The mitotic checkpoint protein Mad2 halts cell division by interfering with MKlp2-mediated relocation of the chromosome passenger complex from centromeres to the mitotic spindle.
Abstract
We identified the mitotic kinesin-like protein 2 (MKlp2), a kinesin required for chromosome passenger complex (CPC)–mediated cytokinesis, as a target of the mitotic checkpoint protein Mad2. MKlp2 possesses a consensus Mad2-binding motif required for Mad2 binding. Mad2 prevents MKlp2 from loading onto the mitotic spindle, a prerequisite step for its function as a mitotic kinesin. Furthermore, Mad2 inhibits the ability of MKlp2 to relocate the CPC from centromeres, an essential step to promote cytokinesis. An MKlp2 mutant that is refractory to Mad2-mediated inhibition prematurely translocates to the mitotic spindle and mislocalizes the CPC component Aurora B from the midbody of dividing cells. This correlates with an increased incidence of cytokinesis failure. Together, these findings reveal that MKlp2 is a novel mitotic target of Mad2 necessary for proper mitotic progression and cytokinesis.
Introduction
The segregation of sister chromatids to daughter cells must be orchestrated with cytokinesis. Mad2 is an essential mitotic checkpoint mediator that arrests cells in metaphase by inhibiting the E3 ubiquitin ligase anaphase-promoting complex (APC) through the formation of an inactive complex with Cdc20 (Li et al., 1997; Fang et al., 1998) until bipolar spindle attachment is completed. Moreover, deregulated Mad2 induces cytokinesis failure at a high frequency. This has been attributed to chromosome segregation errors (Hernando et al., 2004; Sotillo et al., 2007), but it is possible that Mad2 might function directly in orchestrating the mitotic checkpoint with cytokinesis.
A key player in cytokinesis is the chromosome passenger complex (CPC), which consists of Aurora B, INCENP, survivin, and borealin (for review see Ruchaud et al., 2007). Aurora B is a protein kinase that phosphorylates several components involved in cytokinesis, and INCENP is a scaffold protein that activates Aurora B. Although the CPC localizes at centromeres in early mitosis, it relocates to the central spindle at the metaphase to anaphase transition (Cooke et al., 1987; Schumacher et al., 1998). This relocation requires mitotic kinesin-like protein 2 (MKlp2), a kinesin essential for cytokinesis in mammalian cells (Hill et al., 2000; Gruneberg et al., 2004).
In this study, we have identified MKlp2 as a mitotic target of Mad2. We further demonstrate that controlling MKlp2 by Mad2 is important for mediating proper mitotic progression and cytokinesis.
Results and discussion
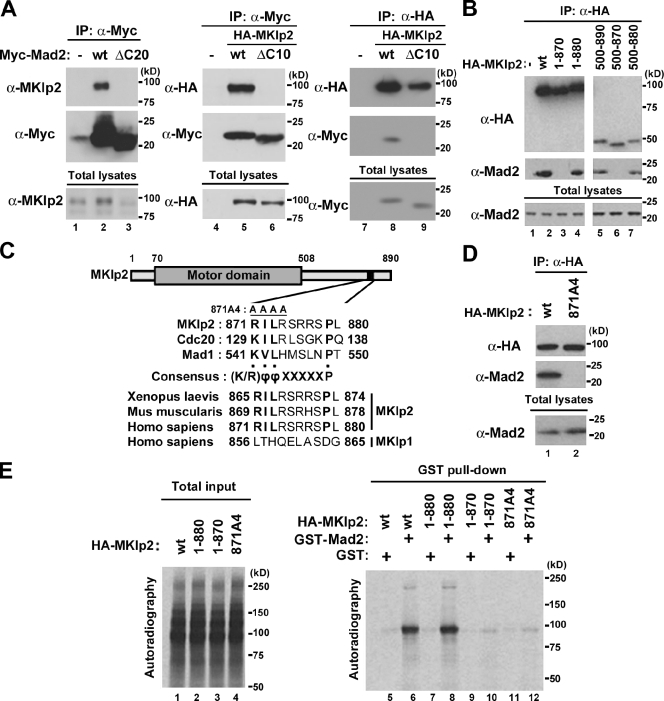
MKlp2 was previously copurified with tandem affinity purification (TAP)–wild type (wt)–Mad2, but not with the nonfunctional deletion mutant, by mass spectrometry analysis after TAP of HEK293 cell lysates expressing TAP-tagged Mad2 (Lee et al., 2008). Indeed, endogenous MKlp2 was coimmunoprecipitated with Myc epitope–tagged Mad2, but not the nonfunctional mutant of Myc-Mad2(ΔC20), using HEK293 cells (Fig. 1 A, lanes 1–3). Similarly, using HeLa (Fig. 1 A, lanes 4–6) and HEK293 cells (Fig. 1 A, lanes 7–9), HA-tagged MKlp2 was coimmunoprecipitated with Myc-Mad2 but not Myc-Mad2(ΔC10), in which the minimal functional region was deleted (Luo et al., 2000). Using a series of deletion mutants of MKlp2 (Fig. 1 B), we found that the C-terminal region of MKlp2 encompassing amino acids 871–880 was required for Mad2 binding. Similar results were obtained using HeLa cells (unpublished data). Mad1 and Cdc20 possess similar Mad2-binding motifs that conform to the consensus sequence (K/R)ϕϕXXXXXP (ϕ, an aliphatic residue; X: any residue; Luo et al., 2002; Sironi et al., 2002). Amino acid residues 871–879 of MKlp2 conform to this consensus sequence, which is conserved from human to Xenopus laevis but not found in its Kinesin-6 family member, MKlp1 (Fig. 1 C; for review see Verhey and Hammond, 2009). Indeed, an MKlp2 mutant in which residues 871–874 were replaced with alanine (MKlp2(871A4)) failed to bind Mad2 (Fig. 1 D). Furthermore, although in vitro–translated HA-MKlp2(wt) and HA-MKlp2(1–880) bound recombinant GST-Mad2, HA-MKlp2(1–870) and HA-MKlp2(871A4) failed to do so (Fig. 1 E). Thus, we conclude that MKlp2 is a direct binding partner of Mad2.
