n (%) |
| Sex | |
| Male | 59 (65.6) |
| Female | 31 (34.4) |
| Age (yr) | |
| Median | 53 |
| Range | 23-75 |
| Tumor site | |
| Colon | 39 (43.3) |
| Rectal | 51 (56.7) |
| Combined chemotherapy | |
| Irinotecan-based | 58 (64.4) |
| Oxaliplatin-based | 29 (32.2) |
| Monotherapy | 3 (3.3) |
| Cetuximab use | |
| First line | 29 (32.2) |
| Second line | 23 (25.6) |
| Third line | 28 (31.1) |
| Fourth line or more | 10 (11.1) |
| Response status | |
| Complete response | 3 (3.3) |
| Partial response | 31 (34.4) |
| Stable disease | 35 (38.9) |
| Progressive disease | 21 (23.3) |
Skin toxic effects were assessed according to the National Cancer Institute Common Toxicity Criteria, version 2. Tumor response to cetuximab was evaluated by computerized tomodensitometry according to the response evaluation criteria in solid tumors (RECIST) and classified as complete response (CR), partial response (PR), stable disease (SD) and progressive disease (PD). Patients with CR or PR and SD or PD were classified as responders and non-responders, respectively, for the analyses.
Nucleotide sequence analysis
KRAS mutation was analyzed by extracting genomic DNA from the paraffin-embedded tissue sections with a QIAamp DNA mini kit (Qiagen, Berlin, Germany) according to its manufacturer’s instructions. Exon 1 of the KRAS gene (GenBank, L00045, nt 102 to 235) was then directly PCR-amplified in a thermal cycler. The sequences of primers used for KRAS analysis (codons 12 and 13) are identical to those used in a previous study[12]. PCR conditions were as follows: 1 cycle at 95°C for 9 min, 45 cycles at 94°C for 1 min, at 55°C for 1 min, and at 72°C for 1 min, followed by 1 cycle at 72°C for 5 min. After confirmed by agarose electrophoresis and ethidium bromide staining, the PCR products were purified and automatically sequenced with an ABI PRISM 3730 (Applied Biosystems, California, USA), then analyzed with Chromas software version 2.0 (Gene Codes Corporation, USA). All sequencing reactions were performed in both forward and reverse directions, by independent PCR.
PTEN protein expression
PTEN protein expression in 3-mm thick tissue sections was evaluated using the anti-PTEN clone 6H2.1 (Millipore Company, Massachusetts, USA). The sections were deparaffinized and hydrated by passing through xylene and a graded series of ethanol. Endogenous peroxidase activity of the sections was blocked by incubating them in 0.3% hydrogen peroxide for 20 min. Antigen was retrieved for 30 min at 98°C in a 0.01 mol/L sodium citrate buffer (pH 6.4) in a microwave oven. After blocked for 30 min in 0.75% normal goat serum, the sections were incubated with 6H2.1 at a dilution of 1:100 overnight at 4°C, washed in PBS, and then incubated with biotinylated goat anti-mouse IgG followed by avidin peroxidase using a Vectastain ABC elite kit (Vector Laboratories, California, USA). The reaction products were counterstained with hematoxylin, and the sections were evaluated under a light microscope. PTEN protein expression was detected mainly in cytoplasm, although nuclear signals were occasionally observed as previously reported[13]. The intensity of reaction was assessed as a score of either 1+, 2+ or 3+, and the percentage of positive cells was classified into three groups (0%-25%, 25%-50% and > 50% of cells) and assigned to 1, 2 or 3 points, respectively. Tumors producing more than 4 points were considered PTEN-positive tumors according to the two values for the products. The evaluation was performed without knowledge of the clinical data or the results of other analyses.
Statistical analysis
Fisher’s exact test was used to calculate the P values for KRAS mutation, PTEN expression, skin toxicity, and response to cetuximab. PFS time was calculated as the period of time from the first day of cetuximab treatment to the date of tumor progression, the date of death due to any factor, or the date of last follow-up. OS time was calculated as the period of time from the first day of cetuximab treatment until death due to any factor, or until the date of last follow-up. Cox proportional hazards regression model was used in survival analysis. PFS curves for PTEN expression and KRAS mutation were plotted using the Kaplan-Meier method, and the difference in biomarkers was evaluated using the log-rank test. Analysis was carried out with the SPSS software version 16.0 (SPSS Company, USA). P < 0.05 was considered statistically significant.
