Phosphorylation of the muscle-specific formin splice variant FHOD3 by CK2 regulates its stability, myofibril targeting, and myofibril integrity.
Abstract
Members of the formin family are important for actin filament nucleation and elongation. We have identified a novel striated muscle–specific splice variant of the formin FHOD3 that introduces a casein kinase 2 (CK2) phosphorylation site. The specific targeting of muscle FHOD3 to the myofibrils in cardiomyocytes is abolished in phosphomutants or by the inhibition of CK2. Phosphorylation of muscle FHOD3 also prevents its interaction with p62/sequestosome 1 and its recruitment to autophagosomes. Furthermore, we show that muscle FHOD3 efficiently promotes the polymerization of actin filaments in cardiomyocytes and that the down-regulation of its expression severely affects myofibril integrity. In murine and human cardiomyopathy, we observe reduced FHOD3 expression with a concomitant isoform switch and change of subcellular targeting. Collectively, our data suggest that a muscle-specific isoform of FHOD3 is required for the maintenance of the contractile structures in heart muscle and that its function is regulated by posttranslational modification.
Introduction
The actin cytoskeleton is involved in a variety of functions from cell adhesion and migration, vesicle transport, and cell division to muscle contraction (Chhabra and Higgs, 2007; Ehler and Gautel, 2008; Campellone and Welch, 2010). Because spontaneous formation of actin filaments happens quite slowly, several actin nucleators have been characterized that enhance this process. The ARP2/3 complex favors the formation of branched actin filaments, whereas members of the formin family promote F-actin assembly at the fast-growing barbed filament end to long unbranched filaments (Campellone and Welch, 2010). Formins are characterized by the presence of an FH2 (formin homology 2) domain. Two FH2 domains form a doughnut-shaped dimer that can then processively move along the actin filaments and promote the addition of further actin monomers (Goode and Eck, 2007; Paul and Pollard, 2009). In addition, formins contain an FH1 domain that binds to profilin and probably serves to deliver G-actin, which is recruited for filament polymerization (Paul and Pollard, 2009). Formin family members have been found from man to yeast and show closest homologies in their FH2 domains. Formin homology domain–containing proteins (FHODs) are members of a formin subfamily that may also play a role in cell signaling and the regulation of transcription (Young and Copeland, 2010).
Striated muscle cells are characterized by a particularly regular arrangement of their actin cytoskeleton in the thin filaments of the sarcomeres, where the barbed end is anchored in the Z disc and the pointed end stretches to the middle of the sarcomere toward the M-band region. Thin filament length is strictly controlled by tropomodulin, which caps the pointed ends (Gregorio et al., 1995). However, the concept that this extremely regular arrangement is totally static is misleading because metabolic labeling experiments have suggested a surprisingly high turnover rate of thin filament proteins, with an average half-life of only 3 d (Martin, 1981). Myofibrillogenesis has been studied extensively over the years; however, the exact mechanism of thin filament assembly and turnover is still unclear (Littlefield and Fowler, 2008; Sparrow and Schöck, 2009). At the earliest stages of myofibrillogenesis in the developing heart, actin filaments assemble in close proximity to the plasma membrane, and they subsequently detach and stretch throughout the cytoplasm (Ehler et al., 2004). Currently, it is unknown whether actin filament nucleation during myofibrillogenesis requires promoting factors and what their nature may be (Sparrow and Schöck, 2009). Recently, it could be shown that leiomodin, which is related to the pointed-end capping protein tropomodulin, has strong actin filament–nucleating activity and aids myofibril maintenance in cultured cardiomyocytes (Chereau et al., 2008). However, no information is available on leiomodin’s function during the initial stages of myofibril assembly. Based on observations on the actin cytoskeleton in other cell types, the construction of actin filaments may depend on multiple actin assembly–promoting factors (Chesarone and Goode, 2009), and formins have been suggested as good potential candidates (Sparrow and Schöck, 2009).
FHOD3, a diaphanous-related formin, is expressed at high levels in the heart, kidney, and brain (Katoh and Katoh, 2004). Tissue-specific splicing in the N-terminal part results in two isoforms, of which only the larger is found in the heart (Kanaya et al., 2005). When cloning full-length FHOD3 from the heart, we identified an additional alternative exon, which leads to the insertion of eight additional amino acids at the C-terminal end of the FH2 domain. Based on its amino acid sequence, we called this additional exon the T(D/E)5XE exon. Inclusion of this exon is specific for striated muscle tissue and adds phosphorylation sites for CK2 (casein kinase 2). Phosphorylation of this site affects subcellular targeting and inhibits the interaction of FHOD3 with p62/SQSTM1 (sequestosome 1), a protein involved in autophagy (for review see Rusten and Stenmark, 2010). Furthermore, we demonstrate a role for FHOD3 in actin polymerization and during myofibrillar maintenance in cultured cardiomyocytes, as well as the down-regulation of its expression in cardiomyopathy.
Results
The FHOD3 gene contains an additional striated muscle–specific exon that is expressed in a tissue-specific fashion
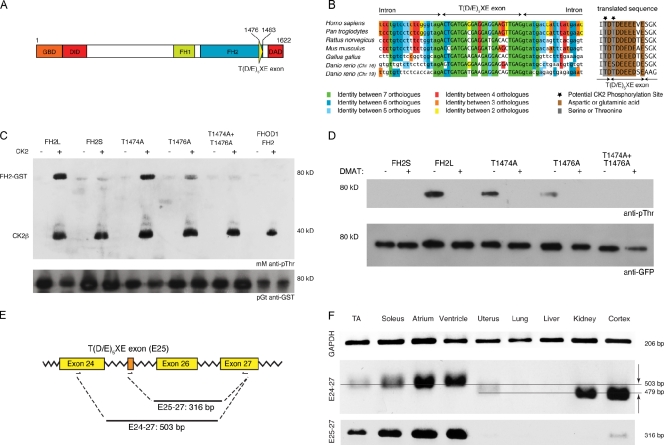
Because FHOD3 is the formin with the highest expression level in the heart, we decided to clone its full-length cDNA from an adult heart library to study its role in cardiomyocytes (Fig. 1 A, schematic representation of FHOD3). During this process, we identified a novel alternative exon, which inserts as exon 26 in humans and as exon 25 in mice (available from GenBank/EMBL/DDBJ under accession no. HM191478; Fig. S1, exon–intron structure of the FHOD3 gene in primates and rodents). This alternative exon is highly conserved between zebrafish and humans and encodes for eight mainly acidic amino acids that insert at the C-terminal end of the FH2 domain (Fig. 1, A [scheme] and B). To avoid confusion caused by the complexity of the FHOD3 gene organization between different species, we called this exon the “T(D/E)5XE exon” based on its characteristic sequence.
Figure 1.
