Lte1 interacts with and inhibits the small GTPases Ras and Bud1, blocking polarization until mitosis is complete.
Abstract
Lte1 is known as a regulator of mitotic progression in budding yeast. Here we demonstrate phosphorylation-dependent inhibition of polarized bud growth during G2/M by Lte1. Cla4 activity first localizes Lte1 to the polarity cap and thus specifically to the bud. This localization is a prerequisite for subsequent Clb–Cdk-dependent phosphorylation of Lte1 and its relocalization to the entire bud cortex. There, Lte1 interferes with activation of the small GTPases, Ras and Bud1. The inhibition of Bud1 prevents untimely polarization until mitosis is completed and Cdc14 phosphatase is released. Inhibition of Bud1 and Ras depends on Lte1’s GEF-like domain, which unexpectedly inhibits these small G proteins. Thus, Lte1 has dual functions for regulation of mitotic progression: it both induces mitotic exit and prevents polarized growth during mitotic arrest, thereby coupling cell cycle progression and morphological development.
Introduction
Coordination of cell cycle progression, cell size, and cell shape is essential to ensure the correct partitioning of genetic material between dividing cells. In Saccharomyces cerevisiae, the bud emerges in late G1, enlarges in S/G2, and in late M it receives a complement of DNA before physically detaching from the mother. As in higher eukaryotes, the Cdc42 Rho-like GTPase and its regulators and effectors activate each phase of morphological development. A Cdc42 complex is initially recruited to the bud site by Bud1–Rsr1. Bud1 is a Ras-like GTPase, which with its regulators, Bud5 (GEF) and Bud2 (GAP), determines the site of the bud. (Chang and Peter, 2003; Pruyne et al., 2004). In late G1, the Cdc42 module primes assembly of both the septin ring, via the effectors Gic1 and Gic2 (Iwase et al., 2006), and the polarity cap (Jaquenoud and Peter, 2000; Rida and Surana, 2005). The multiprotein polarity cap drives polymerization of actin cables by the activated formin, Bni1, another Cdc42 effector (Pring et al., 2003; Zigmond, 2004). This coordinates the secretory pathway with cell cycle–dependent growth and division (Park and Bi, 2007).
Early in bud emergence the actin cytoskeleton is highly polarized toward the polarity cap. G1 cyclin–Cdk directly phosphorylates Cdc24, the Cdc42 GEF, and the Cdc42 GAPs, Rga2, Bem2, and Bem3 (Knaus et al., 2007; McCusker et al., 2007; Sopko et al., 2007) to maintain a correct cycle of GTP/GDP-bound Cdc42 at sites of polarization. At G2/M, Clb–Cdk inhibits apical growth and triggers isotropic bud growth. Cdc42 and Cdc24 then redistribute over the whole bud cortex so that the actin cytoskeleton becomes a diffuse unpolarized network of actin cables. Thus, with no Clb–Cdk activity, the switch from polar to isotropic growth is inhibited and buds become elongated and polarized (Booher et al., 1993; Lew and Reed, 1993; Padmashree and Surana, 2001). Moreover, Clb–Cdk also inhibits bud site assembly so that a new bud is not initiated until Clb–Cdk activity is eliminated at the end of mitosis. However, the molecular targets of Clb–Cdk that mediate these events are unknown (Moseley and Nurse, 2009). After mitotic exit and Clb–Cdk inactivation, Cdc42 and numerous polarity cap components are recruited by septins to the bud neck to reorganize the actin cables for targeted membrane deposition at the site of cytokinesis (Kadota et al., 2004).
Inhibition and inactivation of mitotic cyclins in late anaphase is brought about by the mitotic exit network (MEN), a signaling cascade in which the activated Ras-like Tem1 protein ultimately leads to nuclear release of Cdc14 phosphatase. Cdc14 has multiple substrates whose dephosphorylation terminates mitosis and prepares the divided cells for the next cell cycle (Stegmeier and Amon, 2004). The MEN is inhibited by the spindle positioning checkpoint (SPoC), which arrests the cell cycle if the mitotic spindle is not correctly aligned on the mother–bud axis. Thus, actin polarization and cytoplasmic microtubules contribute to MEN activation by correctly positioning the mitotic spindle.
MEN activation is also connected to cell polarity through the activities of Cdc42 effectors, Cla4, Ste20, Gic1, and Gic2. Gic1 and Gic2 bind to Bub2, promoting Tem1 activation (Höfken and Schiebel, 2002, 2004; Chiroli et al., 2003). Cla4 kinase is essential for the activity of Lte1, a positive regulator of MEN.
Lte1 has long been envisaged as a guanosine nucleotide exchange factor for the Tem1 GTPase. However, there is no direct evidence for this activity, and recently we reported how Lte1 can affect polarity cap behavior and localization of Bfa1, a negative regulator of the MEN (Geymonat et al., 2009). Lte1 localizes specifically at the bud cortex, where it interacts with Ras and the polarity cap component, Kel1. Lte1 depends on Cla4 for its cell cycle–dependent phosphorylation that allows interaction with Ras, which in turn is essential for Lte1 function and localization to the bud cortex (Gavin et al., 2002; Höfken and Schiebel, 2002; Jensen et al., 2002; Seshan et al., 2002; Yoshida et al., 2003; Seshan and Amon, 2005). In addition, Lte1 has several putative Cdk1 phosphorylation sites, it can interact with Clb2 in vivo (Archambault et al., 2004), and it can be phosphorylated in vitro by Cdk1 (Jensen et al., 2002; Ubersax et al., 2003).
Here we report for the first time that a complex phosphorylation process relying on both Cla4 and Clb–Cdk does indeed act on Lte1. This determines Lte1’s correct subcellular localization to allow it to act in a hitherto unknown process that prevents untimely polarized growth by interfering with the activity of the small GTPases, Ras and Bud1, in G2/M. We propose that the parallel roles of Lte1 in MEN activation and inhibition of cell polarization ensures proper coordination between mitosis and cell development.
Results
lte1 mutants display altered bud morphology during mitotic arrest
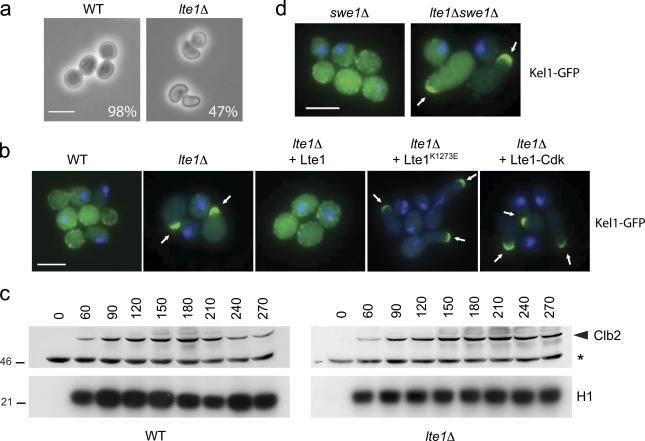
Wild-type yeast treated with the microtubule destabilizer nocodazole activate the SPoC and so arrest in metaphase with high levels of Cdk activity. After a 2-h treatment, large, rounded “dumbbells” accumulate with mother cells attached to enlarged buds (Hoyt et al., 1991). However, almost 50% of lte1Δ mutants similarly treated developed misshapen “hammerhead” buds that expanded transversely to the mother–bud axis (Fig. 1 a). To monitor the time course for development of the polarized bud phenotype, cells were arrested in G1 with α-factor and released into medium containing nocodazole. After 3 h, more than 80% of lte1Δ mutants developed polarized buds whereas wild-type cells maintained the typical dumbbell morphology. By 3 h, the initial hammerheads also became progressively more elongated (Fig. S1 a). The hammerhead phenotype is specific to loss of Lte1 activity because dumbbell formation was restored by reintroducing wild-type LTE1 (Fig. 1 b).
Figure 1.
lte1Δ mutants arrested in mitosis have a polarized bud. (a) Wild-type (WT; 15D) and lte1Δ cells (SY144) were arrested in nocodazole for 2 h; percentage of “hammerhead” phenotype is reported (n > 250). Bar, 5 µm. (b) Localization of Kel1-GFP in WT (SY159), lte1Δ (SY160), and lte1Δ cells complemented by Lte1, Lte1K1273E, or Lte1-Cdk arrested in nocodazole for 2 h. Bar, 5 µm. (c) WT (15D) and lte1Δ (SY144) cells were arrested in α-factor and released in medium containing nocodazole. At the indicated time, cells were harvested and Clb2 levels (top panel) and H1-associated kinase activity (bottom panel) were assayed. (d) Kel1-GFP localization in swe1Δ (MGY340) and swe1Δ lte1Δ (MGY341) cells arrested in nocodazole for 2 h. Bar, 5 µm.
An irreversible mitotic arrest occurs after expressing nondegradable mitotic cyclin, Clb2[Δdb] (Surana et al., 1993). After several hours of Clb2[Δdb] expression in lte1Δ mutants, the initial hammerhead buds developed highly polarized outgrowths that often expanded to one side of the mother–daughter cell axis (Video 1). In contrast, wild-type cells maintained a dumbbell morphology throughout (Video 2). Thus, Lte1 prevents polarization of the daughter cell during mitotic arrest.
In S. cerevisiae, polarized growth is driven by the polarity cap (Moseley and Goode, 2006), whose localization we recently found was affected by Lte1 during normal cell growth (Geymonat et al., 2009). Because cell morphology is clearly perturbed in lte1Δ mutants undergoing mitotic arrest, we asked if Lte1 also affects polarity cap behavior under these conditions. The polarity cap was monitored by tagging Kel1 with GFP. Kel1 is particularly interesting as it interacts with Lte1 (Gavin et al., 2002; Höfken and Schiebel, 2002; Seshan et al., 2002). In mitotically arrested wild-type cells, Kel1-GFP was uniformly distributed in patches on the bud cortex and, like other polarity cap components (Padmashree and Surana, 2001), was often at the bud neck. However, in lte1Δ mutants, a single focus of Kel1 accumulated to one side of the neck in the daughter cell. This phenotype could be complemented by wild-type LTE1 but not by lte1-K1273E with a point mutation in the GEF-like domain that impairs interaction with Ras (Fig. 1 b; Geymonat et al., 2009). Therefore, Lte1 promotes dispersion of the polarity cap during mitotic arrest via its GEF-like domain.
As Lte1 is better known for regulating MEN, we asked if Lte1-dependent MEN activity influenced morphology during mitotic arrest. Lte1-Cdk has five of nine putative (S/T) Cdc28 phosphorylation sites mutated to alanine. In vivo and in vitro, Lte1-Cdk was less phosphorylated than normal (Jensen et al., 2002) (Fig. S1 b) but still rescued the cold sensitivity of lte1Δ mutants and the synthetic lethality between lte1Δ and slk19Δ or spo12Δ (see Fig. 2 e). However, mutants expressing Lte1-Cdk had a single focus of Kel1-GFP and a hammerhead phenotype during mitotic arrest (Fig. 1 b). Thus, lte1-cdk mutants appear proficient in MEN activation but not in suppression of polarized growth. A further indication that the morphological effect of Lte1 is not exerted through MEN activity is that Cdc14 remains sequestered in the nucleoli of lte1 mutants undergoing nocodazole arrest, just as it does in wild-type cells (Fig. S1 c). Thus, as well as contributing to MEN activation, Lte1 has a MEN-independent, phospho-dependent role in the inhibition of polar-ized growth during mitotic arrest.
