Disruption of vesicle trafficking results in distortion of nuclear shape and increased nuclear envelope surface area but doesn’t alter the nuclear/cell volume ratio.
Abstract
The parameters that control nuclear size and shape are poorly understood. In yeast, unregulated membrane proliferation, caused by deletion of the phospholipid biosynthesis inhibitor SPO7, leads to a single nuclear envelope “flare” that protrudes into the cytoplasm. This flare is always associated with the asymmetrically localized nucleolus, which suggests that the site of membrane expansion is spatially confined by an unknown mechanism. Here we show that in spo7Δ cells, mutations in vesicle-trafficking genes lead to multiple flares around the entire nucleus. These mutations also alter the distribution of small nucleolar RNA–associated nucleolar proteins independently of their effect on nuclear shape. Both single- and multi-flared nuclei have increased nuclear envelope surface area, yet they maintain the same nuclear/cell volume ratio as wild-type cells. These data suggest that, upon membrane expansion, the spatial confinement of the single nuclear flare is dependent on vesicle trafficking. Moreover, flares may facilitate maintenance of a constant nuclear/cell volume ratio in the face of altered membrane proliferation.
Introduction
Organelles have distinct morphologies, which are thought to be important for their functions. Yet, little is known about how organelles establish and maintain their shape and size. For example, budding and fission yeast maintain a constant nuclear/cell (N/C) volume ratio, but the mechanism that controls this ratio is unknown (Jorgensen et al., 2007; Neumann and Nurse, 2007). The membrane of the nuclear envelope (NE) is continuous with the ER, yet how membrane is partitioned between the NE and ER such that the nucleus acquires its typical morphology is not understood. Over the past decade, much attention has been given to the link between nuclear morphology and pathology, as altered nuclear shape is associated with both premature and normal aging, and with certain types of cancers (for review see Webster et al., 2009).
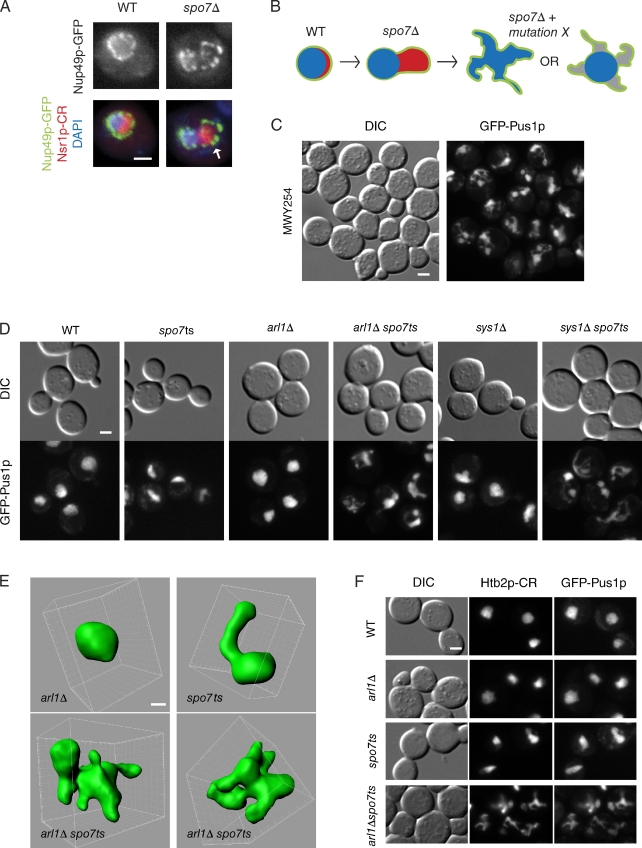
The nucleus of the budding yeast Saccharomyces cerevisiae is spherical in interphase, with the bulk of the chromatin occupying the majority of the nuclear volume. The nucleolus is confined to a crescent-shaped region at the nuclear periphery (Fig. 1 A). Budding yeast that lack the SPO7 gene (spo7Δ) have abnormally shaped nuclei, characterized by expansion of the NE, referred to as a “flare,” which is confined to the NE region associated with the nucleolus (Fig. 1 A, arrow; Siniossoglou et al., 1998; Campbell et al., 2006). The absence of Spo7p results in membrane proliferation, in both the NE and the ER, because of the protein’s role in regulating phospholipid synthesis (Santos-Rosa et al., 2005; O’Hara et al., 2006). Despite this membrane proliferation, the NE associated with the bulk of the chromatin maintains its normal shape (Fig. 1 A; Siniossoglou et al., 1998; Campbell et al., 2006). These observations suggest the existence of a mechanism for maintaining nuclear shape in regions of the NE that are associated with the DNA mass.
Figure 1.
ARL1 and SYS1 affect nuclear morphology in spo7Δ cells. (A) The flare phenotype in spo7Δ cells. Images of fixed wild-type cells (WT) and spo7Δ cells are shown. The flare in spo7Δ cells is marked with an arrow. The NE is identified by the nucleoporin Nup49p fused to GFP (Nup49p-GFP); the nucleolus is identified by the nucleolar protein Nsr1p fused to mCherry red fluorescent protein (Nsr1p-CR); the DNA is stained with DAPI. (B) Diagram of the yeast nucleus showing the NE (green), the nucleolus (red), and the DNA (blue) in wild-type and spo7Δ cells, and hypothetical nuclear phenotypes that would result from a mutation leading to a multi-flare phenotype with (right) or without (left) loss of DNA tethering to the NE. (C) Nuclear phenotype of strain MWY254, carrying a mutation that is synthetically lethal with spo7Δ, observed after a 2-h temperature shift to 37°C. Nuclear morphology was assessed with the nucleoplasmic protein Pus1p fused to GFP (GFP-Pus1p). For comparison to nuclei of wild-type and spo7ts cells, see D or F. (D) Nuclear phenotypes by GFP-Pus1p, associated with arl1Δ and sys1Δ mutations, alone or in combination with spo7ts, after a 2-h temperature shift to 37°C. (E) Three-dimensional reconstruction of nuclei from arl1Δ (top left), spo7ts (top right), and two arl1 spo7ts (bottom) cells. Cells were shifted to 37°C for 2 h. (F) Spatial distribution of chromatin in wild-type, arl1Δ, spo7ts, and arl1Δ spo7ts strains. Chromatin is visualized by the histone H2B fused to mCherry (Htb2p-CR), and nuclear morphology is detected by GFP-Pus1p. Cells were shifted to 37°C for 2 h. Bars: (A, C, D, and F) 2 µm; (E) 1 µm.