FHOD3 contains a novel alternative exon at the end of its FH2 domain. (A) Domain layout of the human FHOD3 splice isoform expressing the T(D/E)5XE exon (exon 26 in humans and exon 25 in rodents; Fig. S1). GBD, GTPase-binding domain; DID, diaphanous inhibitory domain; FH1/FH2, formin homology domains 1 and 2. (B) The alternative T(D/E)5XE exon of FHOD3 encodes for an evolutionarily conserved stretch of eight mainly acidic amino acids that introduce CK2 consensus sites. Lowercase letters are intronic, and capital letters represent exonic sequences. Splice acceptor and donor sites are evolutionarily conserved in all seven orthologues, with the intronic bases AG on the 5′ end and GT on the 3′ end. (C) GST fusion proteins of the FHOD3-FH2 domain containing (FH2L) or excluding (FH2S) the alternative T(D/E)5XE exon, as well as threonine to alanine mutants of the predicted target threonines (T1474, T1476, or both), and the FHOD1-FH2 domain were tested for CK2 in vitro phosphorylation. FH2L, but not FH2S or FHOD1-FH2, was phosphorylated by CK2. Mutation of T1474 or T1476 to alanine reduced phosphorylation slightly or greatly, whereas mutation of both residues completely abolished the phosphorylation. mM, monoclonal mouse. (D) A similar phosphorylation pattern to that seen in vitro could also be demonstrated in COS-1 cells transfected with different GFP-tagged FHOD3 constructs and was abolished by treatment with 10 µM of the CK2 inhibitor DMAT for 24 h before lysis. The signal for GFP is shown as a loading control below. (E) Schematic view of the primer sets used for the RT-PCRs. (F) RT-PCR to test for tissue-specific expression of the alternative T(D/E)5XE exon. The amount of template mouse cDNAs was normalized against glycerine aldehyde 3-phosphate dehydrogenase (GAPDH). The specificity of the T(D/E)5XE exon for striated muscle can either be seen with primer set E25-27 or, because of the difference in the electrophoretic mobility, with primer set E24-27. The top line points out the 503-bp fragment, and the bottom line indicates the 479-bp fragment, which are also indicated by the arrows. TA, tibialis anterior.
Because its high degree of conservation suggested a preserved function, we decided to investigate the role of the T(D/E)5XE exon more closely. The presence of serine or threonine residues flanked by acidic amino acids indicated a potential CK2 consensus sequence motif (Cox et al., 2003). Indeed, in a CK2 in vitro phosphorylation assay, phosphorylation by CK2 was detected only in FHOD3-FH2 domain constructs that contained the alternative exon (FH2L) but not in constructs lacking it (FH2S) or in the FHOD1-FH2 domain. Mutation analysis revealed that the second threonine (T1476) is the main target residue for phosphorylation and that change of both threonines to alanines totally abolishes the reaction (Fig. 1 C). These phosphorylation results could also be confirmed in a cellular environment and were prevented by incubation with the specific CK2 inhibitor DMAT (2-dimethylamino-4,5,6,7-tetrabromo-1H-benzimidazole; Fig. 1 D).
A previously identified larger FHOD3 isoform that is caused by N-terminal alternative splicing was reported to be specific for the mouse heart (Kanaya et al., 2005). We therefore wanted to investigate whether the T(D/E)5XE exon in the FH2 domain of FHOD3 was also spliced in a tissue-specific way. Different murine RNAs (tibialis anterior, soleus, atrium, ventricle, uterus, lung, liver, kidney, and cortex) were tested by RT-PCRs with two sets of primers: the splicing-specific primer set E25-27, with the forward primer located in the T(D/E)5XE exon, and the splicing-independent primer set E24-27 (Fig. 1 E). Confirming the results of a previous study (Kanaya et al., 2005), the strongest expression of the fragment amplified by primer set E24-27 was found in the heart, kidney, and cortex. The decreased electrophoretic mobility with primer set E24-27, caused by the 24 additional bases, as well as the signal obtained with the primer set E25-27, indicated that the expression of the T(D/E)5XE exon is highest in soleus muscle, cardiac ventricles, and atria (Fig. 1 F). This suggests that the novel FHOD3 isoform containing the T(D/E)5XE exon is specifically expressed in striated muscle, where it is subjected to phosphorylation by CK2, predominantly at T1476.
The T(D/E)5XE exon targets FHOD3 to the myofibrils
To assess the effect of the alternative splicing on biological function, we transiently transfected FHOD3, either containing or lacking the alternative T(D/E)5XE exon, into neonatal rat cardiomyocytes (NRCs). Interestingly, we observed a drastically changed targeting pattern depending on the presence of the T(D/E)5XE exon. Constructs that contained the alternative T(D/E)5XE exon localized mainly to the myofibrils, whereas constructs lacking this exon formed mostly cytoplasmic aggregates (Fig. 2 A).
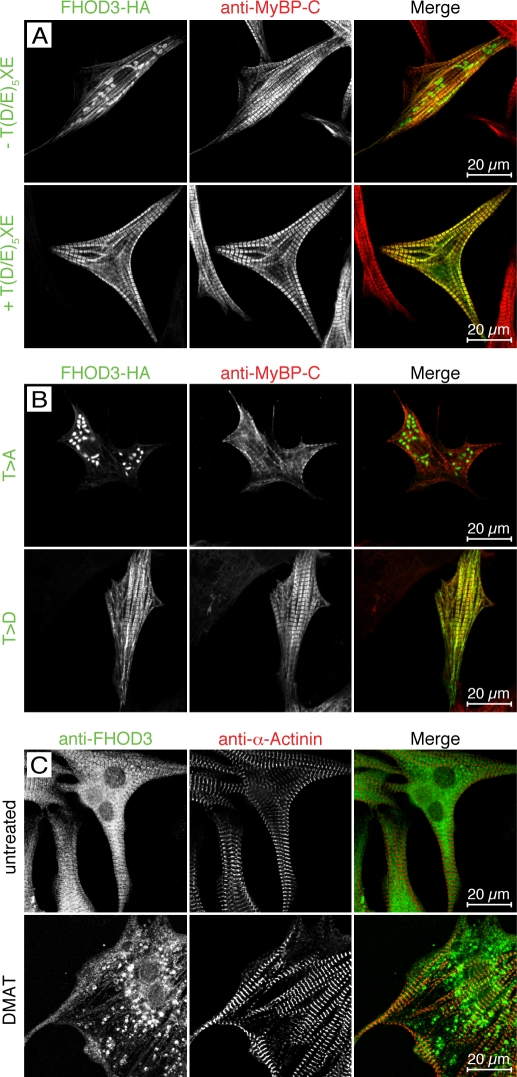
Figure 2.
The muscle-specific FHOD3 isoform is targeted to the sarcomere in a phosphorylation-dependent fashion. (A and B) NRCs were transfected with FHOD3-HA including or lacking the alternative T(D/E)5XE exon (A) or after mutation of residues T1474 and T1476 to alanine (T>A) or aspartate (T>D; B). After 48 h, cells were fixed and stained with monoclonal rat anti-HA and pRb anti–myosin-binding protein C (MyBP-C). Constructs including the T(D/E)5XE exon targeted to the A band, whereas constructs lacking the T(D/E)5XE exon formed cytoplasmic aggregates. Mutation of the threonine residues to alanine induced the formation of cytoplasmic aggregates as well. No change in localization was seen after mutation of the threonine residues to aspartic acid. (C) Untransfected NRCs were treated with 10 µM of the CK2 inhibitor DMAT for 24 h and then fixed and stained with anti-FHOD3 and anti–α-actinin antibodies. Untreated cells were used as a control. Similar to exogenous FHOD3, endogenous FHOD3 localizes to the A band but forms aggregates when phosphorylation by CK2 is inhibited.
To investigate whether this observed change in subcellular localization is caused by the inclusion of the acidic T(D/E)5XE peptide sequence by itself or whether the phosphorylation of the residues T1474 and T1476 by CK2 is responsible, both threonine residues were mutated to alanine or aspartate to mimic the unphosphorylated and phosphorylated state, respectively. Whereas the phosphomimicry T to D mutant of FHOD3 targeted to the myofibrils, constructs with the T to A mutation were found in cytoplasmic aggregates (Fig. 2 B). Analysis of transfected NRCs by phalloidin staining to visualize F-actin revealed colocalization of all FHOD3 constructs with filamentous actin, which was also found in the aggregates (Fig. S2, A and B). The presence or absence of the T(D/E)5XE sequence, as well as its phosphorylation status (as indicated by the phosphomutant constructs), had also no effect on the general ability of transfected FHOD3 to affect actin polymerization in cells (Fig. S2 C). To ensure that these observations are not caused by overexpression of transiently transfected expression constructs, we looked at the localization of endogenous FHOD3 using an affinity-purified polyclonal antibody (for characterization see Fig. S3). Again, FHOD3 was mainly localized at the myofibrils in control NRCs, but a dramatic relocalization to cytoplasmic aggregates was observed after incubation with the CK2 inhibitor DMAT (Fig. 2 C). This suggests that the sarcomeric targeting of FHOD3 constructs containing the T(D/E)5XE sequence, as well as of endogenous FHOD3, is regulated by CK2 phosphorylation.