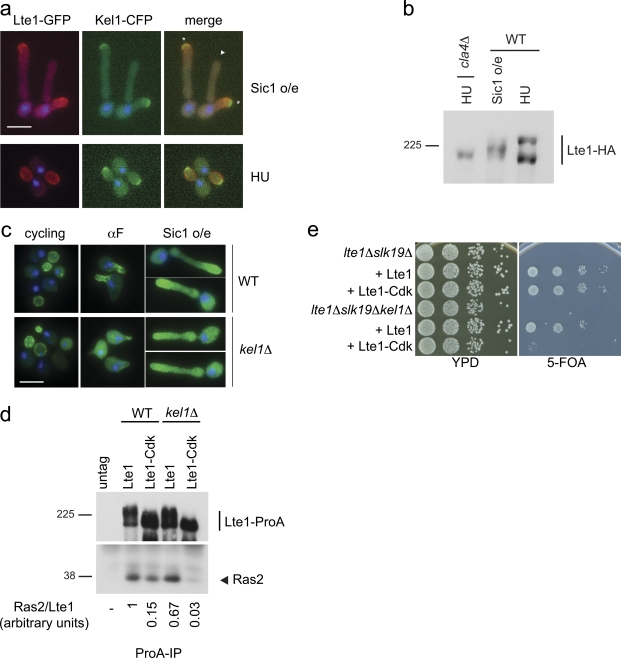
Figure 2.
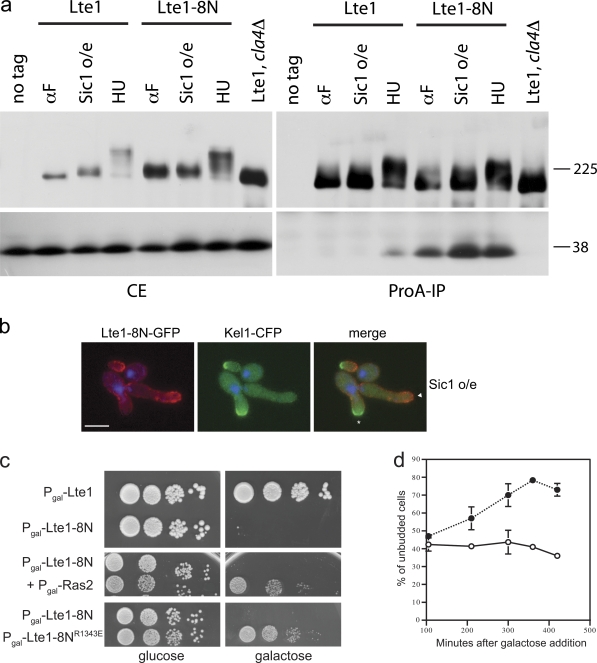
Role of Kel1 in Lte1 localization and function. (a) Localization of Lte1-GFP and Kel1-CFP in cells (MGY305) overexpressing nondegradable Sic1 (top panels) and in cells treated with HU (bottom panels). DAPI was used to stain nuclei. Asterisk indicates actively growing bud; triangle indicates nongrowing bud. Bar, 5 µm. (b) WT (MGY369) and cla4Δ (MGY423) cells expressing Lte1-3HA were arrested by overexpression of nondegradable Sic1 (WT) or by HU treatment (WT and cla4Δ). Lte1-HA was visualized by Western blotting. (c) Localization of Lte1-GFP in WT (MGY593) and kel1Δ (MGY594) cells. Cells expressing Lte1-GFP from a MET3 promoter and Sic1 from the GAL1 promoter were cultivated in minimal medium without methionine and 2% sucrose as carbon source. α-Factor or galactose (2%) was added and Lte1 was observed after 2 h. DAPI was used to stain nuclei. Bar, 5 µm. (d) WT (MGY261 and MGY281) and kel1Δ (MGY479 and MGY480) cells expressing Lte1, Lte1-ProA, or Lte1-Cdk-ProA were arrested in HU and ProA-tagged proteins were immunoprecipitated. Immunoprecipitated Lte1-ProA and coimmunoprecipitated Ras2 proteins were detected (top and bottom panels, respectively). Relative amounts of Ras bound to each form of Lte1were quantified. (e) lte1Δ slk19Δ (MGY212) and lte1Δ slk19Δ kel1Δ (MGY 503) strains kept alive by a centromeric URA3-based plasmid expressing Lte1 were transformed with integrative plasmids expressing Lte1 and Lte1-Cdk. Serial dilutions of transformants were spotted on rich medium and 5-FOA–containing plates and cultivated at 30°C for 3 d.
The aberrant morphology of lte1 mutants is not triggered by activation of the Swe1 checkpoint
Prolonged apical bud growth is normally associated with a delayed switch from axial to isotropic growth (Lew and Reed, 1993). One way this can occur is by the Swe1-dependent checkpoint inhibiting Clb–Cdk kinase in response to morphological perturbation. We therefore asked if Clb–Cdk kinase activity was reduced in mitotically arrested lte1Δ mutants. Wild-type and lte1Δ cells were arrested in G1 with α-factor and released into medium containing nocodazole. In both strains, Clb2 levels and Clb2–Cdk activity increased to similar extents and remained elevated throughout the mitotic arrest (Fig. 1 c). Thus, the morphological changes seen in lte1Δ mutants are not due to inactivation of Clb2–Cdk. Indeed, the hyperpolarized growth phenotype of lte1Δ mutants is independent of Swe1, as lte1Δ and lte1Δ swe1Δ cells both arrested in metaphase with hammerhead buds and a single focus of Kel1-GFP at the daughter cell cortex (Fig. 1 d). Interestingly, the combined deletion of LTE1 and HSL1, a negative regulator of Swe1, exacerbated polarized growth in nocodazole-arrested cells compared with the single mutants (Fig. S1 d), further suggesting that Lte1 and Swe1 act independently.
Role of Kel1 in Lte1 localization
As loss of putative CDK phosphorylation sites on Lte1-Cdk leads to hyperpolarized growth during mitotic arrest, we asked if this was related to changes in Lte1’s known interactions with Kel1 and Ras (Gavin et al., 2002; Höfken and Schiebel, 2002; Seshan et al., 2002).
Cells were first arrested with low levels of Clb–Cdk activity to assess how cell cycle kinase activity affected Lte1 behavior. After expression of Sic1-ΔN, a nondegradable form of the Sic1 inhibitor of mitotic Clb–Cdk kinase, cells accumulated with elongated buds and unreplicated DNA (Fig. 2 a; Singer et al., 1984). In these conditions Lte1 had an intermediate level of phosphorylation (Fig. 2 b) and localized principally to the growing bud tip with the polarity cap instead of being distributed uniformly over the entire bud cortex (Fig. 2 a; Jensen et al., 2002). With prolonged Sic1-ΔN expression, some cells grew a second bud, abandoning the growth of the first. Likewise, the polarity cap, monitored by Kel1-CFP, appeared at the tip of the second bud while disappearing from the first. Importantly, Lte1 followed the polarity cap in relocating to the tip of the second, actively growing, bud (Fig. 2 a). In contrast, highly phosphorylated Lte1 in HU-arrested cells with high levels of Clb–Cdk activity (Fig. 2 b) clearly localized over the whole bud cortex while Kel1-CFP remained as a small crescent at the bud tip (Fig. 2 a). Therefore, when Cdk–Clb activity is low, Lte1 is hypo-phosphorylated and colocalizes with the polarity cap, whereas fully phosphorylated Lte1 was dispersed over the whole bud cortex.
Next, the physiological importance of the Lte1–Kel1 interaction was investigated. As expected, Lte1 was located at the cell cortex in cycling wild-type and kel1Δ cells (Fig. 2 c; Höfken and Schiebel, 2002; Seshan et al., 2002). In contrast, when Cdk activity was low, localization of Lte1 at the tips of mating extensions during α-factor treatment or at the tips of hyperpolarized buds of Sic1-ΔN–arrested cells was reduced in kel1 mutants (Fig. 2 c; Seshan et al., 2002).
In an earlier report (Seshan et al., 2002), loss of Kel1 had no effect on the interaction between wild-type Lte1 and Ras2. Likewise, similar amounts of Ras2 copurified with wild-type Lte1 in KEL1 and kel1Δ cells arrested in S phase with HU (Fig. 2 d). In KEL1 cells, binding of Lte1-Cdk to Ras2 also occurred, albeit at reduced levels, but was undetectable in kel1Δ mutants (Fig. 2 d). Thus, fully phosphorylated Lte1 does not require Kel1 for interaction with Ras2, whereas hypo-phosphorylated Lte1 relies on Kel1 for interaction and proper localization. This apparent dependency was tested functionally in a complementation assay of the synthetic lethality of lte1Δ slk19Δ cells by Lte1 and Lte1-Cdk. Although Lte1 rescued the lethality of the double lte1Δ slk19Δ and triple lte1Δ slk19Δ kel1Δ mutant strains, complementation of lte1Δ slk19Δ synthetic lethality by Lte1-Cdk was KEL1 dependent (Fig. 2 e). Collectively, our results show that hypo-phosphorylated Lte1 colocalizes with the polarity cap and requires Kel1 for both its localization and its activity.
A multi-step phosphorylation process modulates Lte1 interaction with the polarity cap and Ras
A role for Cla4 in Lte1 phosphorylation is already documented (Höfken and Schiebel, 2002; Seshan et al., 2002; Seshan and Amon, 2005), but, as outlined above, Lte1 also has several putative Cdk phosphorylation sites and is an in vitro substrate for Clb/Cdk. Because inhibition of Clb–Cdk by Sic1-ΔN reduced Lte1 phosphorylation in vivo (Fig. 2 b), we considered if Cla4 and Cdk both contribute to Lte1 phosphorylation. In cells overexpressing Sic1-ΔN, Lte1 localized to the growing bud tip via a process requiring Cla4 (Fig. 3 a). As Lte1 was only partially phosphorylated in these cells (Fig. 2 b), Clb–Cdk appears to be required for the full phosphorylation normally observed in S/G2 and mitosis. Importantly, the role of the Cla4-dependent phosphorylation, revealed in the Sic1-ΔN arrest, is to allow Lte1 to colocalize with the polarity cap but not with the cortex as a whole.