RESULTS
The clinical characteristics of mCRC patients treated with cetuximab are summarized in Table 1. The median follow-up time was 13.5 mo. Of the 90 patients, 34 (37.8%) had a response to cetuximab plus chemotherapy. The median PFS and OS time was 22 wk (range 8-129 wk), and 11 mo (range 2-48 mo), respectively. KRAS mutation was found in 30 (33.3%) tumor tissue samples from the 90 patients. Of the 30 tumor tissue samples, 25 and 5 were the single amino acid substitutions in codons 12 and 13, respectively. The KRAS mutations on codon 12 predominantly involved the second base of the codon, with the presence of GaT mutation (GGT-GaT, Gly-Asp, G12D), GtT mutation (GGT-GtT, Gly-Val, G12D), aGT mutation (GGT-aGT, Gly-Ser, G12C), and tGT mutation (GGT-tGT, Gly-Cys, G12C), in 16 (53.3%), 6 (20.0%), 2 (6.7%), and 1 ( 3.3%) out of the 30 patients, respectively. The KRAS mutations on codon 13 corresponded to the transition G-a at the second base of the codon (GGC-GaC, Gly-Asp, G13D). Positive and negative PTEN expression was detected in 58 (64.4%) and 32 (36.6%) of the 90 patients, respectively (Figure 1).
Figure 1.
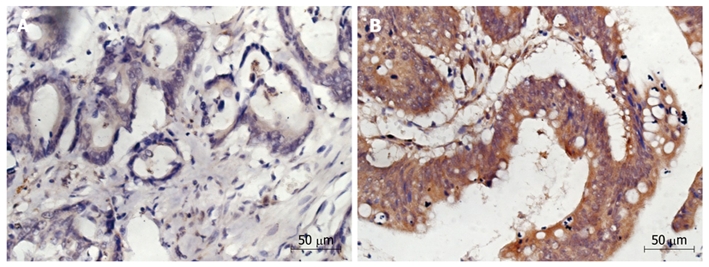
Immunohistochemistry showing phosphatase and tensin protein expression. A: Patients with absent phosphatase and tensin (PTEN) expression (HE stain, × 400); B: Patients showing positive PTEN expression (HE stain, × 400).
Of the 100 mCRC patients used for confirmation of KRAS mutation, 29 (29%) displayed KRAS mutation on codons 12 and 13, which was not significantly different from that in the 90 patients (P = 0.213). The KRAS mutations from GGT to GaT (Gly-Asp) and GtT (Gly-Val) on codon 12 accounted for 58.6% (17 of 29) and 20.7% (6 of 29) of the specified mutations, respectively. Mutations from GGC to GaC (Gly-Asp) occurred in 20.7% (6 of 29) of KRAS mutations on codon 13. The total mutation rate was 31% in 190 patients evaluated for KRAS mutations.
We attempted to assess whether the KRAS mutation, PTEN protein expression or skin toxicity is associated with the clinical response of mCRC to cetuximab. Sixty-one patients treated with cetuximab plus chemotherapy as a second-, third-, or greater-line regimen were enrolled for analysis. The KRAS mutation was detected in 22 patients (36.1%). One of the 22 patients with KRAS mutation responded to cetuximab, and 18 of the 39 patients without KRAS mutation responded to cetuximab, with a response rate of 4.5% and 46.1%, respectively (P = 0.001). Positive PTEN expression was detected in 37 (60.7%) out of the 61 patients. Eighteen of the 37 patients with normal PTEN expression and one of the 24 patients with negative PTEN expression responded to cetuximab, with a response rate of 48.6% and 4.2%, respectively (P = 0.001). The absence of KRAS mutation and the presence of PTEN protein expression were significantly associated with a high response rate to cetuximab (Table 2). Meanwhile, combined KRAS mutation status and PTEN expression analysis showed that 24 (39.3%) of the 61 patients had no KRAS mutation and positive PTEN expression, with a remarkably higher effective rate than other patients (70.8% vs 5.4%, P < 0.001). Fisher’s exact test also showed that the skin toxicity was significantly associated with a high response rate to cetuximab (P < 0.001).
Table 2.