FHOD3 interacts with p62/SQSTM1 in a phosphorylation-dependent manner
We wanted to establish whether the differential subcellular targeting of FHOD3 constructs could be explained by distinct protein–protein interactions. Therefore, we searched for potential novel interaction partners of the FH2 domain by yeast two-hybrid screening of a human heart cDNA library. We identified p62, also referred to as SQSTM1, a protein involved in ubiquitin kinase signaling and proteasomal and autophagic protein degradation (for review see Rusten and Stenmark, 2010), as a putative interaction partner of an FH2-LexA bait construct. A forced yeast two-hybrid assay revealed a stronger interaction with the shorter FH2 (lacking the T(D/E)5XE exon; FH2S) compared with the longer FH2 (containing the T(D/E)5XE exon; FH2L) construct because binding to the latter was already inhibited in the presence of 0.5 mM 3-AT (3-amino-1,2,4-triazole), whereas the construct lacking the T(D/E)5XE exon was still growing in the presence of 5 mM 3-AT (Fig. 3 A).
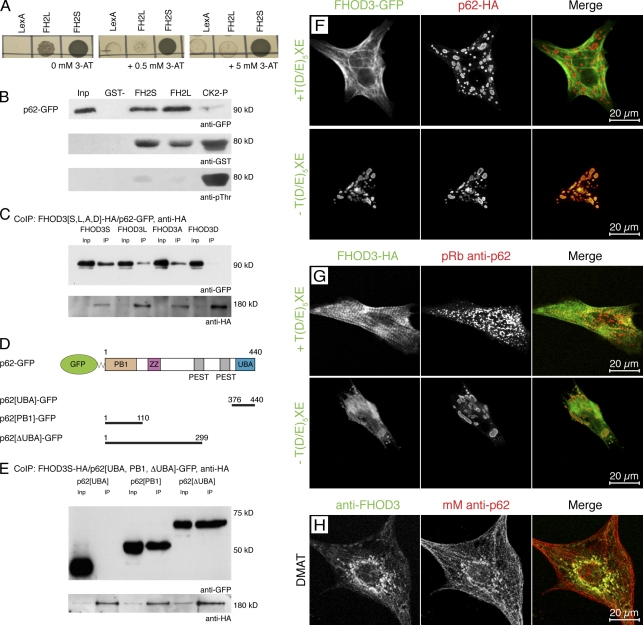
Figure 3.
FHOD3 interacts with the p62-PB1 domain in a phosphorylation-regulated manner. (A) Yeast two-hybrid assay to test for the specificity and strength of the p62 interaction with the FHOD3-FH2 domain. p62-pGAD10 was transformed into yeast together with FH2L and FH2S-LexA, as well as with the empty LexA vector as a control, and grown on medium containing 0, 0.5, and 5.0 mM of 3-AT. p62 strongly interacts with FH2S and interacts much more weakly with FH2L. (B) GST pull-down assays to analyze the FHOD3–p62 interaction. Equal amounts of purified FH2L-GST, FH2S-GST, or CK2-phosphorylated FH2L-GST (CK2-P) were incubated with lysates of p62-GFP–transfected COS-1 cells and subjected to immunoblotting with monoclonal mouse (mM) anti-GFP and, subsequently, polyclonal goat anti-GST. The successful phosphorylation of FH2L-GST was controlled by incubation with monoclonal mouse anti–phospho-threonine (pThr). Although the presence of the unphosphorylated T(D/E)5XE exon has no effect on the interaction with p62, CK2 phosphorylation of FH2L-GST before the incubation strongly reduces the interaction. Inp, input. (C) Coimmunoprecipitation (IP; CoIP) of full-length FHOD3 and p62. Lysates of COS-1 cells, which were transfected with p62-GFP and FHOD3-HA constructs containing (FHOD3L) or excluding (FHOD3S) the T(D/E)5XE exon, as well as with constructs with residues T1474 and T1476 mutated to alanine (FHOD3A) or aspartic acid (FHOD3D), were immunoprecipitated with anti-HA and subjected to immunoblotting with anti-GFP and subsequently anti-HA antibodies. Mutation of the threonine residues to aspartic acid, but not to alanine, greatly reduced the interaction. (D) p62 domain structure and transfection constructs. Domains were constructed according to Simple Modular Architecture Research Tool (SMART) and Geetha and Wooten (2002). ZZ, zinc finger domain (ZZ type). (E) To identify the minimal binding site, COS-1 cells were cotransfected with FHOD3S-HA and p62-GFP constructs containing the PB1 domain, the UBA domain, or the protein without the UBA domain (ΔUBA) as indicated in D. Lysates were treated as described earlier. Binding was found with the ΔUBA and the PB1 domain, indicating that binding of FHOD3 requires presence of the PB1 domain in p62. (F) Cotransfection of NRCs with p62-HA and FHOD3-GFP constructs. Cells were stained with monoclonal rat anti-HA. p62 recruits FHOD3 lacking the T(D/E)5XE exon into aggregates. (G) Transfection of NRCs with FHOD3-GFP constructs and staining with pRb anti-p62 show colocalization of endogenous p62 only with FHOD3 lacking the T(D/E)5XE exon. (H) Visualization of endogenous FHOD3 and p62 by immunofluorescence in NRCs that were treated with DMAT to inhibit CK2 demonstrates colocalization after the redistribution of FHOD3.
GST pull-down and coimmunoprecipitation assays were performed to confirm the interaction of p62 with the FHOD3-FH2 domain and full-length FHOD3 biochemically. Lysates of COS-1 cells that overexpressed p62 were incubated with purified FH2L- or FH2S-GST fusion proteins and FH2L-GST that had been incubated with CK2 before the GST pull-down assay (Fig. 3 B) to obtain information about a potential role of the CK2 phosphorylation on the binding to p62. Indeed, although the interaction with p62 was not affected by the insertion of the alternative T(D/E)5XE exon, it was disrupted by phosphorylation by CK2 (Fig. 3 B). Similarly, the mutation of the threonine residues to aspartic acid inhibited the binding of the full-length FHOD3 construct to p62 in coimmunoprecipitation experiments from COS-1 cell lysates (Fig. 3 C).
p62 is composed of an N-terminal Phox and Bem1p domain (PB1), followed by a zinc finger domain, a PEST region, and a C-terminal ubiquitin-associated (UBA) domain (Fig. 3 D). The PB1 domain is responsible for polymerization and binding to other PB1-containing proteins (Moscat et al., 2006), whereas the UBA domain binds ubiquitin noncovalently, with a preference for polyubiquitin chains (Seibenhener et al., 2007). In an attempt to identify the minimal binding site required for FHOD3 interaction, we cloned and tested different p62 deletion constructs (Fig. 3 D). Coimmunoprecipitation assays with lysates from COS-1 cells overexpressing GFP-tagged constructs of the p62-UBA domain (p62-UBA–GFP), the protein without the UBA domain (p62-ΔUBA–GFP), and the PB1 domain (p62-PB1–GFP), as well as HA-tagged FHOD3S, showed that the UBA-GFP construct did not bind but that interaction was detected with the PB1 domain alone and with p62 lacking the UBA domain. This suggests that the FHOD3 binding site resides in the PB1 domain of p62 (Fig. 3 E).