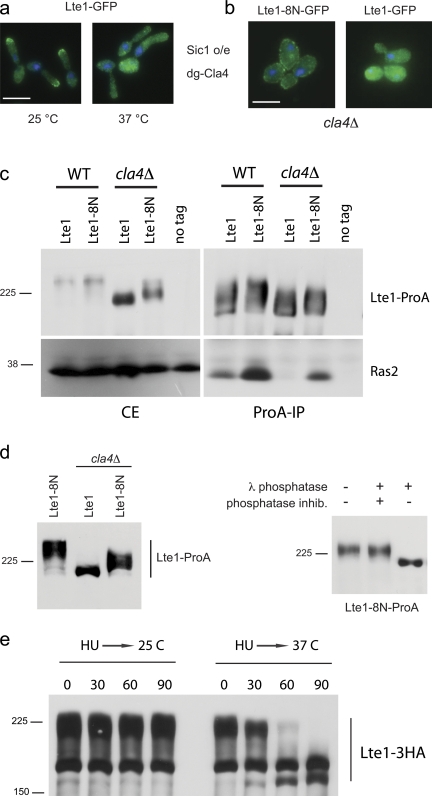
Figure 3.
Multi-step phosphorylation of Lte1. (a) Cells expressing Lte1-GFP and a degron form of Cla4-HA (MGY320) were cultivated at 23°C in YP-sucrose. Galactose was then added to induce nondegradable Sic1 and part of the culture was transferred to 37°C for 2.5 h. (b) Localization of Lte1-8N-GFP (MGY409) and Lte1-GFP (SY158) in a cla4Δ background. (c) Cells expressing Lte1-ProA or Lte1-8N-ProA in a WT (MGY261 and MGY415) or a cla4Δ background (MGY443 and MGY444) were arrested with HU and interaction between Lte1 and Ras2 was analyzed by Co-IP. (d) Lte1-8N-ProA purified from cla4Δ (MGY444) cells arrested in HU was treated with phosphatase buffer, with λ-phosphatase alone or in combination with phosphatase inhibitors and analyzed by Western blot. (e) Cells expressing Lte1-3HA and degron Cla4 (MGY313) were cultivated at 25°C and arrested with HU for 2 h. Half of the culture was then transferred at 37°C. Both cultures were sampled at 30-min intervals and Lte1-3HA was analyzed by Western blotting.
Recently we described a new form of Lte1, Lte1-8N, with a seven-amino acid insertion at residue 212 (Geymonat et al., 2009). Lte1-8N is fully able to complement the MEN defects of an lte1Δ mutant and displays the same pattern of cell cycle–dependent phosphorylation as wild-type Lte1 (Fig. S2). The striking feature of Lte1-8N is its localization to both the mother and daughter cortexes which, as a result, inactivates the SPoC (Bardin et al., 2000; Geymonat et al., 2009). Normally, cortical localization of Lte1 in the bud requires both Cla4 and binding to Ras (Höfken and Schiebel, 2002; Yoshida et al., 2003; Seshan and Amon, 2005). However, Cla4 is not needed for Lte1-8N to localize at the cortex (Fig. 3 b) because this allele interacts constitutively with Ras (see below) and does so even in cla4Δ mutants, although at a reduced level. In contrast, wild-type Lte1 interaction with Ras was completely abolished in cla4 mutants (Fig. 3 c; Yoshida et al., 2003; Seshan and Amon, 2005). Moreover, the partial phosphorylation of Lte1-8N in cla4Δ cells arrested before mitosis (Fig. 3 d) is further evidence that an additional kinase to Cla4 is required for the complete phosphorylation of Lte1.
The role of Cla4 is not limited to the G1/S transition because cells depleted for Cla4 during S/G2 (HU) arrest rapidly accumulated nonphosphorylated Lte1 (Fig. 3 e), which in parallel lost its cortical localization (unpublished data).
As partially phosphorylated Lte1 localizes in a Kel1-dependent manner with the polarity cap, we asked if complete phosphorylation of Lte1 triggers a switch of interaction to Ras. Binding between Lte1 and Ras occurs once cells start budding and peaks in mitosis (Seshan et al., 2002). Interaction was detectable after an HU arrest where Lte1 was highly phosphorylated, but not when Lte1 was only partially phosphorylated after Sic1-ΔN overexpression (Fig. 4 a). This result points to the role of phosphorylation, rather than bud emergence by itself, in Lte1–Ras interaction.
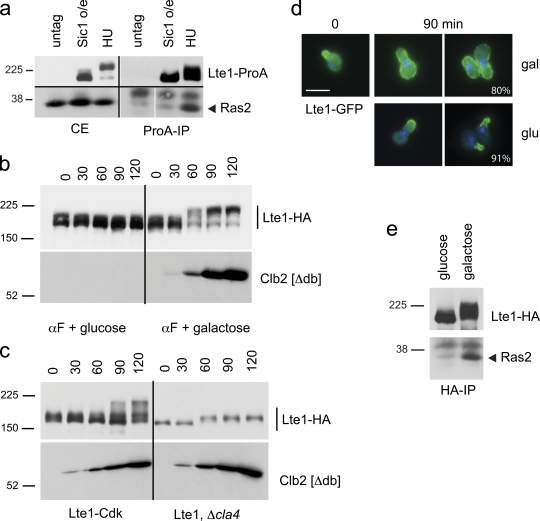
Figure 4.
Phosphorylated Lte1 interacts with Ras and allows cortical distribution of Lte1. (a) Cells expressing Lte1-ProA (MGY415) were arrested by overexpression of nondegradable Sic1 or by HU treatment, and ProA-tagged proteins were immunoprecipitated. Lte1 (top panels) and Ras2 (bottom panels) were detected in crude extracts (CE) and after Lte1-ProA immunoprecipitation (ProA-IP). (b) Cells expressing Lte1-3HA and a nondegradable Clb2 under the control of GAL1 promoter (MGY239) were cultivated in YP-sucrose and arrested in G1 with α-factor. The culture was split and 2% glucose or galactose was added while the arrest was maintained. Cells were harvested at the indicated times and Lte1 and Clb2[Δdb] were detected by Western blotting. (c) WT cells expressing Lte1-Cdk-3HA (MGY240) or cla4Δ mutants expressing Lte1-3HA (MGY592) and a nondegradable Clb2 under the control of GAL1 promoter were treated as in b. Lte1 and Clb2[Δdb] were detected by Western blotting. (d) Cells expressing Lte1-GFP from a MET3 promoter and a nondegradable Clb2 under the control of GAL1 promoter (MGY565) were cultivated at 30°C in minimal medium without methionine and 2% sucrose. Cells were arrested in G1 with α-factor and localization of Lte1 was observed after 90 min (time 0). Then the culture was split and 2% glucose or galactose was added while the arrest was maintained. Lte1 localization was observed after 90 min. The percentage of cells with indicated Lte1-GFP distribution is indicated (n > 120). (e) Cells treated as in b were harvested after 60 min of addition of glucose or galactose and Lte1-HA was immunoprecipitated. Co-immunoprecipitated Ras2 and Lte1-HA were detected by Western blotting.
Clb2’s role in maximal Lte1 phosphorylation and interaction with Ras was tested further by ectopically expressing nondegradable Clb2 in α-factor–arrested cells expressing Lte1-3HA or Lte1-GFP. Lte1 was converted to a slower migrating form in line with the appearance of Clb2, but not if Clb2 was unexpressed (Fig. 4 b). The conclusion that Lte1 is indeed subject to Clb–Cdk modification is further supported by performing the previous experiment with cells expressing Lte1-Cdk. This mutant form of Lte1 lacks some, but not all, putative Cdk phosphorylation sites and displays a reduced level of modification after Clb2 overexpression compared with wild-type Lte1 (Fig. 4 c). Furthermore, the Clb2-dependent modification of Lte1 relies on Cla4 activity because deletion of CLA4 prevented the major modification to Lte1 during Clb2 overexpression (Fig. 4 c). Thus, Cla4 activity is a prerequisite for the subsequent phosphorylation of Lte1 by Clb–Cdk.
The appearance of the phosphorylated Lte1 after ectopic expression of Clb2 in α-factor–arrested cells was matched by relocalization of Lte1 from the shmoo tip to the cortex of the mother cell in 80% of cases (Fig. 4 d) corresponding, in turn, with an increased interaction with Ras2 (Fig. 4 e). In conclusion, phosphorylation of Lte1 is a complex process. Initial phosphorylation by Cla4 allows Lte1 to colocalize with the polarity cap and is a prerequisite for subsequent phosphorylation by Clb–Cdk. This second step increases the affinity of Lte1 for Ras, allowing Lte1 to relocalize over the whole bud cortex.
Unregulated interaction with Ras leads to cortical localization of Lte1-8N in mother and daughter compartments
Lte1-8N localizes to both mother and daughter cell cortexes. As cortical localization normally requires interaction with Ras, we asked if Lte1-8N has an altered cell cycle–dependent pattern for this interaction. Cells were arrested before START with α-factor, or in S phase with high levels of Clb2–Cdk activity in an HU arrest, or after START with Clb–Cdk inhibited by Sic1-ΔN overexpression. As expected, wild-type Lte1 only interacted with Ras2 after arrest with HU, where it underwent full phosphorylation. In contrast, Lte1-8N was bound to Ras in all three arrests regardless of its phosphorylation status (Fig. 5 a). Moreover, after inhibition of Clb–Cdk by Sic1-ΔN overexpression, wild-type Lte1-GFP remained at the actively growing bud tip with Kel1-CFP (Fig. 2 a), whereas Lte1-8N-GFP was distributed over the whole cortex of both daughter and mother cells (Fig. 5 b). Thus, in wild-type cells with low Clb–Cdk activity, Lte1 and Kel1 localization was coupled, whereas Lte1-8N and Kel1 in the same conditions were uncoupled. This difference is consistent with Lte1-8N’s ability to bypass the requirement for phosphorylation for interaction with Ras (Fig. 5 a). This suggests that unregulated and untimely interaction with Ras leads to the localization of Lte1-8N to both mother and daughter cortexes.
Figure 5.
Lte1-8N constantly binds to Ras and interferes with its activity. (a) Cells expressing Lte1-ProA (MGY277) and Lte1-8N-ProA (MGY446) were arrested in α-factor, HU, or by expression of nondegradable Sic1. Lte1-ProA was also expressed in cla4Δ cells (MGY567) arrested in HU. ProA-tagged proteins were immunoprecipitated and Lte1 (top panels) and Ras2 (bottom panels) were detected in crude extracts (CE) and after Lte1-ProA immunoprecipitation (ProA-IP). Cells expressing untagged Lte1 were used as controls. (b) Localization of Lte1-8N-GFP and Kel1-CFP in cells overexpressing nondegradable Sic1 (MGY399). DAPI was used to stain nuclei. Asterisk indicates actively growing bud; triangle indicates nongrowing bud. Bar, 5 µm. (c) Cells expressing Lte1-GFP (SY148) or Lte1-8N-GFP (MGY449) under GAL1 promoter control were spotted on glucose- or galactose-containing medium (top panel). Cells expressing Lte1-8N-GFP alone or in combination with Ras2 (MGY517), both under GAL1 promoter control, were spotted on glucose- or galactose-containing medium (center panel). Cells expressing Lte1-8N-GFP and Lte1-8NR1343E-GFP (MGY527) under GAL1 promoter control were spotted on glucose- or galactose-containing medium (bottom panel). (d) Cells expressing Lte1-GFP (○) and Lte1-8N-GFP (•) under GAL1 promoter control were cultivated in YP-sucrose. 2% galactose was added at time 0 and unbudded cells were counted (n > 200 per point) at the indicated times. Error bars represent SD of three experiments.