Figure 1.
MKlp2 is a novel binding partner of Mad2. (A, B, and D) Immunoblot analysis with the indicated antibodies, and 10% of the input is shown as total lysates. The positions of molecular mass markers (kilodaltons) are indicated. (A) Lysates of HEK293 cells (lanes 1–3) expressing the indicated Myc-Mad2 or HeLa (lanes 4–6) and HEK293 (lanes 7–9) cells coexpressing Myc-Mad2 with HA-MKlp2 were subjected to immunoprecipitation (IP) with antibodies to (α-) Myc or HA. (B) Lysates of HEK293 cells expressing HA-MKlp2 or the indicated mutants were subjected to immunoprecipitation with antibodies to HA. (C) Structural motifs of MKlp2 and alignment of the putative Mad2-binding sequences of MKlp2 with those of human Cdc20 and Mad1 in comparison with human MKlp1. The indicted sequences were aligned using ClustalW (www.ebi.ac.uk/clustalw). (D) Lysates of HEK293 cells expressing HA-MKlp2 or HA-MKlp2(871A4) were subjected to immunoprecipitation with antibodies to HA. (E) Autoradiography of in vitro–translated HA-MKlp2 proteins (left) and precipitates of GST pull-down analysis performed with the indicated proteins and either GST-Mad2 or GST (right). 10% of the input for total in vitro–translated product is shown.
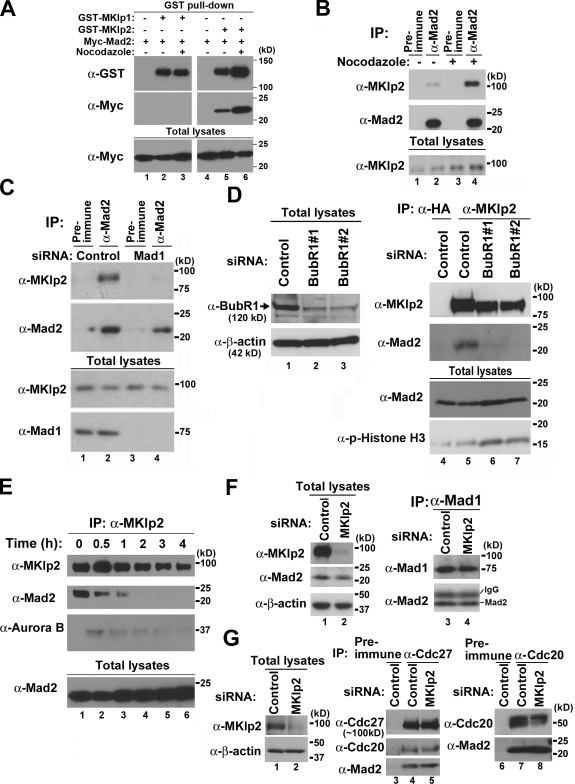
As the C-terminal region of MKlp1 does not have sequence homology with MKlp2 (Fig. 1 C), Myc-Mad2 bound GST-tagged MKlp2 but not GST-MKlp1 (Fig. 2 A). Furthermore, treating with the microtubule destabilizer nocodazole, which activates the mitotic checkpoint, increased the levels of Myc-Mad2 bound to GST-MKlp2 (Fig. 2 A) and the levels of endogenous MKlp2 bound to Mad2 using HeLa cells (Fig. 2 B). Because Mad2 is essential for the mitotic checkpoint, we examined whether formation of the Mad2–MKlp2 complex is regulated by mitotic checkpoint signaling by depleting Mad1 to inactivate Mad2 (Chen et al., 1998, 1999). To avoid the complication of using nocodazole, which increased the levels of MKlp2, HeLa cells were released from the G1–S boundary to mitosis. Endogenous MKlp2 bound Mad2 in the absence of nocodazole, whereas depleting Mad1 decreased the interaction (Fig. 2 C). Likewise, inactivating the mitotic checkpoint by depleting BubR1 decreased the levels of Mad2 bound to MKlp2, where the comparable levels of mitotic marker phospho–Histone H3 (p-Histone H3) were found (Fig. 2 D). Moreover, Mad2 coprecipitated with MKlp2 in early mitotic lysates (Fig. 2 E), whereas Aurora B did so in late mitotic lysates (Gruneberg et al., 2004; Hümmer and Mayer, 2009), suggesting that Mad2 binds MKlp2 in early mitosis and that the mitotic checkpoint also promotes this complex formation. Notably, MKlp2 did not bind Mad1 (not depicted), and depleting MKlp2 had no effect on the levels of Mad2 bound to Mad1, Cdc20, and the APC (Fig. 2, F and G), suggesting that endogenous MKlp2 does not compete with the binding partners of Mad2.
Figure 2.
Mad2 binding to MKlp2 depends on the mitotic checkpoint. Immunoblot analysis with the indicated antibodies, and 10% of the input is shown as total lysates. The positions of molecular mass markers (kilodaltons) are indicated. (A) Lysates of HEK293 cells expressing GST-MKlp1, GST-MKlp2, or together with Myc-Mad2 were subjected to GST pull-down analysis. (B) HeLa cells growing asynchronously were treated with or without nocodazole for 6 h. Cells were then lysed and subjected to immunoprecipitation (IP) with antibodies to Mad2. (C and D) HeLa cells transfected with control or Mad1 siRNAs or two different BubR1 siRNAs (#1 and #2; to ensure reproducibility) were synchronously released from the G1–S boundary. 7 h after release, cells were collected by mitotic shake off. Total cell lysates were subjected to immunoprecipitation analysis with the indicated antibodies. (E) Lysates of HeLa cells harvested at the indicated times after release from nocodazole block were subjected to immunoprecipitation with antibodies to MKlp2. (F and G) 48 h after transfection with control or MKlp2 siRNAs, HeLa cells were treated with nocodazole for 5 h and subjected to immunoprecipitation with antibodies to Mad1 (F), Cdc27, a core component of the APC (G, lanes 4 and 5), or Cdc20 (G, lanes 7 and 8).