Correlation of KRAS gene status and phosphatase and tensin protein expression with clinical response to cetuximab in previously treated colorectal cancer patients n (%)
|
KRAS |
PTEN |
|||||
| Mutation | No mutation | P | Positive | Negative | P | |
| CR | 0 (0) | 2 (5.1) | 0.001 | 2 (5.4) | 0 (0) | 0.001 |
| PR | 1 (4.5) | 16 (41.0) | 16 (43.2) | 1 (4.2) | ||
| SD | 7 (31.8) | 17 (43.6) | 10 (27.0) | 14 (58.3) | ||
| PD | 14 (63.6) | 4 (10.3) | 9 (24.3) | 9 (37.5) | ||
| Total | 22 (100) | 39 (100) | 37 (100) | 24 (100) | ||
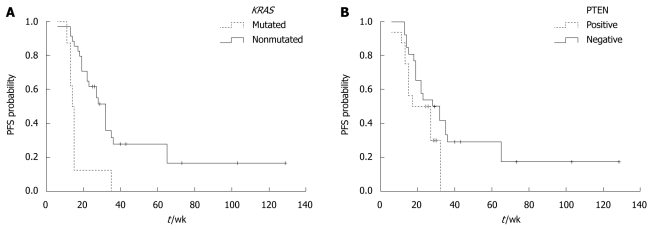
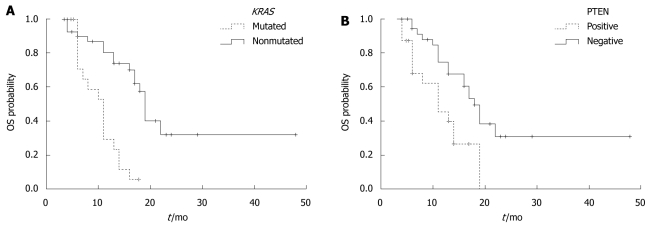
Furthermore, we assessed whether KRAS mutation, PTEN protein expression, or skin toxicity is associated with the PFS and OS time in the 61 patients. Univariate analysis of PFS time showed that KRAS mutation was significantly associated with a short PFS time (P < 0.001). The median PFS time of mCRC patients without and with KRAS mutation was 32 ± 2.5 wk and 14 ± 1.3 wk, respectively. The PFS time was longer in mCRC patients with positive PTEN protein expression than in those with negative PTEN protein expression (28 ± 1.9 wk vs 17 ± 2.0 wk, P = 0.07) (Figure 2). No difference was found in the median PFS time between the patients with and without skin toxicity (27 ± 2.9 wk vs 18 ± 1.7 wk, P = 0.113). The median OS time of mCRC patients without KRAS mutation was significantly longer than that of those with KRAS mutation (19 ± 1.8 mo vs 11 ± 1.2 mo, P < 0.001). The median OS time of mCRC patients with positive PTEN expression was significantly longer than that of those with negative PTEN expression (18 ± 1.9 mo vs 11 ± 1.3 mo, P = 0.004) (Figure 3). The median OS time of mCRC patients with skin toxicity was longer than that of those without skin toxicity (17 ± 1.5 mo vs 11 ± 1.0 mo, P = 0.025). Multivariate analysis of the 61 patients showed that both KRAS mutation and PTEN protein expression were closely related with a shorter OS time (P < 0.001). No correlation was found between skin toxicity and KRAS mutation or PTEN protein expression.
Figure 2.
Progression-free survival time of patients with or without KRAS mutation (A) and phosphatase and tensin protein expression (B). A: P < 0.001; B: P = 0.07. PFS: Progression-free survival.
Figure 3.
Overall survival time of patients with or without KRAS mutation (A) and phosphatase and tensin protein expression and skin toxicity (B). A: P < 0.001; B: P = 0.004. OS: Overall survival.
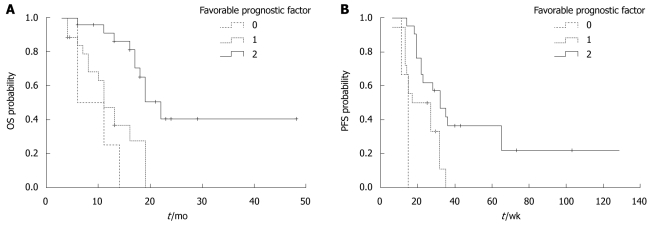
In this study, the absence of KRAS mutation and positive PTEN expression were found to be two favorable prognostic factors for mCRC patients. Combined KRAS mutation and PTEN expression analysis showed that the median PFS time of mCRC patients with the two favorable prognostic factors was longer than that of those with only one favorable prognostic factor or with no favorable prognostic factor (32 ± 2.5 wk vs 17 ± 1.9 wk and 11 ± 1.8 wk, P = 0.001) (Figure 4). The median OS time of these three groups of mCRC patients was 22 ± 2.3 mo, 11 ± 1.5 mo, and 6 ± 1.0 mo, respectively (P < 0.001).