Upon cotransfection of FHOD3 and p62 into NRCs, a differential FHOD3 distribution was observed, depending on whether FHOD3 contained or lacked the T(D/E)5XE exon. Only in the latter case, a colocalization with HA-tagged p62 to cytoplasmic bodies occurred (Fig. 3 F, bottom). Endogenous p62 was detected in a partially overlapping staining pattern in NRCs transfected with FHOD3 lacking the T(D/E)5XE exon (Fig. 3 G, bottom), whereas no colocalization could be detected with T(D/E)5XE exon–containing FHOD3 (Fig. 3 G, top). In addition, endogenous FHOD3 that had lost its association with the myofibrils after DMAT treatment colocalized with p62 as well (Fig. 3 G). To confirm the identity of the p62 and FHOD3 complexes as autophagosomes rather than other types of subcellular aggregates, we stained COS-1 cells and NRCs after double transfection with anti-LC3 antibodies. This revealed that the p62- and FHOD3-containing structures are also positive for LC3, which indicates their identity as autophagosomes (Fig. 4). Collectively, our biochemical analysis, colocalization experiments, and treatment of cells using CK2-specific inhibitors suggest that FHOD3 binds to p62 in a phosphorylation-dependent manner. CK2 phosphorylation of the muscle-specific T(D/E)5XE exon in FHOD3 abolishes the interaction of the FH2 domain of FHOD3 with p62 and its location to autophagosomes and results in myofibrillar targeting. These two FHOD3 isoforms appear to have strikingly diverse functions in muscle cells, and their distinct roles are defined by their divergent posttranslational modification, which regulates the interaction with other proteins such as p62. In addition, the isoforms appear to show a different protein stability as analyzed by transfecting COS-1 cells with different FHOD3 constructs and treating them with cycloheximide after 24 h to inhibit de novo protein synthesis. The muscle isoform of FHOD3, as well as the construct that mimics phosphorylation, can still be detected by immunoblotting after 48 h, whereas constructs that can potentially interact with p62 are no longer seen at that time point (Fig. S4).
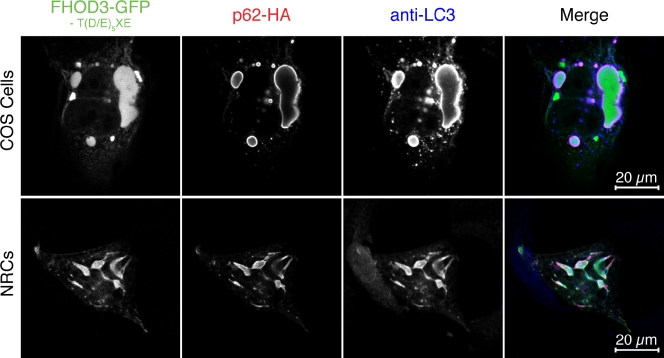
Figure 4.
The interaction with p62 targets FHOD3 to autophagosomes. COS-1 cells (top) and NRCs (bottom) were cotransfected with FHOD3-GFP lacking the T(D/E)5XE exon and p62-HA and stained with monoclonal rat anti-HA and pRb anti-LC3. p62 recruits FHOD3 into aggregates, which colabel for the autophagosomal marker LC3.
FHOD3 localizes to the Z disc in adult heart tissue
To investigate the expression and localization of endogenous FHOD3 in tissue, we generated an antibody against its N-terminal 339 amino acids, which are shared between all known isoforms (for characterization see Fig. S3 A). Attempts to create an antibody that would specifically recognize the T(D/E)5XE exon were unsuccessful, presumably because of the low complexity of this sequence.
Immunostaining of longitudinal sections of mouse left ventricles revealed a cross-striated staining pattern that overlapped with the signal for sarcomeric α-actinin (Fig. S3 B). Preabsorption of the FHOD3 antibodies with the antigen abolished this signal. Interestingly, the targeting of FHOD3 seems to be developmental stage specific and/or affected by culture conditions. In freshly isolated adult rat cardiomyocytes (ARCs), the signal was again concentrated at the Z discs with additional diffuse cytoplasmic signal. However, in NRCs in culture, FHOD3 shows a wider striation pattern that partially overlaps with the A band (Fig. S3 B; and Fig. 2 C, top). Because the same localization pattern is seen with transiently transfected full-length FHOD3 constructs in NRCs, we propose a differentiation stage–specific targeting and that FHOD3 is mainly associated with the Z disc of the sarcomere in fully mature and healthy heart tissue.
FHOD3 aids the polymerization of cardiac thin filaments in cyto
RNAi-mediated knockdown of FHOD3 has been shown to disrupt sarcomeric structure (Taniguchi et al., 2009). We confirmed these observations using a slightly different experimental approach. In our case, the FHOD3-specific short hairpin RNA (shRNA) is transcribed from a vector rather than added directly, and identification of the few transfected NRCs is possible by GFP fluorescence, which is encoded by the same plasmid (Fukuzawa et al., 2008). The shRNA constructs were designed to target conserved regions of the rat FH2 domain. Because of the typically low transfection efficiency of NRCs, we performed the initial characterization in COS-1 cells that were cotransfected with knockdown vectors and FHOD3 constructs containing the target sequences. Analysis by immunoblotting showed that only the specific shRNA construct, but not the construct encoding point-mutated shRNA, was able to reduce the expression levels of the FHOD3-GFP fusion protein (Fig. S5 A). An efficient knockdown was also shown by staining for endogenous FHOD3 in shRNA-transfected NRCs, which indicated 70–90% knockdown after 5 d of transfection (Fig. S5 B).
To investigate FHOD3 knockdown effects on the cardiomyocyte cytoarchitecture, NRCs were transfected with FHOD3-shRNA or the control and incubated for 2, 5, and 8 d. NRCs were subsequently stained for sarcomeric markers (α-actinin, F-actin, myosin-binding protein C, and myomesin) to examine potential changes to their sarcomeres and cytoskeleton. Expression of the shRNA for 2 d did not significantly change the localization patterns of the sarcomeric proteins. Interestingly, the FHOD3 levels, as indicated by anti-FHOD3 staining, were also not significantly altered at that time (unpublished data). After 5 d, however, RNA knockdown resulted in shortening of myofibrils, accompanied by a reduction in the number of mature sarcomeres (Fig. S5, C and D). Patches of mature sarcomeres, as indicated by M-band localization of myomesin, were interspersed with stress fiber–like structures, visualized by alpha-actinin staining. This phenotype may indicate impaired myofibril maintenance rather than defects in sarcomerogenesis and reflects a certain cross talk between thin and thick filaments, i.e., removal of one interferes with the integrity of the other over time in a contracting muscle cell. FHOD3 knockdown for 8 d further aggravated the phenotype in transfected myocytes (Fig. S5, E and F). Therefore, knockdown of FHOD3 expression levels results in a severe myofibrillar phenotype. Our knockdown data suggest that FHOD3 plays an important role for the homeostasis of sarcomeric structures.