Lte1 binds and impairs Ras activity
Two earlier findings interconnect Lte1 and Ras activities: first, Lte1 binds preferentially to GTP- rather than GDP-bound Ras (Yoshida et al., 2003; Seshan and Amon, 2005). Second, increased expression of Lte1 rescues the heat shock sensitivity of hyperactive Ras2 (Shirayama et al., 1994a; Yoshida et al., 2003), suggesting that Lte1 binding reduces Ras activity by preventing activation of adenylate cyclase. We have explored this idea further by asking if overexpression of Lte1-8N, which has an unregulated interaction with Ras (Fig. 5 a), resulted in a concomitant reduction in Ras activity in vivo. Indeed, overexpression of Lte1-8N, but not wild-type Lte1, blocked cell proliferation (Fig. 5 c) and unbudded cells accumulated (Fig. 5 d). This lethality was due to lack of Ras activity, as it was counteracted by compensatory overexpression of Ras2 (Fig. 5 c). Because S. cerevisiae has two RAS genes, RAS1 and RAS, which complement each other for both Lte1 localization and adenylate cyclase activation, we hypothesize that Lte1-8N inhibits both Ras1 and Ras2. Moreover, an Lte1-R1343E point mutation in the GEF-like domain that impairs binding between Lte1 and Ras and prevents cortical localization of Lte1 (Geymonat et al., 2009), abolished the lethality of overexpressed Lte1-8N (Fig. 5 c). Similarly, the lack of any lethal effects of wild-type Lte1 overexpression is explained by essential Ras activity being at START when wild-type Lte1 is unphosphorylated and so unable to bind Ras.
Lte1 negatively regulates Bud1
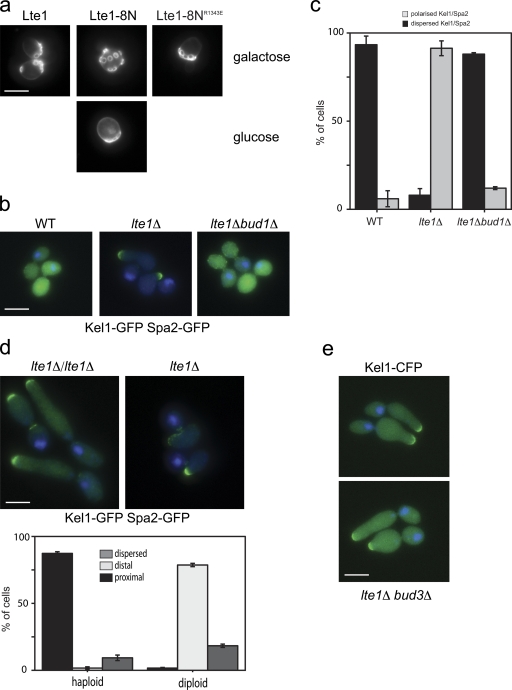
Bud1 is the closest relative of Ras-like G proteins. It is particularly related at the N terminus and in the effector loop (Fig. S3 a), and can even complement a ras1Δras2Δ strain (Ruggieri et al., 1992). We therefore asked if Bud1 too was affected by Lte1 activity. BUD1 deletion is not lethal, but it does randomize bud site selection with bud scars arising over the whole surface rather than accumulating at one pole of haploid cells (Park et al., 1993). The budding patterns of cells overexpressing Lte1-8N or wild-type Lte1 were compared in an exploratory test of Bud1 activity. Ras2 was also overexpressed to maintain viability of cells overexpressing Lte1-8N. Remarkably, overexpression of Lte1-8N, but not wild-type Lte1, randomized the budding pattern. This phenotype was abolished if the R1343E point mutation, which impairs Ras interaction, was introduced into Lte1-8N (Fig. 6 a). The budding pattern was also randomized by Lte1-8N expressed from the weaker MET3 promoter where Lte1 overexpression was not lethal and additional expression of Ras was not required (unpublished data). Because the localization of Bud5 at the neck was not affected in cells overexpressing Lte1-8N (Fig. S4 a), it appears that Lte1 specifically affects Bud1 activity rather than other aspects of the bud site selection process.
Figure 6.
Lte1 binds to and negatively regulates Bud1. (a) Cells expressing Lte1-GFP (MGY518), or Lte1-8N-GFP (MGY517) and Ras2, or cells expressing Lte1-8NR1343E-GFP (MGY527) alone under GAL1 promoter control were cultivated in galactose- or glucose-containing medium. Budding scars were visualized by Calcofluor staining. Bar, 5 µm. (b) WT (MGY308), lte1Δ (MGY309), and lte1Δ bud1Δ (MGY498) cells, expressing Kel1-GFP and Spa2-GFP, were treated with nocodazole for 2.5 h. Bar, 5 µm. (c) Quantification of the cells in b (n > 150). Error bars show the SD of three independent experiments. (d) Haploid (MGY309) and diploid (MGY485) lte1Δ cells, expressing Kel1-GFP and Spa2-GFP, were treated with nocodazole for 2.5 h as in b. Percentages of cells with a proximal, distal, or dispersed polarizome are indicated (n > 100). Error bars show the SD of three independent experiments. Bar, 5 µm. (e) lte1Δ bud3Δ cells expressing Kel1-CFP (MGY603) were treated with nocodazole for 2.5 h as in b. Bar, 5 µm.
If Lte1 does inhibit Bud1 to avoid untimely bud polarization, then deletion of BUD1 should restore polarity cap dispersion in lte1Δ cells arrested in mitosis and also counteract polarization. Indeed, polarity cap localization in bud1Δ lte1Δ mutants, optimally visualized by tagging both Spa2 and Kel1 with GFP, reverted to a wild-type, dispersed pattern rather than focusing at the side of the bud neck of mitotically arrested lte1Δ mutants. Similarly, deletion of BUD1 reduced the hyperpolarization of lte1Δ cells arrested in nocodazole. In contrast, deletion of BUD3 did not reduce hyperpolarization, indicating that the effect of Lte1 on polarity is specific to Bud1 and not the budding process in general (Fig. 6, b and c).
The site of Bud1 activation in a daughter cell depends upon ploidy: haploid cells activate Bud1 next to the last site of cell division marked by the septin ring and Axl2, whereas diploid cells activate Bud1 at the site of previous cell growth site, opposite the neck, and marked by Bud8 (Park and Bi, 2007). As seen in Figs. 1 b and 6 d, 90% of nocodazole-treated haploid lte1Δ cells focused their polarity caps close to the neck where Bud5 is localized (Kang et al., 2001) and from where future buds develop by axial budding. On the contrary, 80% of diploid lte1Δ cells developed a polarized outgrowth, with a concentration of polarity cap components opposite to the neck (Fig. 6 d), as is expected from the diploid bipolar budding pattern and the localization of Bud5 in diploid cells (Kang et al., 2001). Furthermore, when the normal axial pattern of haploid budding is changed to a bipolar, diploid-like budding pattern by deletion of BUD3 (Chant et al., 1995), then lte1Δ bud3Δ cells developed polarized daughters in a metaphase arrest resembling those of diploid cells (Fig. 6 e). These results are entirely consistent with a specific effect of Lte1 on Bud1 rather than on the budding process in general.
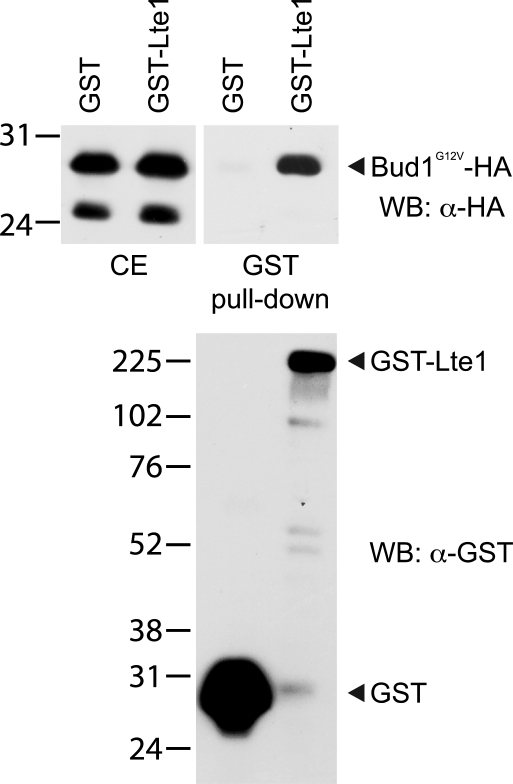
Finally, we asked if Lte1 could bind to Bud1. Using coimmunoprecipitation, no interaction was detectable between physiological levels of the two proteins. In an alternative approach, GST-Lte1 or GST control were coexpressed with a GTP-bound form of Bud1, Bud1G12V-HA, at high levels (Geymonat et al., 2007) in cells treated with HU for 2 h to increase Lte1 phosphorylation. Activated Bud1G12V was used because earlier work had shown the preferential interaction between Lte1 and activated Ras2G19V (Yoshida et al., 2003; Seshan and Amon, 2005). As shown in Fig. 7, Bud1 was eluted from beads containing GST-Lte1 but not from beads carrying GST alone, showing that Lte1 can interact with Bud1G12V.
Figure 7.
Interaction between Lte1 and activated Bud1. Cells coexpressing GST (MGY562) or GST-Lte1 (MGY563) and Bud1G12V-HA under GAL1 promoter control were cultivated for 4 h in rich medium containing 2% galactose, HU was added, and cells were cultivated for another 2.5 h. After GST pull-down, bound proteins were eluted with 20 mM reduced glutathione. Total crude extract (CE) and eluted material (GST pull-down) were blotted and stained with α-HA antibody (top panels) or α-GST (bottom panel).
Discussion
We describe a new, inhibitory activity of the GEF-like domain of Lte1 toward two small G proteins, Ras and Bud1. We find that the interaction of Lte1 for Ras and Bud1 and the concomitant interference with their activities are controlled by Lte1’s Clb–Cdk-dependent phosphorylation. In mitosis, phosphorylated Lte1 therefore contributes to the correct activation of MEN and also prevents untimely polarized growth through the inhibition of Bud1. This inhibitory effect of Lte1 is exerted until Cdc14 is released and Clb–Cdk activity declines at the end of mitosis. Thus, we propose that Lte1 is important for coupling cell cycle progression with timely activation of polarized cell growth. The development of polarized buds during metaphase arrest of lte1Δ mutants is mechanistically different from polarization that occurs when the lack of Clb2–Cdk or its inhibition delays the polar to isotropic switch in bud development (Lew and Reed, 1993). Unlike inhibition of isotropic growth, the untimely activation of Bud1 occurs in the presence of fully active Clb–Cdk (Fig. 1 c) and the polarized growth of lte1Δ mutants arises from initiation of a new growth site at the side of the bud, as determined by the site of bud selection, instead of a continued growth at the bud tip (Fig. 6, d and e).