Figure 4.
Overall survival and progression-free survival time of patients with or without favorable prognostic factors. A: P < 0.001; B: P = 0.001. 0: KRAS mutation and loss of phosphatase and tensin (PTEN); 1: Either no KRAS mutation or normal PTEN expression; 2: No KRAS mutation and normal PTEN expression. OS: Overall survival; PFS: Progression-free survival.
DISCUSSION
KRAS serves as a mediator for the extracellular ligand binding and intracellular transduction signals from EGFR to nuclei. RASCAL study evaluated the KRAS mutational in 2721 CRC patients from 22 centers and demonstrated that KRAS mutation is closely related with the progression and outcome of CRC[14]. In the present study, KRAS mutation was observed in about 40% (20%-50%) of sporadic CRC patients. Up to 90% of activating KRAS mutations were detected at codons 12 and 13, less than 70% of activating KRAS mutations were frequently detected at codon 12, and 70% of activating KRAS mutations were detected at codon 13. It has been shown that the most frequent types of KRAS mutation in CRC patients are GGT-GaT (Gly-Asp) and GGT-GtT (Gly-Val) transitions at codon 12[15]. In our study, 31% of KRAS mutations occurred in 190 CRC patients, with most specified KRAS mutations found at the second base of codon 12. The most common KRAS mutation at codon 12 was GGT to GaT. These findings are consistent with those in a previous study[15], indicating that the frequency or type of KRAS mutations is not different in Chinese and Western CRC patients.
It has been demonstrated that the benefit of cetuximab treatment in combination with first-line chemotherapy is restricted to CRC patients with KRAS wild-type mutations[9,16]. The relation between KRAS mutation and response to anti-EGFR therapy has also been intensively studied[11,13,17]. Lièvre et al[17] found that CRC patients with KRAS mutations are resistant to cetuximab therapy and have an unfavorable prognosis. It was reported that the PFS and OS time are shorter in CRC patients with KRAS mutations than in those with wild-type KRAS mutations[7,8]. In our study, KRAS mutation was found to be a powerful predictor for the resistance to cetuximab, the response rate of CRC patients with KRAS and KRAS wild-type mutations was 4.5% and 46.1%, respectively. The PFS and OS time of CRC patients with KRAS mutations was shorter than that of those without KRAS mutations. It has also been shown that treatment with tyrosine kinase inhibitors is not effective for non small cell lung cancer patients with KRAS mutations[18,19], indicating that KRAS mutations play a fundamental role in the EGFR pathway, thus rendering EGFR inhibitors ineffective[20].
In our study, the response rate of CRC patients with KRAS wild-type mutations was only 46.1%, indicating that there must be other unidentified genetic determinants of resistance to cetuximab therapy for CRC. The PI3K/PTEN/AKT pathway is on the other side of the two EGFR pathways. PTEN is a tumor suppressor protein that regulates the PI3K/AKT signal transduction. Its loss is associated with intrinsic activation of the AKT pathway and confers resistance to inhibitors of the HER family[21]. Thomas and Grandis demonstrated that PTEN is lost in 30% of sporadic CRC patients[22]. PTEN protein expression may be another molecular predictor for the response to cetuximab. Frattini et al[13] reported that loss of PTEN protein expression is associated with the lack of response to cetuximab. Sartore-Bianchi et al[10] showed that loss of PTEN protein expression is associated not only with the lack of objective tumor response, but also with a shorter OS time of mCRC patients treated with cetuximab. Loupakis et al[11] revealed that combined PTEN expression and KRAS mutation analysis helps identify a subgroup of mCRC patients who have a greater chance of benefiting from EGFR inhibition. In our study, positive PTEN expression was detected in 64.4% of mCRC patients, which is consistent with previous reports[10-13]. In this study, the response to cetuximab was significantly correlated with PTEN protein expression. The PFS and OS time of mCRC patients with negative PTEN protein expression was shorter than that of those with positive PTEN expression. The response rate of the 24 mCRC patients with no KRAS mutation and positive PTEN expression was substantially higher than that of those with KRAS mutation and positive PTEN expression. Combined KRAS mutation and PTEN protein expression analysis showed that the PFS and OS time of mCRC patients with two favorable prognostic factors was longer than that of those with one favorable prognostic factor or no favorable prognostic factor, indicating that a comprehensive analysis of KRAS mutation and PTEN protein expression is a better predictor for the clinical outcome of mCRC patients treated with cetuximab, which requires further confirmation in a prospective series.