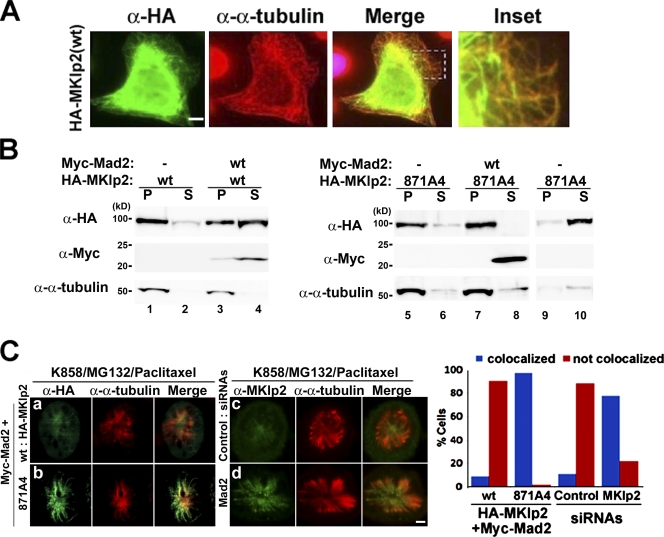
As MKlp2 is a motor that relocates the CPC to the central spindle at the metaphase to anaphase transition (Gruneberg et al., 2004), the motorless MKlp2(500–890) failed to relocate Aurora B (Fig. S1). Given that the interaction of kinesins with microtubules is an important regulatory step (for review see Verhey and Hammond, 2009), we examined whether Mad2 controls the microtubule-binding ability of MKlp2. We chose HeLa cells because of their suitability for immunofluorescence analysis over HEK293 cells. Overexpressed kinesins often show a filamentous staining pattern in interphase (Verhey et al., 1998). Although endogenous MKlp2, which is periodically expressed in mitosis (Hill et al., 2000), was not detected in the cytoplasm of interphasic cells, ectopically expressed HA-MKlp2 revealed such staining and colocalized with microtubules (Fig. 3 A). Furthermore, when HEK293 cells expressing HA-MKlp2 with or without Myc-Mad2 were subjected to microtubule cosedimentation assay using taxol-stabilized microtubules (Fig. 3 B), ∼90% of HA-MKlp2 cosedimented with taxol-stabilized microtubules. In contrast, only ∼30% of HA-MKlp2 coexpressed with Myc-Mad2 did so (Fig. 3 B, lanes 1–4), as quantified by phosphoimager analysis, suggesting that Mad2 inhibits the MKlp2-binding microtubule. Notably, HA-MKlp2(871A4) cosedimented with taxol-stabilized microtubules independent of Myc-Mad2 (Fig. 3 B, lanes 5–8), whereas it did not without taxol-stabilized microtubules (Fig. 3 B, lanes 9 and 10).
Figure 3.
Mad2 inhibits MKlp2 loading onto the mitotic spindle. (A) HeLa cells expressing HA-MKlp2 were subjected to immunofluorescence analysis with the indicated antibodies. Inset shows enlarged view of the boxed area. (B) Lysates of HEK293 cells expressing the indicated HA-MKlp2 or Myc-Mad2 proteins were supplemented with the taxol-stabilized microtubules (except lanes 9 and 10) and subjected to ultracentrifugation. The resulting supernatant (S) and pellet (P) fractions were then subjected to immunoblot analysis with the indicated antibodies. (C) HeLa cells were transfected with expression vectors encoding the indicated HA-MKlp2 together with Myc-Mad2 (a and b) or the indicated siRNAs (c and d). Cells were then treated with K858, MG132, and paclitaxel to form monoasters and were subjected to immunofluorescence analysis. HeLa cells (n > 100) were scored as having MKlp2 colocalized (blue bars) or not colocalized (red bars) with the mitotic spindle. Bars, 5 µm.
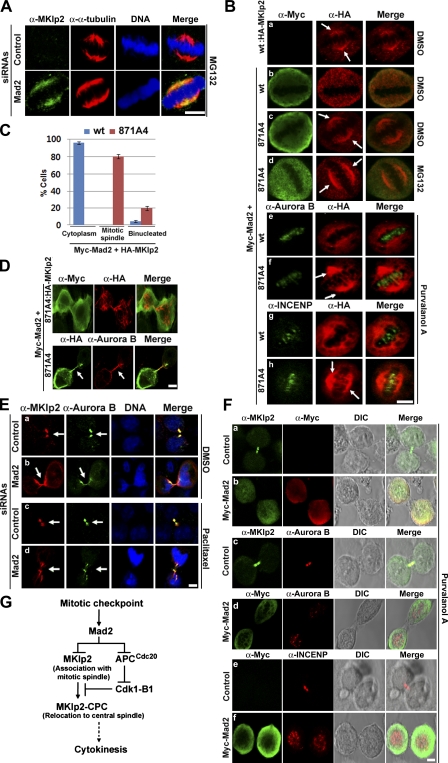
Next, we investigated whether Mad2 also inhibits MKlp2 binding the mitotic spindle. Because the levels of ectopically expressed HA-MKlp2 were typically higher than endogenous MKlp2 by >10-fold (unpublished data), we coexpressed Myc-Mad2 with HA-MKlp2 in HeLa cells. Subsequently, cells were exposed to K858, an inhibitor of the mitotic kinesin Eg5 (Nakai et al., 2009), to induce the formation of monoasters, a proteasome inhibitor MG132 to maintain Cdk1 active by inhibiting cyclin B1 degradation, and paclitaxel to stabilize the spindles (Fig. 3 C). HA-MKlp2 largely localized in the mitotic cytoplasm (Fig. 3 C, a), whereas HA-MKlp2(871A4) localized in the mitotic spindle (Fig. 3 C, b). Moreover, endogenous MKlp2 localized to the mitotic spindle in Mad2-depleted cells (Fig. 3 C, d), whereas it did not in control (Fig. 3 C, c), suggesting Mad2 inhibits MKlp2 loading onto the mitotic spindle.