A two-step process for phosphorylation of Lte1
Lte1 undergoes cell cycle–dependent phosphorylation that accumulates from S phase until mitosis before rapidly disappearing via Cdc14 activity in telophase (Bardin et al., 2000; Höfken and Schiebel, 2002; Jensen et al., 2002; Seshan et al., 2002). We propose that Lte1 is phosphorylated in two steps: the first is Cla4-dependent and is a prerequisite for a second step depending on Clb–Cdk. Direct phosphorylation of Lte1 by Cla4 has been demonstrated in vitro (Geymonat et al., 2007) and overexpression of Cla4 in G1-arrested cells leads to a low level of Lte1 phosphorylation (Seshan and Amon, 2005). This low level of phosphorylation appears to be required for Clb–Cdk-dependent phosphorylation because deletion of CLA4 has an epistatic effect in preventing any Lte1 phosphorylation, whereas inhibition of Clb–Cdk leads to an intermediate, Cla4-dependent level of phosphorylation (Höfken and Schiebel, 2002). There are several other indications that Clb–Cdk acts in Lte1 phosphorylation: phosphorylated Lte1 accumulates when Clb2 is expressed ectopically in G1-arrested cells (Fig. 4 b), Lte1 is a good in vitro substrate for Clb2–Cdk1 (Ubersax et al., 2003), and it interacts with Clb2 in vivo (Archambault et al., 2004). Lte1 also contains several Cdk consensus sites which, when mutated, modify both Lte1’s cell cycle–dependent phosphorylation profile and in vitro reduce its susceptibility to phosphorylation by Clb2–Cdk (Jensen et al., 2002). Moreover, the partial phosphorylation of Lte1-8N in mitotically arrested cla4Δ cells (Fig. 3 d) indicates that kinase(s) in addition to Cla4 can participate in Lte1 phosphorylation.
The two phases of phosphorylation correspond with different phases of cellular localization of Lte1. The first stage of phosphorylation by Cla4 normally promotes colocalization with the polarity cap during early bud emergence but, when Cla4 was ectopically expressed, colocalization was even seen at the bud neck in late anaphase cells or at a cortical crescent in unbudded cells (Fig. S4, b–d; Höfken and Schiebel, 2002). The need for Cla4 in polarized localization of Lte1 in a Sic1-ΔN arrest further indicates that phosphorylation promotes interaction between Lte1 and the polarity cap.
The second step of phosphorylation based on Clb–Cdk promotes Lte1 interaction with Ras and was seen after ectopic expression of Clb2 in G1 cells. This stimulated Lte1 phosphorylation with a concomitant increase in binding to Ras (Fig. 4). Although Clb–Cdk produces the high affinity of Lte1 for Ras, a low yet essential affinity for Ras depends upon Cla4 activity because Ras is essential for the localization of Lte1 at the site of polarization even when Clb–Cdk activity is low (Seshan and Amon, 2005). The requirement for a “priming” event in the bud to permit Clb–Cdk phosphorylation is consistent with the persistence of bud-specific localization after Lte1-GFP was ectopically expressed in cells arrested with high levels of Clb–Cdk activity (Fig. S3 b). Because Lte1 remains bud specific, this indicates that a priming activity in the daughter compartment that allows subsequent full phosphorylation and binding with Ras must exist only in the bud.
Lte1 has a known interaction with the polarity cap protein, Kel1 (Höfken and Schiebel, 2002), which we found is independent of Lte1’s phosphorylation status (Fig. S4 e). Nevertheless, interaction with Kel1 does appear to contribute to the cortical localization of Lte1 if Lte1 is hypo-phosphorylated (Fig. 2). Therefore, we envisage that both Kel1 and phosphorylation independently contribute to the interaction of Lte1 with Ras, the critical step for correct localization of Lte1 at the cortex.
Inhibition of Ras and Bud1 by Lte1
Remarkably, the idea that Lte1 can inhibit Ras activity was embedded in the first ever description of Lte1 in which multicopy LTE1 suppressed the heat shock sensitivity caused by a hyperactive Ras pathway (Shirayama et al., 1994a). However, this property has never been incorporated into any models of Lte1 activity or MEN regulation. Here, the overexpression of Lte1-8N, which has an unregulated high affinity for Ras, caused a cell cycle arrest in G1 (Fig. 5), which could be reversed by a compensatory increase in Ras expression. Inhibition could also be reversed by a mutation in the GEF-like domain of Lte1 that abolishes Lte1 interaction with Ras (Fig. 5 c). We too therefore propose phosphorylated Lte1 binds to Ras and inhibits it.
Bud1 and Ras share very similar N termini with an identical effector domain. Interestingly, this domain in Ras2 is implicated in the interaction with Lte1 (Seshan and Amon, 2005). This homology and the hyperpolarized phenotype of lte1Δ mutants from lateral sites near the bud neck of haploid cells suggested that Lte1 might also affect Bud1 activity. Indeed, Lte1 could bind to an activated form of Bud1 (Fig. 7). In addition, overexpression of Lte1-8N with an intact GEF-like domain randomized bud scar localization (Fig. 6 a) without affecting the localization of other bud site selection proteins such as Bud5. This mimics the effects of BUD1 deletion and suggests that Lte1-8N can inhibit Bud1 just as it inhibits Ras. Furthermore, the relocalization of the polarity cap in mitotically arrested lte1Δ cells was BUD1 dependent (Fig. 6 b), suggesting that Lte1 normally restrains Bud1 activity in mitosis. The different localization of the polarity cap in haploid and diploid lte1Δ mutants arrested in mitosis links the bud site selection machinery with polarization of the bud and further supports the idea that the role of Lte1 is to inhibit Bud1. Indeed, when BUD3 was deleted in an lte1Δ background cells arrested in mitosis still developed polarized buds, indicating that the effect of Lte1 on polarity was specifically exerted on Bud1 rather than on the budding process in general. Moreover, the polarization observed in an lte1Δ bud3Δ mutant reflects the bipolar budding pattern typical of a BUD3 defect (Fig. 6 e).
Any input of Ras inhibition in regulating bud polarity in G2/M is unclear as ras1Δ ras2Δ lte1Δ cells arrested in mitosis still undergo polarization (unpublished data). Nevertheless, Ras activation has been linked to Cdc42-dependent polarized growth (Mösch et al., 1996, 1999), so inhibition of Ras in the regulation of bud polarization cannot be excluded.
If the interactions between Bud1 or Ras with Lte1 are similar, hypo-phosphorylation of Lte1 would decrease Lte1’s affinity for Bud1. Indeed, cells expressing Lte1-Cdk, which is less phosphorylated in vivo and has reduced binding to Ras (Fig. 2 d), underwent inappropriate bud polarization during mitotic arrest like lte1Δ mutants (Fig. 1 b). This is consistent with a role for Lte1 phosphorylation in the inhibition of Bud1 activity. We, like others, were unable to directly demonstrate interactions of physiological levels of Bud1 despite strong indications of associations taking place (Kozminski et al., 2003). This is possibly due to a combination of the low abundance of GTP-bound Bud1 coupled with competition from the more abundant Ras for binding to Lte1. Indeed, this model is supported by the partial alleviation of the lethality of overexpressed Lte1-8N by overexpression of active Bud1 (Fig. S3 c).
A model for Lte1 localization and activity
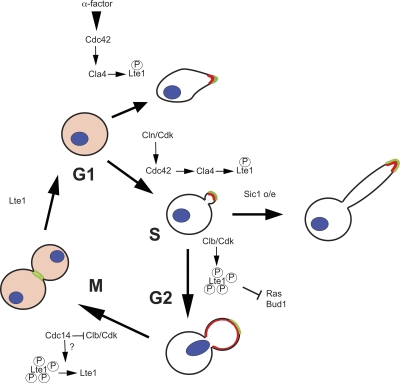
In summary, we propose a model to integrate Lte1 localization and activity (Fig. 8). In G1, Lte1 is not phosphorylated and is dispersed in the cytoplasm, consistent with this period of the cell cycle having the lowest levels of cyclin-dependent kinase activity. After G1 cyclin-dependent activation of Cdc42 through Cdc24 (Gulli et al., 2000), Lte1 is partially phosphorylated by Cla4 and acquires low affinity for Ras and high affinity for elements of the polarity cap. This channels Lte1 to sites of polarization, thereby confining Lte1 to the bud compartment and priming it for subsequent phosphorylation by Clb–Cdk. After START, septin ring deposition further confines Lte1 to the bud. Note that if ectopic phosphorylation occurs before septin ring formation, Lte1 can also localize to the mother cell cortex (Fig. 4 d). Clb–Cdk activity continues to rise and, once phosphorylated at higher levels, Lte1 acquires high affinity for Ras. This interaction is essential to localize Lte1 over the whole bud cortex and hence is vital for Lte1’s subsequent activity. However, the physiological role of the inhibition of Ras by Lte1 is unclear at present. Phosphorylated Lte1 also binds and inhibits Bud1, thereby avoiding any untimely and precocious polarization of the bud. This is particularly evident in cells undergoing a mitotic cell cycle arrest. In late anaphase, Lte1 participates in the MEN to release Cdc14 and abolish Clb–Cdk activity. At this point, Lte1 is dephosphorylated and looses its affinity for Ras, Bud1, and the polarity cap. Lte1 is then released from the cell cortex, thereby freeing Ras and Bud1 to act in the next round of the cell cycle. A role of Clb–Cdk in limiting activation of the bud site has been recognized (Padmashree and Surana, 2001), although the substrates for this kinase activity were not identified. Our results point to Lte1 as at least one target responsible for limiting initiation of a new bud site to a narrow window of the cell cycle after M phase.
Figure 8.
Model for Lte1 phosphorylation and localization. Lte1 is in red and polarizome in green. See text for details.
The novel polarity phenotype of lte1Δ mutants described here is most evident when cell cycle progression is perturbed. Although normally growing lte1Δ mutants display a mild morphological phenotype, they generally grow as well as wild-type cells (Adames et al., 2001). This is reminiscent of other MEN regulators like Bfa1 and Bub2 which, when absent, only display a clear phenotype in arrested conditions (Krishnan et al., 2000). Like Bfa1 and Bub2, which couple chromosome segregation between mother and daughter with cell cycle progression, Lte1 activity contributes to coupling cell morphology with completion of mitosis.
Is the inhibitory action of Lte1 on small GTPases involved in Lte1’s role in mitotic exit? First, the Ras-binding domain is clearly essential for Lte1’s role in mitotic progression (Geymonat et al., 2009), implying that the two are related. However, RAS1/2 deletion mimics lte1Δ in blocking mitosis at low temperatures (Yoshida et al., 2003), suggesting that it is not the inhibition of Ras that promotes mitotic exit. Nevertheless, deletion of BUD1 allows the dispersion of the polarity cap in mitotically arrested lte1Δ cells and partially rescues the cold sensitivity of an lte1Δ strain (Fig. S3 d), implying that Bud1 inhibition can in some way contribute to Lte1’s role in mitotic progression. Because polarization is an integral part of MEN regulation (Caydasi and Pereira, 2009; Monje-Casas and Amon, 2009; Moore et al., 2009), the influence of Lte1 on cell polarity could ultimately contribute to mitotic exit. Alternatively, it is formally possible that Lte1 could exert an inhibitory effect on another small G protein that in someway antagonizes mitotic exit. Whatever the mechanism, the requirement for Lte1 in mitotic exit and in regulation of cell polarity point to a general role of Lte1 in coordinating the correct morphological development of a new cell body with mitotic partition of replicated chromosomes. A fundamental mechanism of this type may well be expected in other eukaryotic systems.