It has been shown that skin toxicity is significantly associated with the response to cetuximab and OS time of mCRC patients[2,5,23,24], which is consistent with the findings in our study. In our study, the response rate of mCRC patients with skin toxicity was higher than that of those without skin toxicity, and the OS time of mCRC patients with skin toxicity was also longer than that of those without skin toxicity. However, univariate analysis showed that skin toxicity was only associated with OS time, while multivariate analysis showed that KRAS mutation and PTEN protein expression were the significant risk factors for OS time, indicating that skin toxicity alone is insufficient to predict the outcome of mCRC patients treated with cetuximab. Moreover, KRAS mutation and PTEN protein expression was detected before cetuximab treatment and can thus be included in the algorithm of treatment decision[17].
To our knowledge, this is the first study on KRAS mutation and PTEN protein expression in Chinese mCRC patients. Other markers were also identified in our study, which can be used to select mCRC patients who are likely to benefit from cetuximab treatment, showing that KRAS mutation and PTEN protein expression in Chinese mCRC patients are similar to those in other populations. In this study, skin toxicity was insufficient to predict the outcome of mCRC patients treated with cetuximab, and KRAS mutation and PTEN protein expression were significantly associated with the response rate to cetuximab and survival time of these patients.
In conclusion, combined KRAS mutation and PTEN protein expression analysis is a better predictor for the clinical outcome of mCRC patients treated with cetuximab. Prospective studies with a large number of patients are required to further confirm the results of our study.
COMMENTS
Background
The incidence of colorectal cancer (CRC) has been increasing in past decades and CRC is presently the third-leading cause of cancer-related deaths in China. During the past few years, several new biological agents have been evaluated in metastatic colorectal cancer (mCRC) with remarkable clinical activity. Cetuximab is an important biological agent used in treatment of mCRC, but it is effective only in a subset of mCRC patients.
Research frontiers
Studies have shown that KRAS mutation and phosphatase and tensin (PTEN) protein expression are associated with the response to cetuximab and may have a prognostic value. However, the situation in Asian patients is unknown. The authors evaluated the prognostic value of KRAS mutation and PTEN protein expression in Chinese mCRC patients treated with cetuximab plus chemotherapy.
Innovations and breakthroughs
To the authors’ knowledge, this is the first study on KRAS mutation and PTEN protein expression in Chinese mCRC patients. The results of this study show that KRAS mutation and PTEN protein expression in Chinese mCRC patients are significantly correlated with the response rate and survival time of patients treated with cetuximab. A comprehensive analysis of KRAS mutation and PTEN protein expression is a better predictor for the clinical outcome of patients treated with cetuximab.
Applications
KRAS mutation and PTEN protein expression can be used to select Chinese mCRC patients who are likely to benefit from cetuximab treatment.
Terminology
Epidermal growth factor receptor (EGFR), one of the most promising targets, can activate the proliferation and prolong the survival time of cancer cells through the Ras/Raf/mitogen-activated protein kinase (MEK)/EPH receptor B2 (ERK) or the phosphoinositide-3-kinase (PI3K)/ PTEN /AKT pathway. KRAS serves as a mediator for the extracellular ligand binding and intracellular signal transduction from EGFR to nuclei. PTEN is a tumor suppressor protein that regulates the PI3K/AKT signal transduction. Its loss is associated with the intrinsic activation of the AKT pathway.
Peer review
The manuscript describes the impact of KRAS mutation and PTEN protein expression, either alone or in combination, on cetuximab-treated Chinese colorectal cancer patients. The manuscript targets a topic that is of scientific and clinical interest.