Here we show that Golgi-associated vesicle trafficking is needed to maintain nuclear shape under conditions of membrane proliferation. When vesicle trafficking is disrupted, nuclei in cells experiencing membrane proliferation have multiple flares and an increase in NE surface area, but the N/C volume ratio remains the same as in wild-type cells. These observations suggest that when faced with membrane proliferation, cells alter nuclear shape by forming projections that increase nuclear surface area without perturbing the N/C volume ratio. Moreover, vesicle trafficking is needed to confine these projections to the NE region associated with the nucleolus.
Results and discussion
A screen to identify proteins and processes that affect nuclear morphology
The flare phenotype of spo7Δ cells suggests that there is a mechanism preventing nuclear membrane expansion in the region of the NE surrounding the chromatin. Thus, introducing a mutation that disrupts this mechanism to spo7Δ cells could result in a multi-flared nucleus (Fig. 1 B). Such a mutation may not affect nuclear shape in an otherwise wild-type cell, where membrane biogenesis is tightly regulated. Additionally, nuclear processes could be disrupted in multi-flared nuclei, compromising cell viability. We therefore conducted a synthetic lethal screen for randomly generated mutations that reduced the viability of spo7Δ cells, followed by a secondary screen for mutations that affected nuclear morphology when Spo7p was inactive.
We obtained roughly 150 strains carrying mutations that were synthetically lethal/sick with spo7Δ. To examine nuclear morphology, these strains were transformed with a plasmid, pspo7-12, expressing a conditional allele of spo7 (spo7ts) that is hypomorphic at 30°C and nonfunctional at 37°C (Campbell et al., 2006). These strains also expressed the nucleoplasmic protein Pus1p fused to GFP (pGFP-PUS1; Hellmuth et al., 1998). Of the synthetic lethal strains, three strains had a multi-flared nuclear phenotype after a 3-h temperature shift to 37°C (Fig. 1 C).
ARL1 and SYS1, which are involved in endosome to late Golgi trafficking, affect nuclear shape
Two of the multi-flared strains carried a premature stop codon in the SYS1 gene at amino acids Q18 or W108 (out of 203 amino acids). The third strain carried a mutation in the ARL1 gene, leading to a G2D substitution. ARL1 and SYS1 code for evolutionarily conserved proteins involved in retrograde vesicle trafficking from the endosome to late Golgi, as well as anterograde trafficking of Gas1p to the plasma membrane (Tsukada and Gallwitz, 1996; Lee et al., 1997; Rosenwald et al., 2002; Panic et al., 2003; Liu et al., 2006). Sys1p is a transmembrane domain protein required for localization of Arl1p to the late Golgi (Behnia et al., 2004; Setty et al., 2004). In mammals, Arl1p’s localization also requires myristoylation of a glycine at position 2 (Lu et al., 2001; Burd et al., 2004), the analogous glycine mutated in our multi-flared strain.
To confirm that mutations in ARL1 or SYS1 caused the multi-flare phenotype, these genes were deleted in wild-type or spo7ts cells and reexamined for genetic interactions with spo7Δ and effects on nuclear morphology. Both arl1Δ and sys1Δ proved to be synthetically lethal with spo7Δ (Fig. S1 and Table 1, group A). Additionally, sys1Δ and arl1Δ caused multi-flared nuclei in spo7ts cells, whereas they had subtle or nonexistent effect on nuclear shape when deleted in an otherwise wild-type cell (Fig. 1, D and E). Three-dimensional reconstruction by confocal microscopy of arl1Δ spo7ts nuclei (n = 29) revealed that multi-flared nuclei are not fragmented, but rather have numerous lobulations (Fig. 1 E). To characterize chromatin distribution in multi-flared nuclei, histone H2B fused to mCherry red fluorescent protein (Htb2p-CR) was expressed in wild-type, arl1Δ, spo7ts, and arl1Δ spo7ts strains expressing GFP-PUS1. After a 2-h temperature shift, the distribution of Htb2p-CR closely resembled that of GFP-Pus1p in all strains (Fig. 1 F), which indicates that in multi-flared nuclei, the chromatin did not detach from the NE.
Table I.