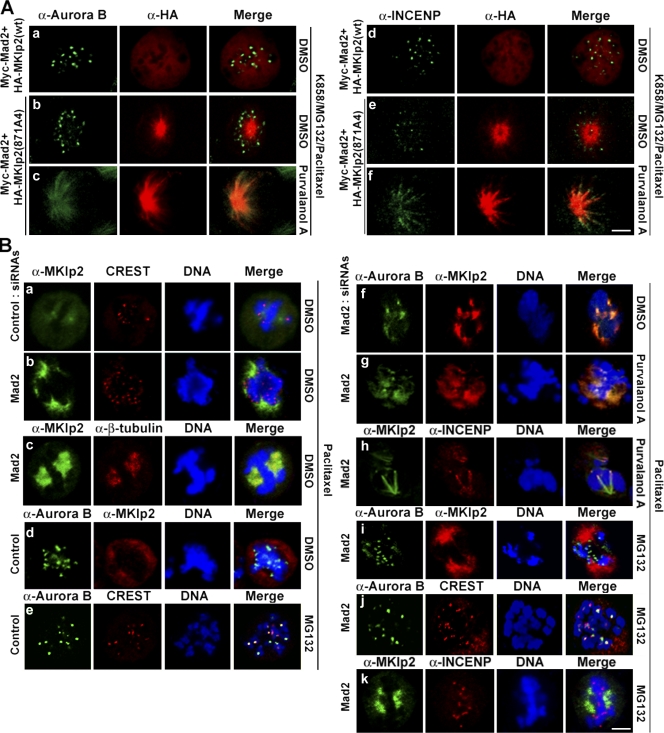
As relocating the CPC by MKlp2 to the central spindle is negatively controlled by Cdk1 (Hümmer and Mayer, 2009), we determined whether Mad2 together with Cdk1 coordinately regulates this event. To address this issue, HeLa cells coexpressing HA-MKlp2 and Myc-Mad2 were forced to form monoasters (Fig. 3 C), and the localizations of Aurora B and INCENP were determined (Fig. 4 A). HA-MKlp2 localized in the mitotic cytoplasm, whereas Aurora B and INCENP showed punctate centromere staining patterns (Fig. 4 A, a and d). In contrast, HA-MKlp2(871A4) localized to the mitotic spindle independent of Aurora B and INCENP (Fig. 4 A, b and e). However, when these cells were treated with a Cdk1 inhibitor purvalanol A, HA-MKlp2(871A4) relocated with Aurora B and INCENP to the mitotic spindle (Fig. 4 A, c and f), suggesting that loading MKlp2 onto the mitotic spindle is a temporally separated step from the MKlp2-mediated relocation of the CPC, the event that Cdk1 negatively controls. Moreover, when HeLa cells were treated with paclitaxel to stabilize the mitotic spindle and to activate the mitotic checkpoint, MKlp2 showed a staining pattern of the paclitaxel-stabilized mitotic spindle in Mad2-depleted cells but not in control (Fig. 4 B, a and b). Indeed, MKlp2 localized to the mitotic spindle in Mad2-depleted cells (Fig. 4 B, c). Furthermore, as determined with the centromere marker CREST, MKlp2 was not found with Aurora B localized at centromeres in control cells (Fig. 4 B, d). In contrast, MKlp2 colocalized with Aurora B and INCENP in Mad2-depleted cells independent of purvalanol A treatment (Fig. 4 B, f–h). Depleting Mad2 prematurely activates the APCCdc20 complex, which inactivates Cdk1 by degrading cyclin B1 (for review see Musacchio and Salmon, 2007). Indeed, when Mad2-depleted cells were treated with MG132 to stabilize cyclin B1, Aurora B and INCENP were at centromeres, whereas MKlp2 showed the mitotic spindle staining patterns (Fig. 4 B, i–k). Together, these results suggest that endogenous Mad2 prevents MKlp2 from loading onto the mitotic spindle when the mitotic checkpoint is active. Furthermore, this regulatory step is temporally separated from the Cdk1-mediated regulation of MKlp2 on relocating the CPC.
Figure 4.
Mad2 and Cdk1 coordinate the mitotic kinesin function of MKlp2 in a spatiotemporal manner. Immunofluorescence analysis using the indicated antibodies is shown. (A) HeLa cells were transfected with expression vectors encoding indicated HA-MKlp2 together with Myc-Mad2. Cells were then treated with K858, MG132, and paclitaxel to form monoasters. (B) HeLa cells transfected with control or Mad2 siRNAs were treated with paclitaxel. Where indicated, cells were treated with DMSO, purvalanol A, or MG132 before fixation. Bars, 5 µm.