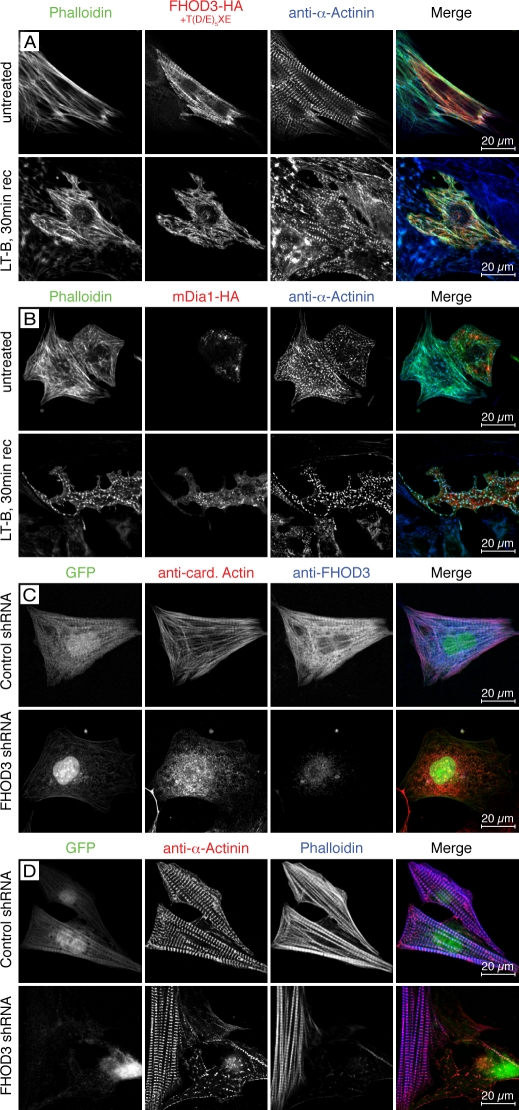
To investigate the importance of FHOD3 for the de novo actin polymerization in cardiomyocytes, we subjected NRCs with increased FHOD3 protein levels (gain of function) or in which endogenous FHOD3 levels were reduced by shRNA (loss of function) to latrunculin B treatment and recovery at different time points. Compared with untransfected controls, NRCs overexpressing FHOD3 display organized sarcomeres and recovered thin filaments already after 30 min, as judged by α-actinin and phalloidin staining, indicating that increased FHOD3 levels partially rescue latrunculin B–induced actin depolymerization (Fig. 5 A, bottom). Transfection of NRCs with HA-tagged mDia1, another member of the formin protein family, is less efficient because only comparatively short actin filaments surrounding Z bodies can be detected after 30 min of recovery (Fig. 5 B, bottom). Knockdown of FHOD3 protein levels in NRCs using the FHOD3-specific shRNA construct results in reduced thin filament and myofibrillar recovery (Fig. 5, C and D). Whereas myofibrils in control cells have fully recovered 2 h after latrunculin B removal, only residual sarcomeric striations were visible in NRCs in which FHOD3 expression was knocked down. Our results suggest that endogenous FHOD3 is required for myofibril maintenance and show for the first time that FHOD3 can aid actin filament polymerization in cardiomyocytes.
Figure 5.
FHOD3 is required for the polymerization of actin filaments in cardiomyocytes. (A–D) NRCs transfected with FHOD3-HA (including the T(D/E)5XE exon; A), mDia1-HA (B), or FHOD3-shRNA constructs (C and D) were treated with 20 µM latrunculin B (LT-B) overnight. Untreated transfected NRCs are shown in A and B (top), whereas control shRNA-transfected NRCs are shown in C and D (top). Latrunculin was removed, and cells were left to recover for 30 min (30 min rec; A and B) or 2 h (C and D). Cells were stained with monoclonal rat anti-HA, monoclonal mouse anti–α-actinin, and phalloidin (A and B) and monoclonal mouse anticardiac actin (anti-card. Actin) and pRb anti-FHOD3 (C) or monoclonal mouse anti–α-actinin and phalloidin (D). (A and B) Only cells transfected with FHOD3 have repolymerized their actin filaments after 30 min. Transfection with mDia1 results in a speckled phalloidin staining, indicating only reduced actin polymerization, whereas untransfected cells display no recovery of actin polymerization. (C) After 2 h, untransfected cells are starting to recover, with the majority of cardiac actin assembled into filaments. Colocalization with cardiac actin indicates an involvement of (endogenous) FHOD3 in the polymerization of F-actin. In contrast, no actin polymerization was detected after knockdown of FHOD3, further stressing the importance of this formin for the assembly of cardiac actin filaments (C and D).
Down-regulation and isoform switch during cardiomyopathy
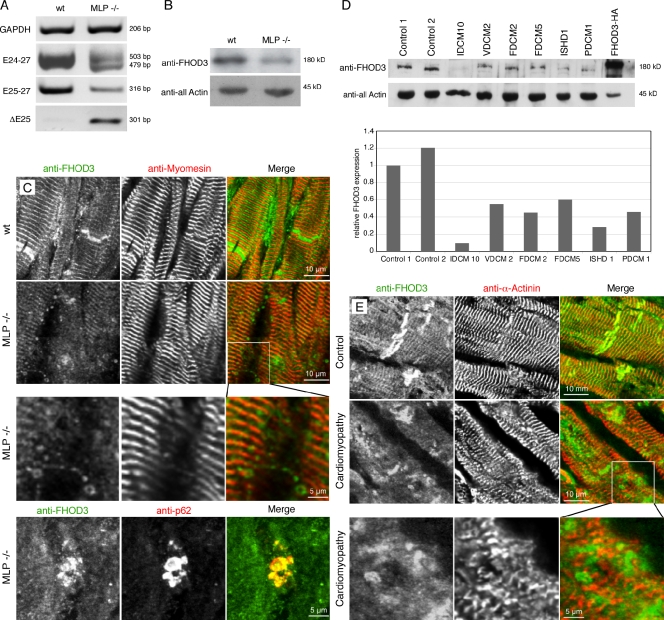
The observation that FHOD3 may be of importance for the assembly and maintenance of cardiac myofibrils led us to investigate the role of FHOD3 in cardiomyopathy. We first studied FHOD3 expression levels in heart tissue from muscle LIM protein (MLP) knockout mice, a model for dilated cardiomyopathy (Arber et al., 1997; Ehler et al., 2001). Analysis of FHOD3 isoforms by RT-PCR revealed a dramatic loss of isoforms containing the T(D/E)5XE exon with a concomitant up-regulation of T(D/E)5XE exon–lacking isoforms (Fig. 6 A). At the protein level, a down-regulation of total FHOD3 expression in MLP knockout compared with wild-type mice was seen (Fig. 6 B). Investigation of the subcellular localization of FHOD3 by immunofluorescence revealed Z-disc and intercalated-disc staining in wild-type tissue (Fig. 6 C). In tissue from MLP knockout mice, these patterns were lost, and only weaker striations could be observed, which tended to overlap with the M-band protein myomesin. Higher magnification analysis revealed, in addition, vesicular staining patterns for FHOD3, which partially colocalized with p62. These results indicate a disease-associated isoform switch from the muscle FHOD3 isoform containing the T(D/E)5XE exon to the isoform lacking the T(D/E)5XE exon. This isoform switch is accompanied by changes to FHOD3 localization from the myofibrils to p62-positive autophagosomes and may account for the reduced FHOD3 protein levels observed in MLP cardiomyocytes.
Figure 6.