Footnotes
Supported by Program for New Century Excellent Talents in University
Peer reviewer: Ulrike S Stein, PhD, Assistant Professor, Max-Delbrück-Center for Molecular Medicine, Robert-Rössle-Straße 10, 13125 Berlin, Germany
S- Editor Wang YR L- Editor Wang XL E- Editor Ma WH
References
- 1.Marshall J. Clinical implications of the mechanism of epidermal growth factor receptor inhibitors. Cancer. 2006;107:1207–1218. doi: 10.1002/cncr.22133. [DOI] [PubMed] [Google Scholar]
- 2.Cunningham D, Humblet Y, Siena S, Khayat D, Bleiberg H, Santoro A, Bets D, Mueser M, Harstrick A, Verslype C, et al. Cetuximab monotherapy and cetuximab plus irinotecan in irinotecan-refractory metastatic colorectal cancer. N Engl J Med. 2004;351:337–345. doi: 10.1056/NEJMoa033025. [DOI] [PubMed] [Google Scholar]
- 3.Lenz HJ, Van Cutsem E, Khambata-Ford S, Mayer RJ, Gold P, Stella P, Mirtsching B, Cohn AL, Pippas AW, Azarnia N, et al. Multicenter phase II and translational study of cetuximab in metastatic colorectal carcinoma refractory to irinotecan, oxaliplatin, and fluoropyrimidines. J Clin Oncol. 2006;24:4914–4921. doi: 10.1200/JCO.2006.06.7595. [DOI] [PubMed] [Google Scholar]
- 4.Van Cutsem E, Peeters M, Siena S, Humblet Y, Hendlisz A, Neyns B, Canon JL, Van Laethem JL, Maurel J, Richardson G, et al. Open-label phase III trial of panitumumab plus best supportive care compared with best supportive care alone in patients with chemotherapy-refractory metastatic colorectal cancer. J Clin Oncol. 2007;25:1658–1664. doi: 10.1200/JCO.2006.08.1620. [DOI] [PubMed] [Google Scholar]
- 5.Zhang W, Gordon M, Press OA, Rhodes K, Vallböhmer D, Yang DY, Park D, Fazzone W, Schultheis A, Sherrod AE, et al. Cyclin D1 and epidermal growth factor polymorphisms associated with survival in patients with advanced colorectal cancer treated with Cetuximab. Pharmacogenet Genomics. 2006;16:475–483. doi: 10.1097/01.fpc.0000220562.67595.a5. [DOI] [PubMed] [Google Scholar]
- 6.Saltz LB, Meropol NJ, Loehrer PJ Sr, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–1208. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 7.Folprecht G, Gruenberger T, Bechstein WO, Raab HR, Lordick F, Hartmann JT, Lang H, Frilling A, Stoehlmacher J, Weitz J, et al. Tumour response and secondary resectability of colorectal liver metastases following neoadjuvant chemotherapy with cetuximab: the CELIM randomised phase 2 trial. Lancet Oncol. 2010;11:38–47. doi: 10.1016/S1470-2045(09)70330-4. [DOI] [PubMed] [Google Scholar]
- 8.Karapetis CS, Khambata-Ford S, Jonker DJ, O'Callaghan CJ, Tu D, Tebbutt NC, Simes RJ, Chalchal H, Shapiro JD, Robitaille S, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359:1757–1765. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 9.Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, D'Haens G, Pintér T, Lim R, Bodoky G, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360:1408–1417. doi: 10.1056/NEJMoa0805019. [DOI] [PubMed] [Google Scholar]
- 10.Sartore-Bianchi A, Martini M, Molinari F, Veronese S, Nichelatti M, Artale S, Di Nicolantonio F, Saletti P, De Dosso S, Mazzucchelli L, et al. PIK3CA mutations in colorectal cancer are associated with clinical resistance to EGFR-targeted monoclonal antibodies. Cancer Res. 2009;69:1851–1857. doi: 10.1158/0008-5472.CAN-08-2466. [DOI] [PubMed] [Google Scholar]
- 11.Loupakis F, Pollina L, Stasi I, Ruzzo A, Scartozzi M, Santini D, Masi G, Graziano F, Cremolini C, Rulli E, et al. PTEN expression and KRAS mutations on primary tumors and metastases in the prediction of benefit from cetuximab plus irinotecan for patients with metastatic colorectal cancer. J Clin Oncol. 2009;27:2622–2629. doi: 10.1200/JCO.2008.20.2796. [DOI] [PubMed] [Google Scholar]