To determine whether Mad2 inhibits MKlp2 loading onto the mitotic spindle without spindle damage, we treated control or Mad2-depleted HeLa cells with MG132 to keep Cdk1 active and subsequently extracted the soluble cytosolic proteins to directly visualize MKlp2 bound to the mitotic spindle. Indeed, depleting Mad2 increased the levels of MKlp2 localized at the mitotic spindle compared with control (Fig. 5 A). Similar results were also observed in the absence of MG132 and Mad1-depleted HeLa cells (Fig. S2). Furthermore, ectopically expressed HA-MKlp2 in metaphasic HeLa cells localized at the mitotic spindle (Fig. 5 B, a), whereas coexpressing Myc-Mad2 abolished it (Fig. 5 B, b). Although a Mad2-independnet effect of MKlp2(871A4) might exist, HA-MKlp2(871A4) localized at the mitotic spindle in all metaphasic cells expressing Myc-Mad2 (Fig. 5 B, c) independent of MG132 treatment (Fig. 5 B, d). Moreover, inhibiting Cdk1 by purvalanol A failed to induce the mitotic spindle localization of HA-MKlp2 in cells coexpressing Myc-Mad2 (Fig. 5 B, e and g), whereas HA-MKlp2(871A4) localized together with Aurora B and INCENP (Fig. 5 B, f and h), further suggesting that Mad2 inhibits MKlp2 binding the mitotic spindle independent of Cdk1 activity. Notably, ∼20% of cells coexpressing HA-MKlp2(871A4) with Myc-Mad2 became binucleated (Fig. 5 C) where HA-MKlp2(871A4) often localized at the entire mitotic cytoskeleton (Fig. 5 D). Furthermore, endogenous MKlp2 and Aurora B specifically accumulated in the midbody of control cells undergoing cytokinesis (Fig. 5 E, a and c), whereas both proteins were also found at the mitotic cytoskeleton of Mad2-depleted cells (Fig. 5 E, b and d), which were evident in Mad2-depleted cells treated with paclitaxel.
Figure 5.
Controlling MKlp2 by Mad2 is important for proper mitotic progression and cytokinesis. (A, B, and D–F) Immunofluorescence analysis with the indicated antibodies is shown. (A) HeLa cells transfected with control or Mad2 siRNA for 48 h were treated with MG132 to arrest cells in metaphase and prepermeabilized before fixation (see Materials and methods). (B and D) HeLa cells were transfected with vectors for HA-MKlp2 alone or together with Myc-Mad2. 24 h after transfection, cells were treated with DMSO, purvalanol A, or MG132 before fixation. Arrows in B indicate HA-MKlp2(871A4) prematurely localized to the mitotic spindle, whereas arrows in D indicate the cleavage furrow of a dividing cell. (C) Mitotic HeLa cells expressing HA-MKlp2(wt) (blue bars) or HA-MKlp2(871A4) (red bars) together with Myc-Mad2 from A were scored as metaphasic cells having HA-MKlp2 in the mitotic cytoplasm or at the mitotic spindle and mitotic cells with cytokinesis defect. Means from three independent experiments (n > 100 each) are shown. Error bars indicate mean ± SD. (E) HeLa cells were transfected with control or Mad2 siRNAs. 48 h after transfection, cells were treated with DMSO (a and b) or paclitaxel (c and d) before fixation. Arrows indicate the midbody. (F) HeLa cells were transfected with control or vector for Myc-Mad2. 24 h after transfection, cells were treated with purvalanol A for 2 h before fixation. (G) Schematic model. As cells progress from interphase to mitosis, Mad2 targets MKlp2 when the mitotic checkpoint is active. Upon completion of the mitotic checkpoint, MKlp2 loads onto the mitotic spindle. Subsequently, Cdk1 is inactivated by the APCCdc20-mediated degradation of cyclin B1 (B1), which permits MKlp2 to bind and relocate the CPC from centromeres to the central spindle to promote cytokinesis. Bars, 5 µm.
Given that Mad2 inhibited MKlp2 binding of the mitotic spindles, we determined whether ectopically expressed Mad2 inhibits the ability of MKlp2 to relocate the CPC to the central spindle in dividing cells. Because overexpressing Mad2 arrested cells in metaphase, HeLa cells expressing Myc-Mad2 were exposed to purvalanol A for 2 h to induce exit from metaphase (Fig. 5 F). Determined by immunofluorescence analysis, MKlp2, Aurora B, and INCENP localized in the midbody of control cells (n > 50 each; Fig. 5 F, a, c, and e) but not in cells expressing Myc-Mad2 (n > 50 each; Fig. 5 F, b, d, and f), where MKlp2 localized with Myc-Mad2 in the mitotic cytoplasm (Fig. 5 F, b). Determined by the punctate staining patterns, Aurora B and INCENP appeared to stay at centromeres of dividing cells expressing Myc-Mad2 (Fig. 5 F, d and f), suggesting that Mad2 negatively regulates the ability of MKlp2 to relocate the CPC for cytokinesis.
In this study, we propose that MKlp2 is a mitotic target of Mad2 and provide several lines of evidence that the mitotic checkpoint promotes complex formation between Mad2 and MKlp2 (Fig. 2). MKlp2 bound Mad2 but not Mad1 in early mitosis, and the mitotic checkpoint increased the levels of Mad2 bound to MKlp2. Conversely, inactivating the mitotic checkpoint by depleting Mad1 or BubR1 decreased the levels of Mad2 bound to MKlp2. Depleting MKlp2 had no effect on the mitotic checkpoint induced by nocodazole treatment (unpublished data), suggesting that it does not regulate the Mad2-mediated checkpoint. Furthermore, depleting MKlp2 had no effect on the complex formations of Mad1–Mad2 and Cdc20–Mad2, suggesting that endogenous MKlp2 does not compete with the binding partners of Mad2. Notably, as kinetochore localization of Mad2 by Mad1 facilitates formation of the Mad2–Cdc20 complex (for review see Nasmyth, 2005), inhibiting MKlp2 by Mad2 might occur in a kinetochore-dependent manner, which remains to be tested. Furthermore, as relocating the CPC from centromeres is important to inhibit the mitotic checkpoint at anaphase (Vázquez-Novelle and Petronczki, 2010), MKlp2 might control the mitotic checkpoint by regulating the CPC.