Vesicle-trafficking mutants’ effects on spo7ts nuclear shape and spo7 viability
| Group | Gene deleted | Percentagea of multi-flared nuclei when combined with spo7ts | Statistical significanceb (P < 0.003) | Genetic interactionc with spo7 | Protein functiond |
| A | ARL1 | 33 ± 5.7 | Yes | ++++ | Soluble GTPase with a role in regulation of membrane traffic |
| A | SYS1 | 34.5 ± 4.9 | Yes | ++++ | Integral membrane protein of the Golgi required for targeting Arl3p to the Golgi |
| B | ARL3 | 30.0 ± 4.2 | Yes | ++++ | GTPase required to recruit Arl1p to the Golgi |
| B | RIC1 | 18.5 ± 2.1 | Yes | ++++ | Forms heterodimer with Rgp1p that acts as a GTP exchange factor for Ypt6p |
| B | YPT6 | 26.5 ± 4.9 | Yes | ++++ | GTPase required for fusion of endosome-derived vesicles with the late Golgi |
| C | ERV14 | 37.5 ± 2.1 | Yes | ++++ | Protein localized to COPII-coated vesicles, involved in vesicle formation and incorporation of specific secretory cargo |
| C | COG8 | 22.5 ± 4.9 | Yes | +++ | Component of the conserved oligomeric Golgi complex that functions in fusion of transport vesicles to Golgi compartments |
| C | VPS52 | 9.5 ± 4.9 | No | ++ | Component of the GARP complex required for the recycling of proteins from endosomes to the late Golgi |
| C | TLG2 | 4.5 ± 0.7 | No | ++ | t-SNARE that mediates fusion of endosome-derived vesicles with the late Golgi |
| C | SWA2 | 8.0 ± 2.8 | No | + | Clathrin-binding protein required for uncoating of clathrin-coated vesicles |
| C | APL5 | 6.5 ± 0.7 | No | + | Delta adaptin-like subunit of the clathrin associated protein complex (AP-3) involved in transport to the vacuole |
| C | GYP1 | 1.0 ± 1.4 | No | − | Cis-golgi GTPase-activating protein (GAP) for Ypt1p, involved in vesicle docking and fusion |
| D | ARF1 | 23.0 ± 4.2 | Yes | ++ | GTPase involved in regulation of coated vesicle formation in intracellular trafficking within the Golgi |
| D | VPS35 | 2.0 ± 1.4 | No | ++ | Endosomal subunit of membrane-associated retromer complex required for retrograde transport |
| D | VPS41 | 1.5 ± 0.7 | No | ++ | Vacuolar membrane protein, subunit of the homotypic vacuole fusion and vacuole protein sorting (HOPS) complex |
| D | GCS1 | 2.0 ± 0.0 | No | + | ARF GAP involved in ER-Golgi transport |
| D | GET2 | 6.0 ± 5.7 | No | + | Subunit of the GET complex; involved in insertion of proteins into the ER membrane |
| D | VPS25 | 2.5 ± 2.1 | No | + | Component of the ESCRT-II complex; involved in ubiquitin-dependent sorting of proteins into the endosome |
Average frequency ± SD of multi-flared nuclei in cells grown at 37°C for 3 h. n = 200 for each strain.
Statistical significance of the multi-flared nuclear phenotype, as calculated by Fisher’s exact test with Bonferroni correction. Values from each deleted gene with spo7ts are compared to the frequency of multi-flared nuclei in spo7ts alone (2 ± 2.8).
The strength of the genetic interaction between spo7Δ and each mutation was assessed by the spot assay (Fig. S1), ranging from synthetic lethality (++++) to no interaction (−).
Description of protein function obtained from Saccharomyces Genome Database (http://www.yeastgenome.org/).
To exclude the possibility that the multi-flare phenotype was a result of cell death, two independent approaches were used: we examined the fraction of dead cells before and after a temperature shift using a vital stain, and we followed the ability of multi-flared cells to recover when returned to the permissive temperature. When the vital stain methylene blue, which stains dead cells, was applied to arl1Δ spo7ts cells before or after a 3-h temperature shift to 37°C, the percentage of dead cells at either temperature was very similar (8.0 ± 1.4% for 30°C and 10.5 ± 0.7% for 37°C), and viable cells with multi-flared nuclei were readily detectable at 37°C. Thus, after a 3-h temperature shift, the formation of multi-flared nuclei is not a result of cell death.
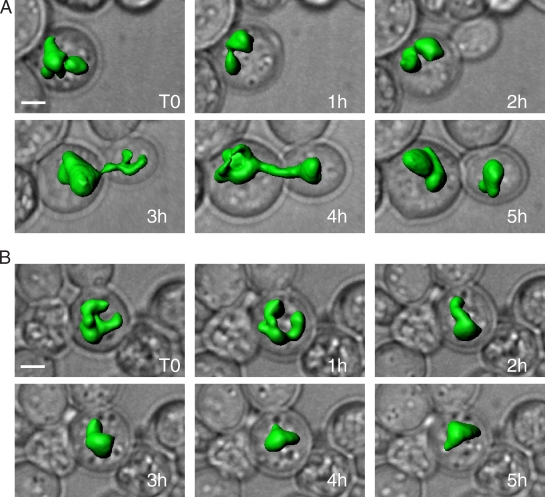
To examine the recovery of cells with multi-flared nuclei, arl1Δ spo7ts cells expressing GFP-Pus1p were returned to 30°C after a 3-h temperature shift to 37°C, then observed by microscopy at 1-h intervals, up to 5 h. For each time point, three-dimensional images of nuclei were overlaid on two-dimensional phase images of the cells to determine cell cycle progression (Fig. 2). Of 11 cells with multi-flare nuclei at t0, there were seven cells that underwent nuclear division (albeit with a prolonged cell division time; e.g., see Fig. 2 A), one cell that began nuclear division, one cell in which the nucleus did not divide (Fig. 2 B), and two cells that died during the 5 h experiment (as determined by the dispersal of the GFP signal). In all cases, the shape of the nucleus changed during the time course (Fig. 2, A and B). Moreover, for cells that did divide, the multi-flared phenotype was lost, and nuclei formed either a single flare (spo7 phenotype; Fig. 2 A and not depicted) or round shape after division (not depicted). Thus, the multi-flare nucleus is not a consequence of cell death but is rather a dynamic structure that can be resolved once cells are returned to permissive conditions.
Figure 2.
arl1Δ spo7ts cells with multi-flared nuclei can recover and divide. 1-h time points of live arl1Δ spo7ts cells grown at 30°C after a 3-h temperature shift to 37°C. Three-dimensional reconstructions of nuclei (with GFP-Pus1) are overlaid on corresponding phase images of cells. (A) A cell with a multi-flared nucleus at t0 divides over the 5-h time course. Mother and daughter cells are seen at the 5-h time point, each with its individual nucleus. (B) A cell with a multi-flared nucleus at t0 does not divide, but undergoes dynamic shape changes. Bars, 2 µm.
Vesicle trafficking via the Golgi affects nuclear shape and distribution of nucleolar proteins
If Arl1p and Sys1p affect nuclear morphology via their known role in vesicle trafficking, then inactivation of other genes in this pathway may have a similar effect. Genetic interactions between the deletion of NEM1, which codes for the phosphatase regulated by Spo7p, and other endosome to late Golgi-trafficking genes, including ARL3, YPT6, and RIC1, have been described previously (Tong et al., 2004; Schuldiner et al., 2005). Consistent with these reports, deletions of ARL3, YPT6, or RIC1 were synthetically lethal with spo7Δ (Fig. S1 and Table I, group B). Importantly, these gene deletions also caused the multi-flared phenotype when combined with spo7ts (Table I, group B), but had only subtle effects on nuclear shape in an otherwise wild-type strain (unpublished data). Thus, the involvement of endosome-to-late Golgi–trafficking proteins in nuclear morphology extends beyond Arl1p and Sys1p.