FHOD3 expression levels are reduced in the failing heart. (A) RT-PCR analysis of samples from MLP knockout hearts and wild type (wt) using primers for glycerine aldehyde 3-phosphate dehydrogenase (GAPDH) for standardization and primer sets for FHOD3, amplifying all splice variants (E24-27), amplifying variants specific for the T(D/E)5XE exon containing (E25-27), or amplifying exclusively the T(D/E)5XE exon excluding isoform (DE25). A dramatic change from splice isoforms containing the T(D/E)5XE exon to the isoform lacking this exon in hearts of MLP knockout mice can be detected. (B) The reduction of total FHOD3 (E24-27) mRNA in MLP hearts is mirrored by reduced FHOD3 protein levels in immunoblot analysis. (C and E) Immunofluorescence with an anti-FHOD3 antibody shows reduced staining in sections of MLP knockout (C) and human failing hearts (E), whereas the intensity of the antimyomesin and anti–α-actinin stainings are comparable. The reduction in muscle FHOD3 is accompanied by the appearance of autophagosome-like structures, which also colabel for p62 (C and E, inset magnifications; C, bottom stained with monoclonal mouse anti-p62). (D) Reduced expression of FHOD3 is also detected by immunoblot analysis in a range of human cardiomyopathies. A quantification of the blotting is shown below. Because of the scarcity of the material, the blotting was only repeated once. IDCM, idiopathic dilated cardiomyopathy; VDCM, ventricular dilated cardiomyopathy; FDCM, familial dilated cardiomyopathy; ISHD, ischemic heart disease; PDCM, perinatal dilated cardiomyopathy.
To see whether the down-regulation of FHOD3 in the failing heart is a peculiarity of a murine model or can be translated to human pathology, we investigated FHOD3 expression levels in different cardiomyopathy samples (Fig. 6 D). Interestingly, down-regulation was observed in all cases of heart disease no matter whether the samples originated from dilated cardiomyopathy or from ischemic heart disease patients. In addition, we stained longitudinal sections of a human left ventricle of an end-stage cardiomyopathy patient, as well as an unaffected control (an organ donor without indications for cardiac disease), with the anti-FHOD3 antibody. FHOD3 was localized to the Z discs and the intercalated discs also in healthy human ventricular cardiomyocytes (Fig. 6 E). In the diseased heart, however, a general down-regulation of anti-FHOD3 staining was observed, whereas the intensity of the α-actinin staining remained comparable with the control.
Collectively, our results suggest that striated muscle expresses a specific splice variant of FHOD3 that introduces CK2 phosphorylation sites at the C-terminal end of its FH2 domain. This posttranslational modification influences both its interaction with p62 and its targeting to the myofibrils. The presence of this isoform is a hallmark of differentiated muscle, and its expression is altered in a mouse model for dilated cardiomyopathy. General knockdown of FHOD3 in NRCs interferes with sarcomere integrity and thin filament organization, suggesting that FHOD3 is crucial for myofibril maintenance. This was confirmed by direct actin polymerization experiments in cardiomyocytes. Observations in failing human hearts suggest that a lack of FHOD3 may also be of relevance in the human setting of cardiomyopathy.
Discussion
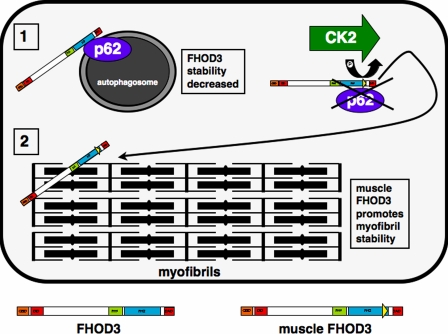
Here, we describe a novel isoform of the formin family member FHOD3, which is specifically expressed in striated muscle. The isoform arises from the inclusion of an additional exon, called the T(D/E)5XE exon, that encodes for eight amino acids at the C-terminal end of the FH2 domain of FHOD3. Inclusion of this exon creates a phosphorylation site for CK2, which in turn regulates the interaction of this FHOD3 isoform with p62/SQSTM1, a protein involved in muscle-specific kinase signaling, proteasomal degradation, and autophagy (Gautel, 2008). In addition, we show that FHOD3 is important for the maintenance of myofibrils and aids the polymerization of actin filaments in cultured cardiomyocytes and that its expression is down-regulated in murine and human samples of failing hearts. Our results are the first to demonstrate the importance of a formin family member in heart muscle and also to correlate distinct subcellular targeting and binding partner interaction of a formin to a posttranslational modification (Fig. 7, model).
Figure 7.
Model for the regulation and subcellular localization of different FHOD3 isoforms. The insertion of the T(D/E)5XE exon into muscle FHOD3 is indicated by the yellow triangle at the end of the FH2 domain. Phosphorylation by CK2 abolishes the interaction of muscle FHOD3 with p62 and promotes targeting to the myofibrils.
Different isoforms of FHOD3 showing expression restricted to striated muscle were already described by Kanaya et al. (2005). However, these arise because of differential splicing in the N-terminal half of the protein. The T(D/E)5XE exon was missed in their work, presumably because a brain library was used for the cloning rather than a heart library as in our case. Although the function of different N-terminal splice variants is presently unclear, our results show the impact of the inclusion of the T(D/E)5XE exon on FHOD3 function, affecting its subcellular localization in muscle cells as well as its interaction with p62. The known major role of the FH2 domain is the nucleation of actin filaments, which requires FH2 dimerization because monomeric FH2 domains inhibit actin polymerization (Shimada et al., 2004). The FH2 domain is the most conserved domain within the formin family and is therefore used to classify formin family members into different categories (Higgs, 2005). From the structural information that is available for the FH2 dimer of a yeast formin, it seems unlikely that the inclusion of the T(D/E)5XE exon would affect dimerization because the main interaction occurs via the N-terminal and central part of the FH2 core domain (Xu et al., 2004). Our results on efficient stimulation of actin polymerization by FHOD3 in cultured cardiomyocytes suggest that the function of the FH2 domain is not impaired by the insertion of the eight amino acids encoded by the T(D/E)5XE exon. Also, comparison of the ability of the different FHOD3 constructs to stimulate F-actin formation in HeLa cells indicates that neither the presence of the T(D/E)5XE exon nor the phosphorylation status (as indicated by phosphomutants) has an effect. Because recombinant FHOD3-FH2 aggregated in our hands, we have been unable to study potential differences in actin-nucleating activity between the FH2 and FH2-T(D/E)5XE isoforms directly so far.
Splice variants have also been identified for other members of the formin family, causing differences in their actin polymerization behavior (Wen and Rubenstein, 2009), their interaction with their activating small GTP-binding proteins of the Rho family (Lammers et al., 2008), and their susceptibility to inhibition by drugs (Gauvin et al., 2009). The isoform switch from the muscle-specific FHOD3 isoform to the isoform lacking the T(D/E)5XE exon in the diseased heart parallels changes that have been observed for many other proteins in the heart, in which embryonic expression patterns are recapitulated during cardiomyopathy (Agarkova et al., 2000; MacLellan and Schneider, 2000; Ahuja et al., 2007). Interestingly, a cardiac-specific knockout of the splicing factor ASF/SF2 results in severe cardiomyopathy, thus further stressing the importance of tissue-specific splicing for the correct function of the heart (Cooper, 2005; Ladd et al., 2005; Xu et al., 2005; Kalsotra et al., 2008).
The interaction of the T(D/E)5XE-lacking FHOD3 with p62 may be of particular importance to the role of this FHOD3 isoform in cells. p62 is a signaling hub that also interacts with multiple other proteins, such as atypical PKC, TRAF6, Nbr1, and caspase 8, and has therefore been implicated in several cellular functions that comprise NF-κB signaling and cell survival, autophagy, apoptosis, and several myopathies (Gautel, 2008). Many of the p62/SQSTM1 interactions occur via its PB1 domain (Moscat et al., 2007), which is also the domain that appears to mediate binding to FHOD3. In addition to a potential effect on FHOD3 protein levels, the interaction between nonmuscle FHOD3 and p62 may also be relevant for the process of autophagy by itself. It has been shown that autophagosomes have to fuse with early endosomes for efficient degradation and that interference with this process inhibits autophagy and leads to the accumulation of p62 (Razi et al., 2009). Other members of the formin protein family, mDia1 and mDia2, were shown to be important for endosome formation, endosome transport, and phagocytosis in macrophages (Wallar and Alberts, 2003; Goode and Eck, 2007). It was also demonstrated that FNBP1L (formin-binding protein 1–like) is necessary for autophagy of bacteria (Huett et al., 2009). Whether the interaction between FHOD3 and p62 could hint at an involvement of formins such as FHOD3 in autophagy will be interesting to investigate.