Together with that inhibiting Cdk1 activity promoted MKlp2 and CPC to the mitotic spindle (Hümmer and Mayer, 2009), we propose that at least two temporally and biochemically separable steps are involved in orchestrating the function of MKlp2. First, Mad2 negatively controls the step of loading MKlp2 onto the mitotic spindle independent of the CPC. Second, Cdk1 activity determines the timing of MKlp2 relocating the CPC. These results do not necessarily contradict that T59 phosphorylation of INCENP by Cdk1 controls the translocation of the CPC and MKlp2 to the midzone during the metaphase to anaphase transition. Instead, we report that the active mutant of MKlp2 constitutively bound the mitotic spindle independent of the CPC, suggesting that loading MKlp2 onto the mitotic spindle does not require the CPC in early mitosis. Completion of bipolar spindle attachment results in the termination of the mitotic checkpoint. Thus, this two-step mechanism of regulating MKlp2 by Mad2 and Cdk1 might orchestrate timing of bipolar spindle attachment, which terminates the Mad2-mediated mitotic checkpoint, allowing MKlp2 to load onto the mitotic spindle. Subsequently, segregation of sister chromatids upon Cdk1 inactivation relocates the CPC by MKlp2 to the central spindle for cytokinesis (Fig. 5 G). Furthermore, as depleting Mad2 mislocalized MKlp2 to the mitotic cytoskeleton, Mad2 might be a part of a mechanism controlling “free” MKlp2 in postmetaphase cells, suggesting a potential role of Mad2 in late mitotic phases.
We also report that Mad2 inhibits microtubule association of MKlp2 through the C-terminal region of MKlp2, which locates outside of its microtubule-binding motor domain. Although kinesins often function as dimers through their nonmotor regions, an excess amount of recombinant Mad2 did not compromise the self-multimerizing ability of MKlp2 (unpublished data). Instead, as folding the tail and motor domains of kinesins together is suggested as a general mechanism for inhibiting kinesin motors (for review see Verhey and Hammond, 2009), Mad2 might keep MKlp2 in a folded inactive state in early mitosis. Furthermore, as the C-terminal region of MKlp2 is evolutionary conserved from Humans to Xenopus, the generality of mechanism controlling MKlp2 by Mad2 also warrants further investigation. Notably, although Mad2 did not bind MKlp1 (Fig. 2 A), Cdk1/cyclin B directly phosphorylates the motor domain of MKlp1 on an evolutionary conserved site within a basic N-terminal region that inhibits its binding to microtubules (Mishima et al., 2004), suggesting a distinctive role of Mad2 in controlling MKlp2.
In summary, MKlp2 is an important mitotic target of Mad2 that controls CPC-mediated cytokinesis. Mad2 is overexpressed in many human cancers, which is thought to be a direct cause of chromosome instability and tumorigenesis (Hernando et al., 2004; Sotillo et al., 2007), raising the possibility that deregulated Mad2 causes cytokinesis failure by misregulating MKlp2 and the CPC, and so contributes to chromosome instability and tumorigenesis.
Materials and methods
Cell lines, culture, transfection, and reagents
HeLa and HEK293 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 10% fetal bovine serum (Invitrogen). Transient transfection with mammalian expression vectors was performed with Lipofectamine 2000 reagents (Invitrogen). Vectors encoding Myc- or GST-tagged human Mad2 were described previously (Lee et al., 2008). Cytomegalovirus-based vectors encoding human MKlp1 or MKlp2 were constructed by insertion of PCR-derived cDNA (NCBI Protein database accession nos. AI910107.1 and AQ196929.1 for MKlp1 and MKlp2, respectively) into pCAN1-HA or pDEST27 (Invitrogen) for GST-tagged proteins using the Gateway system (Invitrogen). Complementary DNAs for deletion mutants of MKlp2 were generated by PCR with appropriate primers and cloned into pCAN1-HA. Site-directed mutagenesis was performed with the use of a site-directed mutagenesis system (GeneTailor; Invitrogen).
In vitro GST pull-down assay
Recombinant GST fusion proteins were expressed in Escherichia coli strain BL21(DE3)pLysS and purified with the use of glutathione agarose beads (GE Healthcare). HA-MKlp2 proteins were translated in vitro in the presence of a mixture of [35S]cysteine and [35S]methionine (PerkinElmer) with the use of a reticulocyte lysate system (Promega). GST and GST-tagged Mad2 were loaded onto glutathione agarose beads for 30 min at 4°C in the presence of NP-40 cell lysis buffer (50 mM Tris-HCl, pH 8.0, 120 mM NaCl, and 1% NP-40) containing 1 mM DTT, protease inhibitor mix (Complete Mini; Roche), and phosphatase inhibitor mixes I and II (Sigma-Aldrich). The beads were washed with the cell lysis buffer and incubated overnight at 4°C with in vitro–translated proteins. The beads were isolated and washed before the addition of SDS sample buffer.
Immunofluorescence analysis
Cells grown on poly-l-lysine–coated coverslips were fixed with 4% paraformaldehyde for 10 min, permeabilized with 0.5% Triton X-100, and exposed to PBS containing 10% fetal bovine serum. For Fig. 5 A, cells were prepermeabilized with 0.005% digitonin in transport buffer (110 mM KOAc, 20 mM Hepes, pH 7.3, 2 mM Mg(OAc)2, 0.5 mM EGTA, 2 mM DTT, and EDTA-free Complete Mini) for 4 min before fixation. They were incubated for at least 2 h with primary antibodies, including those to MKlp2 (B01 [Abnova] and A300-879A [Bethyl Laboratories, Inc.]), to Aurora B (H75), INCENP (H153), HA (Y11), Myc (9E10), and β-tubulin (H235; Santa Cruz Biotechnology, Inc.), to α-tubulin (Sigma-Aldrich), or to CREST (ImmunoVision). Immune complexes were detected by incubation of the cells for 1 h with isotype-specific secondary antibodies coupled to Alexa Fluor 488 or Texas red (Invitrogen). Fixed slides were stained with Hoechst 33342 and mounted (mounting medium; Kirkegaard & Perry Laboratories, Inc.). Images for single optical sections of confocal microscopy were acquired at RT with a camera (AxioCam HRc; Carl Zeiss, Inc.) mounted on a microscope (Axiovert 100M; Carl Zeiss, Inc.) with a Plan Apochromat 63× 1.40 NA oil objective. Images were deconvoluted with image software (LSM 5; Carl Zeiss, Inc.), and contrast enhancement was performed using Photoshop software (Element 8; Adobe). For Fig. 3 C (c and d), images were obtained using a laser-scanning microscope (LSM710; Carl Zeiss, Inc.) and deconvoluted with ZEN image software (2010; Carl Zeiss, Inc.).