To determine if other vesicle-trafficking pathways also affect nuclear morphology, we tested additional genes known to interact genetically with spo7Δ and/or nem1Δ (Table I, group C; Tong et al., 2004; Schuldiner et al., 2005) and genes belonging to well-characterized vesicle-trafficking complexes (Table I, group D). The deleted genes ranged from having no genetic interaction to being synthetically lethal with spo7Δ (Table I and Fig. S1). Not all vesicle-trafficking genes led to a multi-flare phenotype in spo7ts nuclei, although all mutations that led to a multi-flared phenotype had genetic interactions with spo7Δ. Thus, only a subset of vesicle-trafficking complexes and pathways, especially those associated with the Golgi, affect nuclear shape under our experimental conditions.
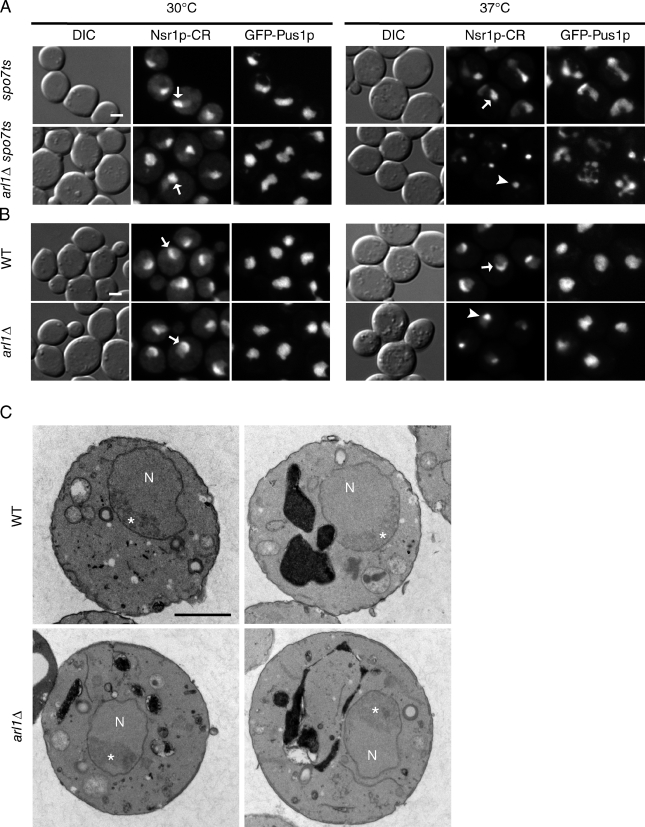
Given the association between the spo7Δ flare and the nucleolus (Fig. 1 A), we examined the localization of the nucleolus in multi-flared nuclei (spo7ts cells deleted for ARL1, YPT6, or COG8) using Nsr1p fused to mCherry red fluorescent protein (Nsr1p-CR). At 30°C, all strains exhibited either a normal crescent nucleolus or single flare filled with Nsr1p-CR (referred to as extended Nsr1p-CR; Fig. 3 A, arrows; and not depicted). After a 3-h shift to 37°C, >80% of cells in all three double mutant strains showed a coalescence of Nsr1p-CR to a single nuclear focus (Fig. 3 A, arrowhead; and not depicted). The temperature-dependent coalescence of Nsr1p-CR was uncommon in wild-type and spo7ts cells (observed in 10.0 ± 2.8% and 7.0 ± 1.4% of cells, respectively), but was prevalent in arl1Δ, ypt6Δ, and cog8Δ single mutants (55.0 ± 5.7%, 97.0 ± 0%, and 94.5 ± 2.1%, respectively; Fig. 3 B and not depicted). This focal appearance was also observed with two other common nucleolar markers: Gar1p and Utp10p (unpublished data). However, by electron microscopy, overall nucleolar structure remained normal in the arl1Δ cells at 37°C (n = 23; Fig. 3 C). Though they were coincidental, the focal appearance of these nucleolar markers (all of which are small nucleolar RNA–associated proteins) and the multi-flared phenotype were independent of each other: at early time points after the temperature shift, we detected both multi-flare nuclei with extended Nsr1-CR (Fig. S2 B) and single-flare nuclei with an Nsr1-CR focus (Fig. S2 C). It is possible that the focal appearance of certain nucleolar proteins in vesicle-trafficking mutants is related to the defect in ribosome synthesis reported for ypt6 and ric1 mutant cells (Li and Warner, 1996; Mizuta et al., 1997).
Figure 3.
Deleting ARL1 causes the coalescence of Nsr1p-CR to a single focus but does not affect overall nucleolar structure. (A) spo7ts cells, with and without ARL1, were fixed either before (left) or after a 3-h temperature shift to 37°C (right), then examined for nuclear morphology (with GFP-Pus1p) and Nsr1p-CR distribution. Arrows show examples of Nsr1p-CR in single flares; the arrowhead shows an example of focal Nsr1p-CR. (B) Wild-type (WT) and arl1Δ strains were treated the same as in A. Arrows show examples of Nsr1p-CR in crescent form; the arrowhead shows an example of focal Nsr1p-CR. (C) Electron micrographs of wild-type and arl1Δ cells fixed after a 2-h temperature shift to 37°C. Nucleoli (labeled with asterisks) are the darker, more electron-dense regions within the nuclei (N). Bars, 2 µm.