Our results indicate that FHOD3 plays a major role in the maintenance of myofibrils in NRCs. This confirms recent observations by Taniguchi et al. (2009), who also reported a disruption of sarcomeric structure after RNAi-mediated knockdown of FHOD3. However, in these knockdown experiments, an effect is already seen after 2 d. The delayed knockdown in our system may potentially be because the shRNA is transcribed from a vector rather than added directly by transfection. The mechanisms of thin filament assembly and maintenance are still under debate (Sparrow and Schöck, 2009). Although it is known that the average half-life of thin filament proteins such as actin and troponin is ∼3–5 d (Martin, 1981), it is still not entirely clear where in the sarcomere and by which mechanism the exchange could happen in the contracting myocyte (Littlefield and Fowler, 2008). Littlefield et al. (2001) have performed elegant experiments that show that actin exchange at the pointed end (toward the M band) is 1.6 times greater than at the barbed end (located in the Z disc), which would be the major candidate for actin incorporation based on actin filament dynamics in fibroblasts. We could show that FHOD3, which is a predicted barbed-end actin filament nucleator based on its analogy with other formins, is located mainly at the Z disc in the mature heart and may thus assist actin turnover or reduce barbed-end depolymerization. Another actin filament nucleator that has recently been identified is leiomodin (Chereau et al., 2008). Leiomodin is related to the pointed-end capping protein tropomodulin but additionally contains a WH2 (Wiskott-Aldrich syndrome homology 2) domain, which is a hallmark of actin-nucleating proteins. Knockdown of leiomodin similarly affects sarcomere maintenance in NRCs (Chereau et al., 2008), which suggests that nucleators at both the pointed end (leiomodin) and the barbed end (FHOD3) may be needed to assemble and maintain functional thin filaments.
Lastly, our observations on isoform switches and general down-regulation of FHOD3 expression in the human heart suggest its crucial involvement in myofibril maintenance in vivo and not just in cultured cardiomyocytes. There is a certain redundancy in the regulation of actin polymerization, as amply documented for nonmuscle cells (Chesarone and Goode, 2009), because thin filaments do not disappear completely. Nevertheless, other actin assembly factors cannot fully compensate for the down-regulation of FHOD3, or only to a limited extent as, for example, mDia1 in our de novo actin polymerization experiments in NRCs. Therefore, the loss of the adult heart isoform of FHOD3 might contribute to the progress of cardiomyopathies.
Materials and methods
RNA extraction and RT-PCRs
Mouse RNA was extracted with TRIZOL (Invitrogen) according to the manufacturer’s instructions. Adult human total heart RNA was obtained from Agilent Technologies. cDNA pools were prepared using Moloney murine leukemia virus reverse transcriptase (Promega).
Murine cDNAs were amplified with DNA polymerase (Phusion; Finnzymes) and splicing-specific (E25-27 forward, 5′-ACCGATGACGAGGATGAAGCTGAG-3′, and reverse, 5′-CTCTCTGACTGGGCACTTGGGTGG-3′; and ΔE25 forward, 5′-ATCACTGACTCTGGCAAGTTC-3′, and reverse, 5′-CTCTCTGACTGGGCACTTGGGTGG-3′) or splicing-independent (E24-27 forward, 5′-CTCATTTTTACTGTTTATGGGCCATCC-3′, and reverse, 5′-CTCTCTGACTGGGCACTTGGGTGG-3′) primer sets. Amplification of glycerine aldehyde 3-phosphate dehydrogenase (forward, 5′-CTCAAGATTGTCAGCAATGCATCC-3′, and reverse, 5′-CCAGTGGATGCAGGGATGATGTTC-3′) was used to normalize the amount of cDNA.
Preparation of transfection constructs
The FHOD3-FH2 domain was amplified from adult human heart cDNA using 5′-GAAGATCTGCCACCATGGTTCCTCCTCCTCCAGTG-3′ forward and 5′-CCCCGCGGCGGCTGGCTTGGGGGC-3′ reverse primers and cloned into the BglII and SacII restriction sites of a pEGFP-C1 vector (Takara Bio Inc.). Constructs containing or excluding the alternative T(D/E)5XE exon were selected after sequencing. CK2 receptor residues were mutated to alanine and aspartic acid by standard site-directed mutagenesis using 5′-GACCAGAGGGAAGATGATCGCCGATACTGATGAGGAGGAGG-3′ (T1474A), 5′-GGGAAGATGATCACCGATGCTGATGAGGAGGAGGAAGTTG-3′ (T1476A), 5′-CAGAGGGAAGATGATCGCCGATGCTGATGAGGAGGAGGAAG-3′ (T1474A + T1476A), and 5′-CAGAGGGAAGATGATCGATGATGATGATGAGGAGGAGGAAG-3′ (T1474D + T1476D) primers. FH2-GST constructs were created by subcloning the respective fragments into the BglII and SacII restriction sites of a pGEX-2TK vector (GE Healthcare).
Full-length FHOD3 was amplified from human heart cDNA with LA-taq polymerase (Takara Bio Inc.) using 5′-ATAACCGGTGCCACCATGGCCACGCTGGCTTGCC-3′ forward and 5′-TCCCCGCGGCAGCTGCAACTCCGAGG-3′ reverse primers, digested with AgeI and SacII restriction enzymes, and cloned into the BspEI and SacII restriction sites of a pEGFP-C1 or HA-C1 vector. Splice isoforms were selected after sequencing, and phosphomutants were created as described earlier for the FH2 domain constructs. Diaphanous autoregulation domain (DAD) deletion constructs were amplified from full-length FHOD3, and the respective splice isoforms and mutant constructs were amplified by using the full-length forward and the FH2 reverse oligos and cloned as described for the full-length constructs.
The FHOD1-FH2 domain was amplified from adult human skeletal muscle cDNA using 5′-GGAATTCGACAGCTCAGCCCTCCCC-3′ forward and 5′-GGGGTACCGGCGATTGTGTGTGGTG-3′ reverse primers and cloned into the EcoRI and KpnI restriction sites of a pEGFP-C2 vector. mDia1 was amplified from adult human heart cDNA using 5′-CCGCTCGAGCCACCATGGAGCCGCCCGGCGG-3′ forward and 5′-GGGGTACCGTGCTTGCACGGCCAACC-3′ reverse primers and cloned into the XhoI and KpnI restriction sites of a HA-C1 vector. All p62 constructs were a gift from M. Gautel (King’s College London, London, England, UK).