Cell synchronization
A control nonsilencing siRNA and siRNAs specific for MKlp2 (SI02654064 or SI03060015), Mad1 (SI00052808), Mad2 (SI02653847), or BubR1 (SI00605010 or SI00605017, to ensure reproducibility) mRNAs were obtained from QIAGEN. Cells were synchronized at the G1–S boundary by exposure to 2 mM thymidine for 16 h, incubation in fresh medium for 10 h (during which time they were transfected with 100 nM siRNAs with the use of Lipofectamine 2000), and exposure again to 2 mM thymidine for 14 h. The cells were subsequently washed, incubated in fresh medium, and harvested at the indicated times.
Immunoprecipitation and immunoblot analysis
For Fig. 2 (C and D), HeLa cells released synchronously from the G1–S boundary were harvested by mitotic shake off, and cell lysates were subjected to immunoprecipitation with rabbit antibodies to Mad2 (FL205; Santa Cruz Biotechnology, Inc.) and MKlp2 (A300-878A; Bethyl Laboratories, Inc.), respectively. For Fig. 2 E, nocodazole-arrested HeLa cells that were collected by mitotic shake off were washed with PBS, released in fresh medium, and harvested at the indicated times. For Fig. 2 (F and G), 48 h after siRNA transfections, cells were treated with nocodazole for 5 h before being lysed in NP-40 cell lysis buffer. Cell lysates were then subjected to immunoprecipitation with antibodies to Mad1 (9B10), Cdc27 (C4), or Cdc20 (H175) obtained from Santa Cruz Biotechnology, Inc. For transient transfection, HeLa or HEK293 cells were transfected with equal amounts of indicated expression vectors (2 µg each). About 24–48 h after transfections, cells were lysed in NP-40 cell lysis buffer and subjected to immunoprecipitation with 2 µg agarose-conjugated antibodies to HA (Y11) or Myc (9E10; Santa Cruz Biotechnology, Inc.). For Fig. 2 A, 12 h after transfection, cells were treated with 200 ng/ml nocodazole or DMSO for 12 h and subjected to GST pull-down analysis. For immunoblot analysis, antibodies to Mad2 (17D10 or FL205), Myc (9E10), HA (Y11 or F7), GST (B14), Mad1 (9B10), BubR1 (H23) or Aurora B (H75) were obtained from Santa Cruz Biotechnology, Inc., antibody to β-actin was obtained from Sigma-Aldrich, and antibody to MKlp2 was obtained from Bethyl Laboratories and Abnova. Immune complexes were detected with the use of ECL reagents (GE Healthcare).
Microtubule-binding and mitotic spindle–binding assays
For Fig. 3 B, HEK293 cells expressing HA-MKlp2 or with Myc-Mad2 were lysed in NP-40 cell lysis buffer, and equal amounts of lysates were subjected to a microtubule-binding assay with the use of the Microtubule Binding Protein Spin-Down Assay kit (Cytoskeleton, Inc.). Cell lysates supplemented with taxol-stabilized microtubules were centrifuged at 40,000 rpm for 40 min at RT. After immunoblot analysis, each blot was quantified by phosphorimager using ImageQuant software (Molecular Dynamics). For Fig. 3 C, HeLa cells transfected with siRNAs were treated with 10 µM K858 and 10 µM MG132 for 2 h. Cells were treated with10 µM paclitaxel for 30 min before fixation. For Figs. 3 C and 4 A, after transfection with vectors for Myc-Mad2 and HA-MKlp2, HeLa cells were treated with 10 µM K858 for 12 h and fixed for immunofluorescence analysis. Where indicated in Figs. 4 and 5 B, cells were treated with 10 µM purvalanol A for 30 min before fixation. For Fig. 5 A, cells were treated with 10 µM MG132 for 4 h. For Fig. 4 B and the panels indicated in Fig. 5 E, 48 h after siRNA transfections, cells were treated with 10 µM paclitaxel for 2 h before fixation. For Fig. 5 F, 24 h after transfections, cells were treated with purvalanol A for 2 h before fixation.
Online supplemental material
Fig. S1 shows HeLa cells transfected with MKlp2 siRNA specific for the N terminus of MKlp2 mRNA (SI02654064; QIAGEN) to deplete endogenous MKlp2. 24 h after transfection, cells were transfected with pCAN1 plasmid encoding the motorless HA-MKlp2(500–890) proteins, fixed, and subjected to immunofluorescence analysis with antibodies to HA (F7; Santa Cruz Biotechnology, Inc.) or Aurora B (H75; Santa Cruz Biotechnology, Inc.). Fig. S2 shows HeLa cells transfected with control, Mad1 (SI00052808), or Mad2 (SI02653847) siRNAs (QIAGEN). 48 h after transfection, cells were prepermeabilized with digitonin in transport buffer before fixation and subjected to immunofluorescence analysis using antibodies to α-tubulin (Sigma-Aldrich) and MKlp2 (A300-879A; Bethyl Laboratories, Inc.). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201003095/DC1.
Supplementary Material
Acknowledgments
We thank M. Kitagawa and T. Hirota for comments on the manuscript and the University of California San Francisco Comprehensive Cancer Center cytometry core facility and the Keio Global Center of Excellence flow cytometry core facility for assistance with confocal microscopy and flow cytometry.
This work was partly supported by grants from the Ministry of Education, Science, Sports, and Culture of Japan (to H. Saya). S.H. Lee is a recipient of the National Research Foundation research fellowship (NRF-RF2010-02) in Singapore, which also supported this work.