- 12.Aoki Y, Hosaka S, Tachibana N, Karasawa Y, Kawa S, Kiyosawa K. Reassessment of K-ras mutations at codon 12 by direct PCR and sequencing from tissue microdissection in human pancreatic adenocarcinomas. Pancreas. 2000;21:152–157. doi: 10.1097/00006676-200008000-00008. [DOI] [PubMed] [Google Scholar]
- 13.Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, Camponovo A, Etienne LL, Cavalli F, Mazzucchelli L. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97:1139–1145. doi: 10.1038/sj.bjc.6604009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter "RASCAL" study. J Natl Cancer Inst. 1998;90:675–684. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 15.Heinemann V, Stintzing S, Kirchner T, Boeck S, Jung A. Clinical relevance of EGFR- and KRAS-status in colorectal cancer patients treated with monoclonal antibodies directed against the EGFR. Cancer Treat Rev. 2009;35:262–271. doi: 10.1016/j.ctrv.2008.11.005. [DOI] [PubMed] [Google Scholar]
- 16.Bokemeyer C, Bondarenko I, Makhson A, Hartmann JT, Aparicio J, de Braud F, Donea S, Ludwig H, Schuch G, Stroh C, et al. Fluorouracil, leucovorin, and oxaliplatin with and without cetuximab in the first-line treatment of metastatic colorectal cancer. J Clin Oncol. 2009;27:663–671. doi: 10.1200/JCO.2008.20.8397. [DOI] [PubMed] [Google Scholar]
- 17.Lièvre A, Bachet JB, Boige V, Cayre A, Le Corre D, Buc E, Ychou M, Bouché O, Landi B, Louvet C, et al. KRAS mutations as an independent prognostic factor in patients with advanced colorectal cancer treated with cetuximab. J Clin Oncol. 2008;26:374–379. doi: 10.1200/JCO.2007.12.5906. [DOI] [PubMed] [Google Scholar]
- 18.van Zandwijk N, Mathy A, Boerrigter L, Ruijter H, Tielen I, de Jong D, Baas P, Burgers S, Nederlof P. EGFR and KRAS mutations as criteria for treatment with tyrosine kinase inhibitors: retro- and prospective observations in non-small-cell lung cancer. Ann Oncol. 2007;18:99–103. doi: 10.1093/annonc/mdl323. [DOI] [PubMed] [Google Scholar]
- 19.Massarelli E, Varella-Garcia M, Tang X, Xavier AC, Ozburn NC, Liu DD, Bekele BN, Herbst RS, Wistuba II. KRAS mutation is an important predictor of resistance to therapy with epidermal growth factor receptor tyrosine kinase inhibitors in non-small-cell lung cancer. Clin Cancer Res. 2007;13:2890–2896. doi: 10.1158/1078-0432.CCR-06-3043. [DOI] [PubMed] [Google Scholar]
- 20.Baselga J. The EGFR as a target for anticancer therapy--focus on cetuximab. Eur J Cancer. 2001;37 Suppl 4:S16–S22. doi: 10.1016/s0959-8049(01)00233-7. [DOI] [PubMed] [Google Scholar]
- 21.Pandolfi PP. Breast cancer--loss of PTEN predicts resistance to treatment. N Engl J Med. 2004;351:2337–2338. doi: 10.1056/NEJMcibr043143. [DOI] [PubMed] [Google Scholar]
- 22.Thomas SM, Grandis JR. Pharmacokinetic and pharmacodynamic properties of EGFR inhibitors under clinical investigation. Cancer Treat Rev. 2004;30:255–268. doi: 10.1016/j.ctrv.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Saltz LB, Meropol NJ, Loehrer PJ Sr, Needle MN, Kopit J, Mayer RJ. Phase II trial of cetuximab in patients with refractory colorectal cancer that expresses the epidermal growth factor receptor. J Clin Oncol. 2004;22:1201–1208. doi: 10.1200/JCO.2004.10.182. [DOI] [PubMed] [Google Scholar]
- 24.Vallböhmer D, Zhang W, Gordon M, Yang DY, Yun J, Press OA, Rhodes KE, Sherrod AE, Iqbal S, Danenberg KD, et al. Molecular determinants of cetuximab efficacy. J Clin Oncol. 2005;23:3536–3544. doi: 10.1200/JCO.2005.09.100. [DOI] [PubMed] [Google Scholar]