Single- and multi-flared nuclei maintain a normal N/C volume ratio despite an increase in NE surface area
The size of the yeast nucleus scales in proportion to cell size (Jorgensen et al., 2007; Neumann and Nurse, 2007). In theory, perturbations to nuclear morphology caused by membrane proliferation could alter the N/C volume ratio. Alternatively, the N/C volume ratio could remain constant, for example, if the excess nuclear membrane is sequestered in protrusions or invaginations. In the case of spo7ts and arl1Δ spo7ts cells, we found the latter to be true. Nuclear surface area and volume were obtained using three-dimensional reconstruction (see Materials and methods), as shown in Fig. 1 E. The N/C ratio was not affected by the temperature at which cells were grown, growth media, or imaging fixed versus live cells (unpublished data, and see Materials and methods). For reasons that are currently unclear, spo7Δ cells are significantly larger than the three other strains tested (Table II). Nonetheless, the N/C volume ratios of wild-type, spo7ts, and arl1Δ spo7ts strains were nearly the same (Table II). However, nuclei of both spo7ts and arl1Δ spo7ts cells had greater surface area than nuclei of wild-type cells (Table III). To compare nuclear surface areas between cells of potentially different sizes, we normalized the measured surface area for each nucleus to a calculated surface area had the nucleus been a perfect sphere (see Materials and methods). For wild-type cells, the mean measured and calculated nuclear surface areas were very similar (measured/calculated = 1.08 ± 0.04; Table III), as expected from the nearly spherical shape of wild-type nuclei. In contrast, the surface areas of spo7ts and arl1Δ spo7ts nuclei were on average 1.27 and 1.48 times greater than a sphere, respectively (Table III; P < 0.0001). Because the N/C volume ratios for wild-type, spo7ts, and arl1Δ spo7ts are the same, this means that for a given cell size, the nucleus of a multi-flared cell will have, on average, a surface area that is 1.48 times greater than that of the spherical wild-type nucleus. We speculate that by sequestering excess nuclear membrane to one or more flares, spo7ts and arl1Δ spo7ts cells can maintain a normal N/C volume ratio.
Table II.
The N/C volume ratios of spo7ts and arl1Δ spo7ts cells are the same as wild-type cells
| Genotype | Cell volume | Nuclear volume | Percentage of nuclear volume/cell volumea |
| µm3 | µm3 | ||
| WT | 30.3 ± 17.8 | 4.3 ± 1.4 | 16 ± 4 |
| arl1Δ | 36.3 ± 15.8 | 4.1 ± 0.9 | 12 ± 3* |
| spo7ts | 53.6 ± 19.0 | 6.6 ± 1.6 | 14 ± 4 |
| arl1Δ spo7ts | 37.9 ± 16.4 | 6.0 ± 1.8 | 18 ± 7 |
Asterisk denotes significance (Student’s t test, P = 0.0016) compared to the mean percentage of nuclear volume of cell volume in wild-type cells.
Table III.
Nuclei of spo7ts and arl1Δ spo7ts cells have greater surface area than nuclei of wild-type cells
| Genotype | Actual surface area | Calculated (spherical) surface area | Fold increase over spherea |
| µm2 | µm2 | ||
| WT | 13.6 ± 2.8 | 12.6 ± 2.8 | 1.08 ± 0.04 |
| arl1Δ | 13.9 ± 2.6 | 12.4 ± 1.8 | 1.12 ± 0.09 |
| spo7ts | 21.6 ± 4.6 | 17.0 ± 2.7 | 1.27 ± 0.12* |
| arl1Δ spo7ts | 23.7 ± 6.2 | 15.9 ± 3.1 | 1.48 ± 0.13* |
Asterisks denote significance (Student’s t test, P < 0.0001) compared to mean fold increase of wild-type nuclear surface area over a perfect sphere.
It is likely that internal nuclear organization is important for nuclear function. This is true in metazoans, where mutations in nuclear lamina proteins affect both nuclear shape and cell fitness (for review see Webster et al., 2009). Yeast and plants do not have genes homologous to nuclear lamina genes, and thus it is of interest to understand what determines nuclear organization in these organisms, findings that could extend to mammalian cells. Here, we identified vesicle-trafficking genes that affect nuclear morphology. Mutations in these trafficking genes are synthetically lethal with spo7Δ. Although it is tempting to speculate that gross alteration in nuclear structure, and consequently abnormal chromatin distribution (Fig. 1 F), is the underlying cause, at this point we cannot rule out the possibility that synthetic lethality is caused by an uncharacterized defect. Many of the genes that we have identified as affecting nuclear morphology are involved in trafficking to and through the Golgi. It is possible that yet unknown proteins must get modified as they traffic through the Golgi before they can function in nuclear shape maintenance. Alternatively, membrane trafficking may be important for the redistribution of membrane or specific lipids to maintain homeostasis of the endomembrane system. For example, defects in trafficking membrane from the ER lead to ER expansion and abnormal nuclear morphology (Kimata et al., 1999). In principle, defects in Golgi trafficking could lead to membrane expansion through the induction of the unfolded protein response (Mousley et al., 2008). However, induction of the unfolded protein response in spo7Δ cells did not cause a multi-flare phenotype (unpublished data). Moreover, arl1Δ alone did not cause expansion of the ER, nor did it exacerbate the ER defect of spo7 mutant cells (Fig. 3 C and not depicted). Lipid trafficking from the Golgi is also necessary for the membrane expansion leading to the formation of autophagosomes (Abudugupur et al., 2002; Reggiori et al., 2004; Geng et al., 2010; Ohashi and Munro, 2010; van der Vaart et al., 2010), which illustrates that the Golgi can function not only in protein sorting, but also in endo-membrane distribution. We did observe in arl1Δ and arl1Δ spo7ts cells a significant increase in the punctate distribution of the endosome marker, Vps24p fused to GFP, as well as a modest increase in the number of early Golgi structures, labeled by Vrg4p fused to GFP (unpublished data). Additionally, mutations in arl1 have been found to cause fragmentation of the vacuole (Abudugupur et al., 2002; Rosenwald et al., 2002). These observations indicate that disrupting endosome-to-Golgi trafficking leads to expansion and/or fragmentation of organelles within the endo-membrane system, which suggests that the Golgi could affect the NE through its role in membrane distribution. In this study, we observed a relative increase in the surface area of the nucleus in arl1Δ spo7ts cells (Tables II and III). Collectively, our data provide a novel link between vesicle trafficking and nuclear morphology, and raise the possibility that the Golgi regulates the availability of nuclear membrane, either directly or indirectly.