Footnotes
Abbreviations used in this paper:
- APC
- anaphase-promoting complex
- CPC
- chromosome passenger complex
- MKlp2
- mitotic kinesin-like protein 2
- TAP
- tandem affinity purification
References
- Chen R.H., Shevchenko A., Mann M., Murray A.W.. 1998. Spindle checkpoint protein Xmad1 recruits Xmad2 to unattached kinetochores. J. Cell Biol. 143:283–295. 10.1083/jcb.143.2.283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R.H., Brady D.M., Smith D., Murray A.W., Hardwick K.G.. 1999. The spindle checkpoint of budding yeast depends on a tight complex between the Mad1 and Mad2 proteins. Mol. Biol. Cell. 10:2607–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooke C.A., Heck M.M., Earnshaw W.C.. 1987. The inner centromere protein (INCENP) antigens: movement from inner centromere to midbody during mitosis. J. Cell Biol. 105:2053–2067. 10.1083/jcb.105.5.2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang G., Yu H., Kirschner M.W.. 1998. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes Dev. 12:1871–1883. 10.1101/gad.12.12.1871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Neef R., Honda R., Nigg E.A., Barr F.A.. 2004. Relocation of Aurora B from centromeres to the central spindle at the metaphase to anaphase transition requires MKlp2. J. Cell Biol. 166:167–172. 10.1083/jcb.200403084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernando E., Nahlé Z., Juan G., Diaz-Rodriguez E., Alaminos M., Hemann M., Michel L., Mittal V., Gerald W., Benezra R., et al. 2004. Rb inactivation promotes genomic instability by uncoupling cell cycle progression from mitotic control. Nature. 430:797–802. 10.1038/nature02820 [DOI] [PubMed] [Google Scholar]
- Hill E., Clarke M., Barr F.A.. 2000. The Rab6-binding kinesin, Rab6-KIFL, is required for cytokinesis. EMBO J. 19:5711–5719. 10.1093/emboj/19.21.5711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hümmer S., Mayer T.U.. 2009. Cdk1 negatively regulates midzone localization of the mitotic kinesin Mklp2 and the chromosomal passenger complex. Curr. Biol. 19:607–612. 10.1016/j.cub.2009.02.046 [DOI] [PubMed] [Google Scholar]
- Lee S.H., Sterling H., Burlingame A., McCormick F.. 2008. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 22:2926–2931. 10.1101/gad.1677208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Gorbea C., Mahaffey D., Rechsteiner M., Benezra R.. 1997. MAD2 associates with the cyclosome/anaphase-promoting complex and inhibits its activity. Proc. Natl. Acad. Sci. USA. 94:12431–12436. 10.1073/pnas.94.23.12431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo X., Fang G., Coldiron M., Lin Y., Yu H., Kirschner M.W., Wagner G.. 2000. Structure of the Mad2 spindle assembly checkpoint protein and its interaction with Cdc20. Nat. Struct. Biol. 7:224–229. 10.1038/73338 [DOI] [PubMed] [Google Scholar]
- Luo X., Tang Z., Rizo J., Yu H.. 2002. The Mad2 spindle checkpoint protein undergoes similar major conformational changes upon binding to either Mad1 or Cdc20. Mol. Cell. 9:59–71. 10.1016/S1097-2765(01)00435-X [DOI] [PubMed] [Google Scholar]
- Mishima M., Pavicic V., Grüneberg U., Nigg E.A., Glotzer M.. 2004. Cell cycle regulation of central spindle assembly. Nature. 430:908–913. 10.1038/nature02767 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D.. 2007. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 8:379–393. 10.1038/nrm2163 [DOI] [PubMed] [Google Scholar]
- Nakai R., Iida S., Takahashi T., Tsujita T., Okamoto S., Takada C., Akasaka K., Ichikawa S., Ishida H., Kusaka H., et al. 2009. K858, a novel inhibitor of mitotic kinesin Eg5 and antitumor agent, induces cell death in cancer cells. Cancer Res. 69:3901–3909. 10.1158/0008-5472.CAN-08-4373 [DOI] [PubMed] [Google Scholar]
- Nasmyth K. 2005. How do so few control so many? Cell. 120:739–746. 10.1016/j.cell.2005.03.006 [DOI] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W.C.. 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8:798–812. 10.1038/nrm2257 [DOI] [PubMed] [Google Scholar]
- Schumacher J.M., Golden A., Donovan P.J.. 1998. AIR-2: an Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J. Cell Biol. 143:1635–1646. 10.1083/jcb.143.6.1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sironi L., Mapelli M., Knapp S., De Antoni A., Jeang K.T., Musacchio A.. 2002. Crystal structure of the tetrameric Mad1-Mad2 core complex: implications of a ‘safety belt’ binding mechanism for the spindle checkpoint. EMBO J. 21:2496–2506. 10.1093/emboj/21.10.2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sotillo R., Hernando E., Díaz-Rodríguez E., Teruya-Feldstein J., Cordón-Cardo C., Lowe S.W., Benezra R.. 2007. Mad2 overexpression promotes aneuploidy and tumorigenesis in mice. Cancer Cell. 11:9–23. 10.1016/j.ccr.2006.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vázquez-Novelle M.D., Petronczki M.. 2010. Relocation of the chromosomal passenger complex prevents mitotic checkpoint engagement at anaphase. Curr. Biol. 20:1402–1407. 10.1016/j.cub.2010.06.036 [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Hammond J.W.. 2009. Traffic control: regulation of kinesin motors. Nat. Rev. Mol. Cell Biol. 10:765–777. 10.1038/nrm2782 [DOI] [PubMed] [Google Scholar]
- Verhey K.J., Lizotte D.L., Abramson T., Barenboim L., Schnapp B.J., Rapoport T.A.. 1998. Light chain–dependent regulation of kinesin’s interaction with microtubules. J. Cell Biol. 143:1053–1066. 10.1083/jcb.143.4.1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.