The GEF-like domain of Lte1 is a new inhibitory module for small G proteins
Lte1 has long been considered a GEF for Tem1 because the need for Lte1 at low temperatures can be complemented by increased Tem1 activity (Shirayama et al., 1994b) and because of the similarities of Lte1’s C terminus and the well-characterized Cdc25 and Sos GEF domains (Shirayama et al., 1994a). However, we recently demonstrated that Lte1 does not rely on GEF activity for MEN regulation (Geymonat et al., 2009), and here we show that Lte1’s putative GEF domain has an inhibitory rather than activating effect on Ras and Bud1. This makes Lte1’s GEF-like domain the first reported with this novel activity and prompts a reevaluation of the definition of GEF domain based solely on sequence similarity, at least until the residues responsible for the inhibitory effect are identified.
Materials and methods
Yeast techniques
Yeast strains and plasmids are described in Tables I and II. Most strains are derivatives of BF264-15DU: a ura3Δns ade1 his2 leu2-3, 112 trp1-1a (Richardson et al., 1989). Gene disruptions were performed by a PCR-targeting technique (Berben et al., 1991; Longtine et al., 1998). Yeast media and genetic procedures were performed as described previously (Guthrie and Fink, 1991). α-Factor, hydroxy urea (HU), and nocodazole were used at concentrations of 5 µg/ml, 100 mM, and 15 µg/ml, respectively.
Table I.
Yeast strains
| Strain name | Relevant genotype | Source or reference |
| 15D | MATa, ura3Δns ade1 his2 leu2-3,112 trp1-1 | Richardson et al., 1989 |
| SY144 | MATa lte1::KANR | Derived from 15D; Jensen et al., 2002 |
| SY148 | MATa lte1::KANR ura3:: PGAL1-10-LTE1-GFP(URA3) | Derived from 15D |
| SY157 | MATa LTE1::LTE1-3HA cla4::LEU2 | Derived from 15D |
| SY158 | MATa LTE1::LTE1-GFP cla4::LEU2 | Derived from 15D |
| SY159 | MATa KEL1::KEL1-GFP(KANR) | Derived from 15D |
| SY160 | MATa lte1::KANR KEL1::KEL1-GFP(KANR) | Derived from 15D |
| MGY205 | MATa lte1::KANR leu2::LTE1-3HA(LEU2) | Derived from 15D |
| MGY212 | MATa lte1::KANR slk19:: KANR + YCplac33-LTE1 | Derived from 15D |
| MGY218 | MATa lte1::KANR ura3::PGAL1-10-CLB2[Δcdb](URA3) | Derived from 15D |
| MGY232 | MATa ura3::PGAL1-10-CLB2[Δcdb](URA3) | Derived from 15D |
| MGY239 | MATa lte1::KANR leu2::LTE1-3HA(LEU2) ura3::PGAL1-10-CLB2[Δcdb](URA3) | Derived from 15D |
| MGY240 | MATa lte1::KANR leu2::LTE1-Cdk-3HA(LEU2) ura3::PGAL1-10-CLB2[Δcdb](URA3) | Derived from 15D |
| MGY261 | MATa lte1::KANR leu2::LTE1-ProA(LEU2) | Derived from 15D |
| MGY277 | MATa lte1::KANR leu2::LTE1-ProA(LEU2) KEL1::KEL1-3HA(TRP1) + pLD1 | Derived from 15D |
| MGY281 | MATa lte1::KANR leu2::LTE1-Cdk-ProA(LEU2) | Derived from 15D |
| MGY296 | MATa lte1::KANR leu2::GFP-CDC14(LEU2) | Derived from 15D |
| MGY302 | MATa leu2::GFP-CDC14(LEU2) | Derived from 15D |
| MGY305 | MATa LTE1::LTE1-GFP(KANR) KEL1::KEL1-CFP(TRP1) + pLD1 | Derived from 15D |
| MGY308 | MATa KEL1::KEL1-GFP(KANR) SPA2::SPA2-GFP(URA3) | Derived from 15D; Geymonat et al., 2009 |
| MGY309 | MATa lte1::KANR KEL1::KEL1-GFP(KANR) SPA2::SPA2-GFP(URA3) | Derived from 15D; Geymonat et al., 2009 |
| MGY313 | MATa LTE1::LTE1-3HA(TRP1) cla4::LEU2 ura3::cla4-75dg(URA3) | Derived from 15D |
| MGY319 | MAT lte1::KANR hsl1::LEU2 | Derived from 15D |
| MGY320 | MATa LTE1::LTE1-GFP(KANR) cla4::LEU2 ura3::cla4-75dg(URA3) trp1::PGAL1-10-SIC1[ΔN](TRP1) | Derived from 15D |
| MGY321 | MATa hsl1::LEU2 | Derived from 15D |
| MGY340 | MATa swe1::URA3 KEL1::KEL1-GFP(KANR) | Derived from 15D |
| MGY341 | MATa lte1::KANR swe1::URA3 KEL1::KEL1-GFP(KANR) | Derived from 15D |
| MGY369 | MATa lte1::KANR leu2::LTE1-3HA(LEU2) + pLD1 | Derived from 15D |
| MGY370 | MATa lte1::KANR leu2::LTE1-Cdk-3HA(LEU2)) + pLD1 | Derived from 15D |
| MGY399 | MATa lte1::KANR leu2::LTE1-8N-GFP(LUE2) KEL1::KEL1-CFP(TRP1) + pLD1 | Derived from 15D |
| MGY423 | MATa lte1::KANR cla4::URA3 leu2::LTE1-3HA(LEU2) | Derived from 15D |
| MGY409 | MATa cla4::URA3 leu2::LTE1-8N-GFP(LEU2) | Derived from 15D |
| MGY415 | MATa lte1::KANR leu2::LTE1-8N-PraA(LUE2) | Derived from 15D |
| MGY443 | MATa lte1::KANR cla4::URA3 leu2::LTE1-PraA(LUE2) | Derived from 15D |
| MGY444 | MATa lte1::KANR cla4::URA3 leu2::LTE1-8N-PraA(LUE2) | Derived from 15D |
| MGY446 | MATa lte1::KANR leu2::LTE1-8N-ProA(LEU2) KEL1::KEL1-3HA + pLD1 | Derived from 15D |
| MGY449 | MATa lte1::KANR leu2:: PGAL1-10-LTE1-8N-GFP(LEU2) | Derived from 15D |
| MGY465 | MATa lte1::KANR leu2::LTE1-GFP(LEU2) + pLD1 | Derived from 15D |
| MGY466 | MATa lte1::KANR leu2::LTE1-GFP(LEU2) kel1::TRP1 + pLD1 | Derived from 15D |
| MGY479 | MATa lte1::KANR leu2::LTE1-ProA(LEU2) kel1::TRP1 | Derived from 15D |
| MGY480 | MATa lte1::KANR leu2::LTE1-cdk-ProA(LEU2) kel1::TRP1 | Derived from 15D |
| MGY485 | MATa/α lte1::KANR/lte1::KANR KEL1::KEL1-GFP(KANR)/KEL1 SPA2::SPA2-GFP(URA3)/SPA2 | Derived from 15D |
| MGY498 | MATa lte1::KANR bud1::LEU2 KEL1::KEL1-GFP(KANR) SPA2::SPA2-GFP(URA3) | Derived from 15D |
| MGY503 | MATa lte1::KANR slk19:: KANR kel1::TRP1 + YCplac33-LTE1 | Derived from 15D |
| MGY516 | MATa lte1::KANR bud1::TRP1 | Derived from 15D |
| MGY517 | MATa lte1::KANR leu2:: PGAL1-10-LTE1-8N-GFP(LEU2) trp1:: PGAL1-10-RAS2 (TRP1) | Derived from 15D |
| MGY518 | MATa lte1::KANR ura3:: PGAL1-10-LTE1-GFP(URA3) trp1:: PGAL1-10-RAS2 (TRP1) | Derived from 15D |
| MGY527 | MATa lte1::KANR leu2:: PGAL1-10-LTE1-8N-R1343E-GFP(LEU2) | Derived from 15D |
| MGY530 | MATa lte1::KANR bud1::TRP1 ura3::BUD1-HA(URA3) | Derived from 15D |
| MGY533 | MATa lte1::KANR bud1::TRP1 ura3::BUD1-HA(URA3) leu2::LTE1(LEU2) | Derived from 15D |
| MGY555 | MATa lte1::KANR leu2:: PGAL1-10-LTE1-8N-GFP(LEU2) ura3:: PGAL1-10-BUD1G12V(URA3) | Derived from 15D |
| MGY562 | MATa ura3-1 trp1-289 leu2Δ0 lys2Δ0 his3 mob1::kanMX4 cdc28::LEU2 pep4::LYS2 +pMH919-GST/pMH14-Bud1G12VHA | Derived from MGY140; Geymonat et al., 2007 |
| MGY563 | MATa ura3-1 trp1-289 leu2Δ0 lys2Δ0 his3 mob1::kanMX4 cdc28::LEU2 pep4::LYS2 +pMG1-LTE1/pMH14-Bud1G12VHA | Derived from MGY140; Geymonat et al., 2007 |
| MGY565 | MATa lte1::KANR leu2::PMET3-LTE1-GFP(LEU2) trp1::PGAL1-10-CLB2[Δcdb](TRP1) | Derived from 15D |
| MGY567 | MATa lte1::KANR cla4::URA3 leu2::LTE1-ProA(LEU2) KEL1::KEL1-3HA(TRP1) | Derived from 15D |
| MGY574 | MATa lte1::KANR leu2::PMET3-LTE1-GFP(LEU2) KEL1::KEL1-CFP(TRP1) ura3:: PGAL1-10-CDC42G12V(URA3) | Derived from 15D |
| MGY583 | MATα dbf2-2 leu2::LTE1-3HA(LEU2) | Derived from CG378 |
| MGY584 | MATα dbf2-2 leu2::LTE1-8N-3HA(LEU2) | Derived from CG378 |
| MGY589 | MATa lte1::KANR leu2:: PGAL1-10-LTE1-8N-3HA BUD5::BUD5-GFP(URA3) trp1:: PGAL1-10-RAS2 (TRP1) | Derived from 15D |
| MGY592 | MATa LTE1::LTE1-3HA(TRP1) cla4::LEU2 ura3::PGAL1-10-CLB2[Δcdb](URA3) | Derived from 15D |
| MGY593 | MATa lte1:: KANR leu2::PMET3-LTE1-GFP(LEU2) + pLD1 | Derived from 15D |
| MGY594 | MATa lte1:: KANR kel1::TRP1 leu2::PMET3-LTE1-GFP(LEU2) + pLD1 | Derived from 15D |
| MGY603 | MATa lte1::KANR bud3::KANR KEL1::KEL1-CFP(TRP1) ura3::GFP-Tub1(URA3) | Derived from 15D |
Table II.