Materials and methods
Media and growth conditions
Cells containing plasmids were initially grown in synthetic complete liquid media (2% glucose, 0.17% yeast nitrogen base, 2.5% ammonium sulfate, and the appropriate amino acid and nucleoside mix) to ensure plasmid maintenance. Before fixation, cells were grown overnight at 30°C in YPD (1% yeast extract, 2% peptone, and 2% glucose) to mid-log phase and then shifted to 37°C for the times indicated. For the vital stain assay, cells were grown to mid-log phase in YPD with or without a 3-h temperature shift, resuspended in 0.2 µg/ml methylene blue (Sigma-Aldrich) in 0.05 M KH2PO4 for 5 min, and scored for dye accumulation.
Strains
All strains were in the W303 background (see Table S1 for strain list). Strains containing spo7Δ pspo7-12 (see the following paragraph) were referred to in the text as spo7ts. Gene disruptions and fusions were done according to Longtine et al. (1998) and Goldstein and McCusker (1999). Genomic fusions of the gene coding Discosoma sp. red fluorescent protein, mCherry, were performed by transformation of a PCR product from the plasmid pPW57, a gift from J. Thorner (University of California, Berkeley, Berkeley, CA), modified from pPW58 (Westfall and Thorner, 2006) by switching URA3 with his5+.
Plasmids
pGFP-PUS1: pASZ11-ADE2-PUS1-GFP, encoding GFP-PUS1 (Hellmuth et al., 1998), was modified by PCR-mediated replacement of the ADE2 gene with LEU2. pNUP49-GFP: pASZ11-ADE2-NUP49-GFP, encoding NUP49-GFP (Belgareh and Doye, 1997), was modified by PCR-mediated replacement of the ADE2 gene with URA3. Both original plasmids were gifts from E. Hurt (University of Heidelberg, Heidelberg, Germany).
pSPO7 encodes the SPO7 and ADE3 genes cloned into the polylinker of a TRP1 CEN plasmid (pRS314; Sikorski and Hieter, 1989). pspo7-12 encodes a temperature-sensitive allele of the SPO7 gene spo7-12 (Campbell et al., 2006) cloned into the polylinker of a URA3 CEN plasmid (pRS316; Sikorski and Hieter, 1989).
Isolation of synthetic lethal mutations and identification of strains with abnormal nuclear morphology
Strains JCY607 and TH4201 were mutagenized with ethyl methanesulfonate to 50% cell viability, as described previously (Ross and Cohen-Fix, 2003), and screened for mutations causing synthetically lethality with spo7Δ via a sectoring assay (Bender and Pringle, 1991). Synthetic lethality was confirmed by checking sensitivity of strains to growth on synthetic media plates containing 0.05% 2-amino-5-fluorobenzoic acid (FAA; Sigma-Aldrich), which is toxic to cells expressing Trp1p from the pSPO7 plasmid (Toyn et al., 2000). To score nuclear morphology, the pSPO7 plasmid was swapped with pspo7-12. Strains were then transformed with pGFP-PUS1 and examined for nuclear morphology after a 2-h temperature shift to 37°C. Strains showing a >20% penetrance of an abnormal nuclear phenotype were backcrossed and rescored to ensure that the synthetic lethality and nuclear phenotype were linked and caused by a mutation in a single locus.
Cloning mutations by complementation of synthetic lethality
Strains were transformed with a yeast genomic library and cloned on a LEU CEN plasmid (ATCC 37323), then grown on plates containing 5-fluoroorotic acid (5-FOA; Zymo Research) to select for clones that allowed growth in the absence of the pspo7-12 plasmid (which also codes for URA3; Boeke et al., 1984). Genes present on the suppressing genomic clones were sequenced in the mutagenized genomes to identify the ethyl methanesulfonate–generated mutations.
Fluorescence microscopy and measurements
Cells were fixed in YPD containing 4% paraformaldehyde (Electron Microscopy Sciences) for 1 h at 23°C, washed with 1× PBS, and stored at 4°C for no more than 72 h. For fluorescent and differential interference contrast imaging, fixed cells were incubated briefly in 0.1% Triton X-100 immediately before observation and mixed with an equal volume of Vectashield with DAPI (Vector Laboratories). Images were captured with a microscope (E800; Nikon), equipped with a 100× Plan Fluor DIC H objective lens, using a charge-coupled device camera (CCD; C4742-95; Hamamatsu Photonics) and operated by IPLab 3.0 software (Scanalytics, Inc.). Image overlays in Fig. 1 A were done with the pseudo-colored images using the IPLab software. For three-dimensional reconstruction and volume analyses of cells in G1 phase of the cell cycle, fixed cells were incubated for 5 min in 100 µg/ml calcofluor white (Sigma-Aldrich) at room temperature, washed with 1× PBS, mixed with an equal volume of Vectashield mounting medium, and aliquoted onto a 3% agarose pad. For live cell recovery from 3 h at 37°C, cells were mounted on 3% agarose pads containing synthetic complete media and maintained at 30°C with an objective heater controller (Bioptechs). Images were captured by spinning-disk confocal microscopy using a microscope (Eclipse TE2000U; Nikon) equipped with a 100× Plan-Apochromat objective lens. The imaging system also included a spinning-disk unit (CSU10; Yokogawa) and an electron microscope CCD camera (C9100-13; Hamamatsu). Confocal images at 0.2-µm intervals were acquired using IPlab 4.0 software. Cell and nuclear volumes, as well as nuclear surface area, were acquired with Imaris 7.0.0 software (Bitplane). Images were adjusted using “Auto blend” followed by smoothing using “Median filter 3 × 3 × 1.” A surface was then created using a threshold of absolute intensity. Nuclear volume and surface area of the created surfaces were calculated by Imaris 7.0.0. For cell volume, cells were considered to be prolate spheroids with a volume equal to (4/3)πa2b, where a is the short radii and b is the long radius. Radii were determined by taking half of the longest diameters for each cell as measured in an individual stack using the “slice” view in Imaris 7.0.0. Surface areas of hypothetical “spherical” nuclei were calculated for each nucleus from the measured nuclear volume. Measurements were taken from 21 cells of each genotype after a 3-h temperature shift to 37°C. We calculated averages for cell volumes, nuclear volumes, and the percentage of nuclear volume out of the cell volume. For each cell, the actual nuclear surface area was divided by a calculated nuclear surface area (deduced from nuclear volume measurements and assuming the object is a sphere) to determine the fold increase in actual surface area over that of a sphere. Statistical analyses in Tables II and III were done using an unpaired Student’s t test with Bonferroni correction. Note that in our hands, the nucleus of wild-type cells occupies a greater fraction of the cell volume than described previously (∼16% here compared with ∼7% in Jorgensen et al., 2007). This difference is unlikely to be the result of the temperature at which cells are grown, the growth media, or the imaging of fixed versus live cells (see main text). Additionally, the range for our measured nuclear volumes, 4.3 ± 1.4 µm3, is close to that described by Jorgensen et al. (2007), 2.91 ± 0.85 µm3, and our cell volumes are consistent with data described previously for G1 cells (Jorgensen et al., 2004).
Electron microscopy
Cells were fixed and processed for electron microscopy as described previously (Rieder et al., 1996). The grids were examined on a transmission electron microscope (EM 420; Philips), and images were collected with a Soft Imaging System MegaView III CCD camera (Olympus). Figures were assembled in Photoshop (Adobe) with only linear adjustments in brightness and contrast.
Online supplemental material
Figs. S1 shows testing genetic interactions between spo7Δ and deletions of vesicle-trafficking genes. Fig. S2 shows that formation of Nsr1p-CR foci and multi-flared nuclei in arl1Δ spo7ts cells are independent of each other. Table S1 lists the strains used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201006083/DC1.
Acknowledgments
We thank Will Prinz, Manu Hegde, and members of the Cohen-Fix laboratory for helpful discussions and comments on the manuscript. We also thank Ed Hurt and Jeremy Thorner for plasmids, Joe Campbell for technical assistance, and Alan Fix for assistance with statistical analysis.
M.T. Webster and O. Cohen-Fix were funded by an intramural National Institute of Diabetes and Digestive and Kidney Diseases grant.
Footnotes
Abbreviations used in this paper:
- N/C
- nuclear/cell
- NE
- nuclear envelope
References
- Abudugupur A., Mitsui K., Yokota S., Tsurugi K. 2002. An ARL1 mutation affected autophagic cell death in yeast, causing a defect in central vacuole formation. Cell Death Differ. 9:158–168 10.1038/sj.cdd.4400942 [DOI] [PubMed] [Google Scholar]
- Behnia R., Panic B., Whyte J.R., Munro S. 2004. Targeting of the Arf-like GTPase Arl3p to the Golgi requires N-terminal acetylation and the membrane protein Sys1p. Nat. Cell Biol. 6:405–413 10.1038/ncb1120 [DOI] [PubMed] [Google Scholar]
- Belgareh N., Doye V. 1997. Dynamics of nuclear pore distribution in nucleoporin mutant yeast cells. J. Cell Biol. 136:747–759 10.1083/jcb.136.4.747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender A., Pringle J.R. 1991. Use of a screen for synthetic lethal and multicopy suppressee mutants to identify two new genes involved in morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 11:1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J.D., LaCroute F., Fink G.R. 1984. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol. Gen. Genet. 197:345–346 10.1007/BF00330984 [DOI] [PubMed] [Google Scholar]
- Burd C.G., Strochlic T.I., Gangi Setty S.R. 2004. Arf-like GTPases: not so Arf-like after all. Trends Cell Biol. 14:687–694 10.1016/j.tcb.2004.10.004 [DOI] [PubMed] [Google Scholar]
- Campbell J.L., Lorenz A., Witkin K.L., Hays T., Loidl J., Cohen-Fix O. 2006. Yeast nuclear envelope subdomains with distinct abilities to resist membrane expansion. Mol. Biol. Cell. 17:1768–1778 10.1091/mbc.E05-09-0839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng J., Nair U., Yasumura-Yorimitsu K., Klionsky D.J. 2010. Post-golgi sec proteins are required for autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell. 21:2257–2269 10.1091/mbc.E09-11-0969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein A.L., McCusker J.H. 1999. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 15:1541–1553 [DOI] [PubMed] [Google Scholar]
- Hellmuth K., Lau D.M., Bischoff F.R., Künzler M., Hurt E., Simos G. 1998. Yeast Los1p has properties of an exportin-like nucleocytoplasmic transport factor for tRNA. Mol. Cell. Biol. 18:6374–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Rupes I., Sharom J.R., Schneper L., Broach J.R., Tyers M. 2004. A dynamic transcriptional network communicates growth potential to ribosome synthesis and critical cell size. Genes Dev. 18:2491–2505 10.1101/gad.1228804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P., Edgington N.P., Schneider B.L., Rupes I., Tyers M., Futcher B. 2007. The size of the nucleus increases as yeast cells grow. Mol. Biol. Cell. 18:3523–3532 10.1091/mbc.E06-10-0973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y., Lim C.R., Kiriyama T., Nara A., Hirata A., Kohno K. 1999. Mutation of the yeast epsilon-COP gene ANU2 causes abnormal nuclear morphology and defects in intracellular vesicular transport. Cell Struct. Funct. 24:197–208 10.1247/csf.24.197 [DOI] [PubMed] [Google Scholar]
- Lee F.J., Huang C.F., Yu W.L., Buu L.M., Lin C.Y., Huang M.C., Moss J., Vaughan M. 1997. Characterization of an ADP-ribosylation factor-like 1 protein in Saccharomyces cerevisiae. J. Biol. Chem. 272:30998–31005 10.1074/jbc.272.49.30998 [DOI] [PubMed] [Google Scholar]
- Li B., Warner J.R. 1996. Mutation of the Rab6 homologue of Saccharomyces cerevisiae, YPT6, inhibits both early Golgi function and ribosome biosynthesis. J. Biol. Chem. 271:16813–16819 10.1074/jbc.271.28.16813 [DOI] [PubMed] [Google Scholar]
- Liu Y.W., Lee S.W., Lee F.J. 2006. Arl1p is involved in transport of the GPI-anchored protein Gas1p from the late Golgi to the plasma membrane. J. Cell Sci. 119:3845–3855 10.1242/jcs.03148 [DOI] [PubMed] [Google Scholar]
- Longtine M.S., McKenzie A., III, Demarini D.J., Shah N.G., Wach A., Brachat A., Philippsen P., Pringle J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 14:953–961 [DOI] [PubMed] [Google Scholar]