Plasmids used
| Name | Backbone | Content | Source or reference |
| pLD1 | pMHTGal | Sic1 Δaa2-50; under GAL1-10 promoter control | Noton and Diffley, 2000 |
| pGal-CLB2[ΔDB] | YIplac211 | Clb2-Δcdb; under GAL1-10 promoter control | A. Amon |
| pGal-LTE1-GFP | pRS406 | LTE1-GFP; under GAL1-10 promoter control | Jensen et al., 2002 |
| pcla4-75-degron | cla4-75-HA degron allele | T. Yamamoto | |
| pHP659 | pRS306 | BUD1-HA | H.O. Park |
| pMG-Lte1 | pMG1 | GST-LTE1; under GAL1-10 promoter control | This study; Geymonat et al., 2007 |
| pMH919-GST | pMH919 | GST; under GAL1-10 promoter control | This study; Geymonat et al., 2007 |
| pMH14 | pRS423 | bud1G12V-HA; under GAL1-10 promoter control | This study; Geymonat et al., 2007 |
| pKGFP-Lte1 | pKGFP | Last 600 bp of LTE1 in frame with GFP | Jensen et al., 2002 |
| pKGFP-Kel1 | pKGFP | Last 674 bp of KEL1 in frame with GFP | This study |
| pMG97 | pRS304 | Last 674 bp of KEL1 in frame with CFP | This study |
| pMG52 | YIplac128 | LTE1-HA3 | Geymonat et al., 2009 |
| pMG58 | YIplac128 | LTE1-GFP | Geymonat et al., 2009 |
| pMG69 | pRS406 | cdc42G12V; under GAL1-10 promoter control | This study |
| pMG108 | YIplac128 | lte1-cdk-HA3 | Jensen et al., 2002 |
| pMG110 | pRS306 | Last 947 bp of SPA2 in frame with GFP | Geymonat et al., 2009 |
| pMG119 | YIplac128 | LTE1-ProA | This study |
| pMG120 | YIplac128 | lte1-cdk-ProA | This study |
| pMG180 | YIplac128 | lte1-8N-GFP | Geymonat et al., 2009 |
| pMG183 | YIplac128 | lte1-8N-HA3 | Geymonat et al., 2009 |
| pMG184 | YIplac128 | lte1-8N-ProA | This study |
| pMG191 | YIplac128 | lte1-8N-GFP; under GAL1-10 promoter control | This study |
| pMG213 | pRS304 | RAS2; under GAL1-10 promoter control | This study |
| pMG220 | YIplac128 | lte1-8N-R1343E-GFP; under GAL1-10 promoter control | This study |
| pMG221 | YIplac128 | LTE1-GFP; under MET3 promoter | This study |
| pMG237 | pRS306 | bud1G12V-HA; under GAL1-10 promoter control | This study |
| pMG246 | pRS306 | Last 570 bp of BUD5 in frame with GFP | This study |
| pMG247 | YIplac128 | lte1-8N-3HA; under GAL1-10 promoter control | This study |
Protein preparation and coimmunoprecipitation
Whole-cell extracts were obtained as described previously (Geymonat et al., 2009) or using a modified trichloroacetic acid (TCA) method (Foiani et al., 1994). In brief, cells (108) were washed and resuspended in 20% TCA. An equal volume of glass beads was added for cell disruption on a Fast-Prep beater (MP Biomedicals). After centrifugation of the lysate at 4,000 rpm for 10 min, the pellet was resuspended in one volume of 0.5 M Tris and two volumes of Laemmli buffer and subjected to SDS-PAGE.
For coimmunoprecipitation, cells treated according to figure legends were processed as described previously (Geymonat et al., 2009). In brief, cells were resuspended in IP buffer (50 mM Tris, pH 7.5, 1% NP-40, 150 mM NaCl, 1mM DTT, 10 mM NaF, 50 mM β-glycerophosphate, 0.1 mM VaV03, 10 mM p-nitrophenyl phosphate, complete protease inhibitors [Roche], and 1 mM PMSF). Equal amounts of crude extract were incubated with 50 µl of rabbit IgG-Agarose (Sigma-Aldrich) for 2 h at 4°C. Beads were washed four times with IP buffer and twice with wash buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 1 mM DTT, and 0.1% Triton X-100). Proteins were recovered from the beads and separated on an SDS-PAGE gel for subsequent Western blot analysis. Peroxidase anti-peroxidase antibody (Sigma-Aldrich) was used at a dilution of 1:500 to detect Lte1-ProA. Anti-Ras2 antibody (yC-19; Santa Cruz Biotechnology, Inc.) and monoclonal 12CA5 (anti-HA) were used at a dilution of 1:1,000. ImageJ 1.43u (National Institutes of Health, Bethesda, MD) was used to quantify Western blot signals.
GST pull-down
Cells coexpressing GST or GST-Lte1 and Bud1G12V-HA in the dual vector system (Geymonat et al., 2007) were grown in YP 2% gal for 4 h, 100 mM HU was then added, and cells were cultivated for another 2.5 h before harvesting. Cells were resuspended in IP buffer containing 5 mM MgCl2 and disrupted using glass beads (see above). Equal amounts of protein (2 mg) were added to 50 µl of glutathione–Sepharose beads (GE Healthcare) and incubated at 4°C for 2 h.
The beads were washed 4x with IP buffer and twice with IP buffer, without protease inhibitors. GST and GST-Lte1 were eluted with 20 mM reduced glutathione (Sigma-Aldrich) in 0.1% Triton X-100, 150 mM NaCl, 50 mM Tris-HCl, pH 7.5, and 1 mM DTT. Samples were analyzed by Western blot using rabbit anti-GST antibody (G7781; Sigma-Aldrich) and mouse anti-HA (12CA5).
Kinase and phosphatase assays
Clb2-associated kinase activity was assayed as described previously (Jensen et al., 2002). In brief, protein extracts (200 µg) were immunoprecipitated with 1 µg of anti-Clb2 (y-180; Santa Cruz Biotechnology, Inc.). Protein A–Sepharose beads (GE Healthcare) were added, and after 1 h at 4°C the immunocomplex was washed three times with breaking buffer and twice with kinase buffer (25 mM MOPS, pH 7.2, and 10 mM MgCl2). Beads were then incubated for 20 min at room temperature with 10 µl of kinase buffer containing 5 µg of histone H1 (Sigma-Aldrich), 50 µM ATP and 0.1 µl γ-[32P]ATP (10 mCi/ml). The reaction was stopped and radioactive H1 was analyzed by SDS-PAGE and autoradiography. Dephosphorylation of Lte1-ProA was performed using lambda protein phosphatase (LPP) from New England Biolabs, Inc. Proteins were purified on IgG beads and washed 2x with phosphatase buffer and incubated with or without LPP (500 U) and LPP with phosphatase inhibitor (5 mM Na3VO3).
Microscopy and imaging
GFP- and CFP-labeled proteins were analyzed by fluorescence microscopy after fixation with 3.7% formaldehyde added directly to the medium for 10 min at room temperature. For bud scar staining, fixed cells were treated with Calcofluor White stain (Fluka) diluted 1/10 for 1 h in the dark. DNA was stained with DAPI contained in the mounting medium (2 µg/ml) Vectashield (Vector Laboratories). Fluorescence microscopy used a liquid cooled CCD camera (model CH350L; Photometrics) on an inverted microscope (model IX70; Olympus), with a 100x F1.4 objective. Cell images were captured and manipulated using SoftWoRx software (Applied Precision) and Photoshop CS (Adobe). For time-lapse experiments, images were taken at 10-min intervals of cells growing at 30°C on agar plugs in a temperature-controlled chamber.
Online supplemental material
Fig. S1 shows polarization and localization of Cdc14 in synchronized lte1Δ cells arrested in mitosis and polarization of hsl1Δ, lte1Δ, and hsl1Δlte1Δ cells arrested in mitosis. Fig. S2 shows the cell cycle phosphorylation patterns of wild-type Lte1 and Lte1-8N. Fig. S3 shows the alignment of Ras2 and Bud1 N termini, the ability of Bud1G12V to partially rescue the overexpression of Lte1-8N, and the partial complementation of the cold sensitivity of an lte1Δ strain by deletion of BUD1. Fig. S4 shows that Lte1-8N does not perturb Bud5 localization and that expression of Cdc42G12V promotes cortical localization of Lte1 and Kel1 in unbudded cells. Fig. S4 e examines the interaction of Lte1 and Kel1 at different cell cycle points. Videos 1 and 2 show lte1Δ and wild-type cells expressing nondegradable Clb2 for 10–14 h. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201005070/DC1.
Acknowledgments
We are very grateful to Hay-Oak Park, Marisa Segal, and Simonetta Piatti for experimental materials and advice.
This work was funded by the MRC (file reference: u117531949).
Footnotes
Abbreviations used in this paper:
- GAP
- GTPase-activating protein
- GEF
- guanosine exchange factor
- MEN
- mitotic exit network
- SPoC
- spindle position checkpoint
References
- Adames N.R., Oberle J.R., Cooper J.A. 2001. The surveillance mechanism of the spindle position checkpoint in yeast. J. Cell Biol. 153:159–168 10.1083/jcb.153.1.159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archambault V., Chang E.J., Drapkin B.J., Cross F.R., Chait B.T., Rout M.P. 2004. Targeted proteomic study of the cyclin-Cdk module. Mol. Cell. 14:699–711 10.1016/j.molcel.2004.05.025 [DOI] [PubMed] [Google Scholar]
- Bardin A.J., Visintin R., Amon A. 2000. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 102:21–31 10.1016/S0092-8674(00)00007-6 [DOI] [PubMed] [Google Scholar]
- Berben G., Dumont J., Gilliquet V., Bolle P.A., Hilger F. 1991. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 7:475–477 10.1002/yea.320070506 [DOI] [PubMed] [Google Scholar]
- Booher R.N., Deshaies R.J., Kirschner M.W. 1993. Properties of Saccharomyces cerevisiae wee1 and its differential regulation of p34CDC28 in response to G1 and G2 cyclins. EMBO J. 12:3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caydasi A.K., Pereira G. 2009. Spindle alignment regulates the dynamic association of checkpoint proteins with yeast spindle pole bodies. Dev. Cell. 16:146–156 10.1016/j.devcel.2008.10.013 [DOI] [PubMed] [Google Scholar]
- Chang F., Peter M. 2003. Yeasts make their mark. Nat. Cell Biol. 5:294–299 10.1038/ncb0403-294 [DOI] [PubMed] [Google Scholar]
- Chant J., Mischke M., Mitchell E., Herskowitz I., Pringle J.R. 1995. Role of Bud3p in producing the axial budding pattern of yeast. J. Cell Biol. 129:767–778 10.1083/jcb.129.3.767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiroli E., Fraschini R., Beretta A., Tonelli M., Lucchini G., Piatti S. 2003. Budding yeast PAK kinases regulate mitotic exit by two different mechanisms. J. Cell Biol. 160:857–874 10.1083/jcb.200209097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foiani M., Marini F., Gamba D., Lucchini G., Plevani P. 1994. The B subunit of the DNA polymerase alpha-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14:923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin A.C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J.M., Michon A.M., Cruciat C.M., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 415:141–147 10.1038/415141a [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., Sedgwick S.G. 2007. A Saccharomyces cerevisiae autoselection system for optimised recombinant protein expression. Gene. 399:120–128 10.1016/j.gene.2007.05.001 [DOI] [PubMed] [Google Scholar]
- Geymonat M., Spanos A., de Bettignies G., Sedgwick S.G. 2009. Lte1 contributes to Bfa1 localization rather than stimulating nucleotide exchange by Tem1. J. Cell Biol. 187:497–511 10.1083/jcb.200905114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulli M.P., Jaquenoud M., Shimada Y., Niederhäuser G., Wiget P., Peter M. 2000. Phosphorylation of the Cdc42 exchange factor Cdc24 by the PAK-like kinase Cla4 may regulate polarized growth in yeast. Mol. Cell. 6:1155–1167 10.1016/S1097-2765(00)00113-1 [DOI] [PubMed] [Google Scholar]
- Guthrie C., Fink G.R. 1991. Guide to Yeast Genetics and Molecular Biology. Methods in Enzymology. Vol. 194 Academic Press, Inc., San Diego, California: [PubMed] [Google Scholar]
- Höfken T., Schiebel E. 2002. A role for cell polarity proteins in mitotic exit. EMBO J. 21:4851–4862 10.1093/emboj/cdf481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfken T., Schiebel E. 2004. Novel regulation of mitotic exit by the Cdc42 effectors Gic1 and Gic2. J. Cell Biol. 164:219–231 10.1083/jcb.200309080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., Totis L., Roberts B.T. 1991. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 66:507–517 10.1016/0092-8674(81)90014-3 [DOI] [PubMed] [Google Scholar]
- Iwase M., Luo J., Nagaraj S., Longtine M., Kim H.B., Haarer B.K., Caruso C., Tong Z., Pringle J.R., Bi E. 2006. Role of a Cdc42p effector pathway in recruitment of the yeast septins to the presumptive bud site. Mol. Biol. Cell. 17:1110–1125 10.1091/mbc.E05-08-0793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaquenoud M., Peter M. 2000. Gic2p may link activated Cdc42p to components involved in actin polarization, including Bni1p and Bud6p (Aip3p). Mol. Cell. Biol. 20:6244–6258 10.1128/MCB.20.17.6244-6258.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen S., Geymonat M., Johnson A.L., Segal M., Johnston L.H. 2002. Spatial regulation of the guanine nucleotide exchange factor Lte1 in Saccharomyces cerevisiae. J. Cell Sci. 115:4977–4991 10.1242/jcs.00189 [DOI] [PubMed] [Google Scholar]
- Kadota J., Yamamoto T., Yoshiuchi S., Bi E., Tanaka K. 2004. Septin ring assembly requires concerted action of polarisome components, a PAK kinase Cla4p, and the actin cytoskeleton in Saccharomyces cerevisiae. Mol. Biol. Cell. 15:5329–5345 10.1091/mbc.E04-03-0254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang P.J., Sanson A., Lee B., Park H.O. 2001. A GDP/GTP exchange factor involved in linking a spatial landmark to cell polarity. Science. 292:1376–1378 10.1126/science.1060360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knaus M., Pelli-Gulli M.P., van Drogen F., Springer S., Jaquenoud M., Peter M. 2007. Phosphorylation of Bem2p and Bem3p may contribute to local activation of Cdc42p at bud emergence. EMBO J. 26:4501–4513 10.1038/sj.emboj.7601873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozminski K.G., Beven L., Angerman E., Tong A.H., Boone C., Park H.O. 2003. Interaction between a Ras and a Rho GTPase couples selection of a growth site to the development of cell polarity in yeast. Mol. Biol. Cell. 14:4958–4970 10.1091/mbc.E03-06-0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan R., Pangilinan F., Lee C., Spencer F. 2000. Saccharomyces cerevisiae BUB2 prevents mitotic exit in response to both spindle and kinetochore damage. Genetics. 156:489–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D.J., Reed S.I. 1993. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J. Cell Biol. 120:1305–1320 10.1083/jcb.120.6.1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- McCusker D., Denison C., Anderson S., Egelhofer T.A., Yates J.R., III, Gygi S.P., Kellogg D.R. 2007. Cdk1 coordinates cell-surface growth with the cell cycle. Nat. Cell Biol. 9:506–515 10.1038/ncb1568 [DOI] [PubMed] [Google Scholar]
- Monje-Casas F., Amon A. 2009. Cell polarity determinants establish asymmetry in MEN signaling. Dev. Cell. 16:132–145 10.1016/j.devcel.2008.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore J.K., Magidson V., Khodjakov A., Cooper J.A. 2009. The spindle position checkpoint requires positional feedback from cytoplasmic microtubules. Curr. Biol. 19:2026–2030 10.1016/j.cub.2009.10.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch H.U., Roberts R.L., Fink G.R. 1996. Ras2 signals via the Cdc42/Ste20/mitogen-activated protein kinase module to induce filamentous growth in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA. 93:5352–5356 10.1073/pnas.93.11.5352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mösch H.U., Kübler E., Krappmann S., Fink G.R., Braus G.H. 1999. Crosstalk between the Ras2p-controlled mitogen-activated protein kinase and cAMP pathways during invasive growth of Saccharomyces cerevisiae. Mol. Biol. Cell. 10:1325–1335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.B., Goode B.L. 2006. The yeast actin cytoskeleton: from cellular function to biochemical mechanism. Microbiol. Mol. Biol. Rev. 70:605–645 10.1128/MMBR.00013-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley J.B., Nurse P. 2009. Cdk1 and cell morphology: connections and directions. Curr. Opin. Cell Biol. 21:82–88 10.1016/j.ceb.2008.12.005 [DOI] [PubMed] [Google Scholar]
- Noton E., Diffley J.F. 2000. CDK inactivation is the only essential function of the APC/C and the mitotic exit network proteins for origin resetting during mitosis. Mol. Cell. 5:85–95 10.1016/S1097-2765(00)80405-0 [DOI] [PubMed] [Google Scholar]
- Padmashree C.G., Surana U. 2001. Cdc28-Clb mitotic kinase negatively regulates bud site assembly in the budding yeast. J. Cell Sci. 114:207–218 [DOI] [PubMed] [Google Scholar]
- Park H.O., Bi E. 2007. Central roles of small GTPases in the development of cell polarity in yeast and beyond. Microbiol. Mol. Biol. Rev. 71:48–96 10.1128/MMBR.00028-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.O., Chant J., Herskowitz I. 1993. BUD2 encodes a GTPase-activating protein for Bud1/Rsr1 necessary for proper bud-site selection in yeast. Nature. 365:269–274 10.1038/365269a0 [DOI] [PubMed] [Google Scholar]
- Pring M., Evangelista M., Boone C., Yang C., Zigmond S.H. 2003. Mechanism of formin-induced nucleation of actin filaments. Biochemistry. 42:486–496 10.1021/bi026520j [DOI] [PubMed] [Google Scholar]
- Pruyne D., Legesse-Miller A., Gao L., Dong Y., Bretscher A. 2004. Mechanisms of polarized growth and organelle segregation in yeast. Annu. Rev. Cell Dev. Biol. 20:559–591 10.1146/annurev.cellbio.20.010403.103108 [DOI] [PubMed] [Google Scholar]
- Richardson H.E., Wittenberg C., Cross F., Reed S.I. 1989. An essential G1 function for cyclin-like proteins in yeast. Cell. 59:1127–1133 10.1016/0092-8674(89)90768-X [DOI] [PubMed] [Google Scholar]
- Rida P.C., Surana U. 2005. Cdc42-dependent localization of polarisome component Spa2 to the incipient bud site is independent of the GDP/GTP exchange factor Cdc24. Eur. J. Cell Biol. 84:939–949 10.1016/j.ejcb.2005.07.004 [DOI] [PubMed] [Google Scholar]
- Ruggieri R., Bender A., Matsui Y., Powers S., Takai Y., Pringle J.R., Matsumoto K. 1992. RSR1, a ras-like gene homologous to Krev-1 (smg21A/rap1A): role in the development of cell polarity and interactions with the Ras pathway in Saccharomyces cerevisiae. Mol. Cell. Biol. 12:758–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seshan A., Amon A. 2005. Ras and the Rho effector Cla4 collaborate to target and anchor Lte1 at the bud cortex. Cell Cycle. 4:940–946 [DOI] [PubMed] [Google Scholar]
- Seshan A., Bardin A.J., Amon A. 2002. Control of Lte1 localization by cell polarity determinants and Cdc14. Curr. Biol. 12:2098–2110 10.1016/S0960-9822(02)01388-X [DOI] [PubMed] [Google Scholar]
- Shirayama M., Matsui Y., Tanaka K., Toh-e A. 1994a. Isolation of a CDC25 family gene, MSI2/LTE1, as a multicopy suppressor of ira1. Yeast. 10:451–461 10.1002/yea.320100404 [DOI] [PubMed] [Google Scholar]
- Shirayama M., Matsui Y., Toh-E A. 1994b. The yeast TEM1 gene, which encodes a GTP-binding protein, is involved in termination of M phase. Mol. Cell. Biol. 14:7476–7482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer R.A., Bedard D.P., Johnston G.C. 1984. Bud formation by the yeast Saccharomyces cerevisiae is directly dependent on “start”. J. Cell Biol. 98:678–684 10.1083/jcb.98.2.678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sopko R., Huang D., Smith J.C., Figeys D., Andrews B.J. 2007. Activation of the Cdc42p GTPase by cyclin-dependent protein kinases in budding yeast. EMBO J. 26:4487–4500 10.1038/sj.emboj.7601847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. 2004. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 38:203–232 10.1146/annurev.genet.38.072902.093051 [DOI] [PubMed] [Google Scholar]
- Surana U., Amon A., Dowzer C., McGrew J., Byers B., Nasmyth K. 1993. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 12:1969–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ubersax J.A., Woodbury E.L., Quang P.N., Paraz M., Blethrow J.D., Shah K., Shokat K.M., Morgan D.O. 2003. Targets of the cyclin-dependent kinase Cdk1. Nature. 425:859–864 10.1038/nature02062 [DOI] [PubMed] [Google Scholar]
- Yoshida S., Ichihashi R., Toh-e A. 2003. Ras recruits mitotic exit regulator Lte1 to the bud cortex in budding yeast. J. Cell Biol. 161:889–897 10.1083/jcb.200301128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zigmond S.H. 2004. Formin-induced nucleation of actin filaments. Curr. Opin. Cell Biol. 16:99–105 10.1016/j.ceb.2003.10.019 [DOI] [PubMed] [Google Scholar]