- Lu L., Horstmann H., Ng C., Hong W. 2001. Regulation of Golgi structure and function by ARF-like protein 1 (Arl1). J. Cell Sci. 114:4543–4555 [DOI] [PubMed] [Google Scholar]
- Mizuta K., Park J.S., Sugiyama M., Nishiyama M., Warner J.R. 1997. RIC1, a novel gene required for ribosome synthesis in Saccharomyces cerevisiae. Gene. 187:171–178 10.1016/S0378-1119(96)00740-8 [DOI] [PubMed] [Google Scholar]
- Mousley C.J., Tyeryar K., Ile K.E., Schaaf G., Brost R.L., Boone C., Guan X., Wenk M.R., Bankaitis V.A. 2008. Trans-Golgi network and endosome dynamics connect ceramide homeostasis with regulation of the unfolded protein response and TOR signaling in yeast. Mol. Biol. Cell. 19:4785–4803 10.1091/mbc.E08-04-0426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann F.R., Nurse P. 2007. Nuclear size control in fission yeast. J. Cell Biol. 179:593–600 10.1083/jcb.200708054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara L., Han G.S., Peak-Chew S., Grimsey N., Carman G.M., Siniossoglou S. 2006. Control of phospholipid synthesis by phosphorylation of the yeast lipin Pah1p/Smp2p Mg2+-dependent phosphatidate phosphatase. J. Biol. Chem. 281:34537–34548 10.1074/jbc.M606654200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y., Munro S. 2010. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol. Biol. Cell. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panic B., Whyte J.R., Munro S. 2003. The ARF-like GTPases Arl1p and Arl3p act in a pathway that interacts with vesicle-tethering factors at the Golgi apparatus. Curr. Biol. 13:405–410 10.1016/S0960-9822(03)00091-5 [DOI] [PubMed] [Google Scholar]
- Reggiori F., Wang C.W., Nair U., Shintani T., Abeliovich H., Klionsky D.J. 2004. Early stages of the secretory pathway, but not endosomes, are required for Cvt vesicle and autophagosome assembly in Saccharomyces cerevisiae. Mol. Biol. Cell. 15:2189–2204 10.1091/mbc.E03-07-0479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder S.E., Banta L.M., Köhrer K., McCaffery J.M., Emr S.D. 1996. Multilamellar endosome-like compartment accumulates in the yeast vps28 vacuolar protein sorting mutant. Mol. Biol. Cell. 7:985–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenwald A.G., Rhodes M.A., Van Valkenburgh H., Palanivel V., Chapman G., Boman A., Zhang C.J., Kahn R.A. 2002. ARL1 and membrane traffic in Saccharomyces cerevisiae. Yeast. 19:1039–1056 10.1002/yea.897 [DOI] [PubMed] [Google Scholar]
- Ross K.E., Cohen-Fix O. 2003. The role of Cdh1p in maintaining genomic stability in budding yeast. Genetics. 165:489–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Rosa H., Leung J., Grimsey N., Peak-Chew S., Siniossoglou S. 2005. The yeast lipin Smp2 couples phospholipid biosynthesis to nuclear membrane growth. EMBO J. 24:1931–1941 10.1038/sj.emboj.7600672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M., Collins S.R., Thompson N.J., Denic V., Bhamidipati A., Punna T., Ihmels J., Andrews B., Boone C., Greenblatt J.F., et al. 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell. 123:507–519 10.1016/j.cell.2005.08.031 [DOI] [PubMed] [Google Scholar]
- Setty S.R., Strochlic T.I., Tong A.H., Boone C., Burd C.G. 2004. Golgi targeting of ARF-like GTPase Arl3p requires its Nalpha-acetylation and the integral membrane protein Sys1p. Nat. Cell Biol. 6:414–419 10.1038/ncb1121 [DOI] [PubMed] [Google Scholar]
- Sikorski R.S., Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 122:19–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siniossoglou S., Santos-Rosa H., Rappsilber J., Mann M., Hurt E. 1998. A novel complex of membrane proteins required for formation of a spherical nucleus. EMBO J. 17:6449–6464 10.1093/emboj/17.22.6449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong A.H., Lesage G., Bader G.D., Ding H., Xu H., Xin X., Young J., Berriz G.F., Brost R.L., Chang M., et al. 2004. Global mapping of the yeast genetic interaction network. Science. 303:808–813 10.1126/science.1091317 [DOI] [PubMed] [Google Scholar]
- Toyn J.H., Gunyuzlu P.L., White W.H., Thompson L.A., Hollis G.F. 2000. A counterselection for the tryptophan pathway in yeast: 5-fluoroanthranilic acid resistance. Yeast. 16:553–560 [DOI] [PubMed] [Google Scholar]
- Tsukada M., Gallwitz D. 1996. Isolation and characterization of SYS genes from yeast, multicopy suppressors of the functional loss of the transport GTPase Ypt6p. J. Cell Sci. 109:2471–2481 [DOI] [PubMed] [Google Scholar]
- van der Vaart A., Griffith J., Reggiori F. 2010. Exit from the golgi is required for the expansion of the autophagosomal phagophore in yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 21:2270–2284 10.1091/mbc.E09-04-0345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M., Witkin K.L., Cohen-Fix O. 2009. Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 122:1477–1486 10.1242/jcs.037333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westfall P.J., Thorner J. 2006. Analysis of mitogen-activated protein kinase signaling specificity in response to hyperosmotic stress: use of an analog-sensitive HOG1 allele. Eukaryot. Cell. 5:1215–1228 10.1128/EC.00037-06 [DOI] [PMC free article] [PubMed] [Google Scholar]