NRCs
Newborn rat hearts were digested using the neonatal cardiomyocyte isolation system (Worthington Biochemical Corp.) according to the manufacturer and cultured basically as described previously (Lange et al., 2002). Cells were plated onto collagen-coated dishes (PureCol; Inamed Biomaterials) with a density of 0.4 × 106 cells per 35-mm culture dish in plating medium (68% DME [Sigma-Aldrich], 16% medium M199 [Sigma-Aldrich], 10% horse serum [Sigma-Aldrich], 5% fetal calf serum [PAA], 4 mM glutamine [Sigma-Aldrich], and 1% penicillin–streptomycin [Sigma-Aldrich]). After 24 h in culture, the cells were transfected for 6 h with ∼1 µg of plasmid DNA per 35-mm dish with Escort III (Sigma-Aldrich) or jetPRIME (Polyplus-transfection via Source BioScience) in antibiotics-free medium and then changed to maintenance medium (78% DME, 20% medium M199, 4% horse serum, 1% penicillin–streptomycin, 4 mM glutamine, and 0.1 mM phenylephrine [Sigma-Aldrich]). After 48 h, cells were fixed with 4% paraformaldehyde (Agar Scientific) in PBS for 10 min. For long-term maintenance, the cells were kept in long-term maintenance medium (20% medium M199, 75% DBSSK [116 mM NaCl, 1 mM NaH2PO4, 0.8 mM MgSO4, 5.5 mM glucose, 32.1 mM NaHCO3, and 1.8 mM CaCl2, pH 7.2], 4% horse serum, and 4 mM glutamine). Freshly isolated ARCs were donated by F. Cuello (King’s College London, London, England, UK). For fixation, cells were incubated with 4% paraformaldehyde (Agar Scientific) in PBS for 10 min. To permeabilize the membrane, cells were incubated for 5 min with 0.2% Triton X-100/PBS and then washed three times in PBS. Primary and secondary antibodies were diluted in antibody dilution buffer (1% BSA, 20 mM Tris-base, 155 mM NaCl, 2 mM EGTA, and 2 mM MgCl2, pH 7.5). Blocking reagent (MAXblock; Active Motif) was used before stainings with the FHOD3 antiserum according to the manufacturer’s instructions. The cells were incubated in the primary antibody solution for 1 h at RT (overnight at 4°C for ARCs) and then washed three times in PBS, followed by incubation with the secondary antibody solution for 1 h at RT (5 h at RT for ARCs). For triple-immunofluorescence staining, Cy2/Alexa Fluor 488–, Cy3-, and Cy5/Alexa Fluor 633–conjugated secondary antibodies and phalloidin (Jackson ImmunoResearch Laboratories, Inc.), as well as DAPI (Sigma-Aldrich), were used. Cells were mounted in 0.1 M Tris-HCl/glycerol (3:7) and 50 mg/ml N-propyl-gallate, pH 9.5.
Cryosections
For immunohistochemistry, tissues were cut on a cryostat (Jung CM1800; Leica) into 12-µm sections, mounted on poly-l-lysine–coated microscope slides, fixed with precooled acetone for 5 min at −20°C, and stained as described for the NRCs.
Human tissue
Human heart samples were obtained from the heart tissue bank set up by C. dos Remedios at the University of Sydney (Hospital Research Ethical Committee approval 09-2009-12146). Human materials were used in accordance with the ethical guidelines of King’s College London (College Research Ethical Committee 04/05-74) and the current UK law.
COS-1 and HeLa cells
COS-1 or HeLa cells were cultured in maintenance medium (10% fetal calf serum, 1% penicillin–streptomycin, and 4 mM glutamine in DME) at 37°C in 5% CO2. Cells were passaged at 70–80% confluency by standard trypsinization. For transfection, cells were plated onto 35-mm dishes and grown to 50–70% confluency. COS-1 and HeLa cells were transfected with 1 µg of plasmid DNA using transfection reagent (Escort IV; Sigma-Aldrich) or FuGENE HD (Roche), respectively. After 48 h, cells were fixed for immunocytochemistry as described for the NRCs or lysed with 200 mM NaCl, 10 mM Tris, 0.5% NP-40, 1 mM DTT, and protease inhibitor (Complete, Mini; Roche), pH 7.4, for pull-down assays and coimmunoprecipitations. For phosphorylation-sensitive experiments, 0.05 µM calyculin A, 0.2 mM NaVO4, and 30 mM sodium pyrophosphate were added to the lysis buffer.
Confocal microscopy
All confocal microscopy was performed at RT on a laser-scanning upright confocal microscope (LSM510; Carl Zeiss, Inc.) with solid-state (diode) laser excitation at 405 nm and emission at 420–480 nm, Ar laser excitation at 488 nm and emission at 505–530 nm, or HeNe laser excitation at 563 and 633 nm and emission at 560–615 nm and >650 nm using LCI Plan-Neofluar 25×/0.8 NA, Plan-Apochromat 63×/1.4 NA, or Plan-Neofluar 100×/1.3 NA oil immersion objectives.
Bacterial protein expression
Expression plasmids were transformed into the Escherichia coli expression strain BL21-RIL (EMD). Bacteria were incubated until the OD600 reached 0.6, and protein expression was induced with 0.4 mM IPTG. After 3 h, bacterial cells were harvested by centrifugation, and cell pellets were frozen in liquid nitrogen. Cells were lysed with lysozyme and DNase in 10 mM Tris, 200 mM NaCl, and 1 mM DTT, pH 7.4. GST fusion proteins were purified with glutathione Sepharose 4 fast flow beads (GE Healthcare) according to the manufacturer’s instructions.
GST pull-down assays and coimmunoprecipitations
The amount of GST fusion proteins bound to glutathione beads was normalized by SDS-PAGE and Coomassie staining. Equal amounts of fusion proteins were then added to lysates of COS-1 cells, previously transfected with GFP fusion constructs as described earlier. Unmodified GST peptides were used as a control. Samples were incubated for 90 min at 4°C on a shaking platform before beads were centrifuged and washed with PBS. Supernatant was collected in new tubes. SDS samples of beads and COS-1 lysates were prepared and applied to SDS-PAGE and immunoblotting as described previously (Lange et al., 2002). For coimmunoprecipitation assays, lysates were incubated with anti-HA or anti-GFP antibodies for 4 h at 4°C before addition of BSA-blocked protein G–Sepharose (Sigma-Aldrich) for 4 h. Beads were separated by centrifugation, washed with PBS, boiled in SDS buffer, and applied to SDS-PAGE and immunoblotting. For tissue immunoprecipitation and in cyto phosphorylation assays, the antibodies were covalently cross-linked to protein G–Sepharose using dimethylpimelimidate as a coupling agent. Samples were prepared as described earlier and applied to SDS-PAGE and immunoblotting. After incubation with the primary antibody, blots were incubated with immunoprecipitation detection reagent (Clean-Blot; Thermo Fisher Scientific) instead of the secondary antibody for detection of the bands with ECL.
CK2 phosphorylation assays and CK2 inhibitor experiments
For in vitro phosphorylation assays, 2.5 µg of purified GST fusion proteins were incubated with 50 U CK2 (New England Biolabs, Inc.) in 20 mM Tris, 50 mM KCl, 10 mM MgCl2, and 25 µM ATP, pH 7.5, for 30 min at 30°C and subjected to SDS-PAGE and immunoblotting. For in cyto experiments, COS-1 cells or NRCs were grown for 24 h and then treated with 10 µM of the CK2 Inhibitor DMAT (InSolution; Merck Chemicals) for 24 h before cell lysis or fixation and antibody staining as described earlier.
Yeast two-hybrid assay
FHOD3-FH2 (including or excluding the alternative T(D/E)5XE exon) was amplified by PCR and cloned into the NcoI and BamHI sites of a modified pLexA vector (provided by M. Gautel). Yeast two-hybrid screening procedures and β-galactosidase filter assays were performed as described by the manufacturer. The plasmid was transferred into Saccharomyces cerevisiae reporter strains (NMY51; Dualsystems), which were then transformed with an adult human cardiac cDNA library (Takara Bio Inc.). 105 primary transformants were screened by plating on selection plates lacking the amino acids histidine, tryptophan, and leucine. NMY51 clones were transferred to more restricted selection plates, additionally lacking adenine for further exclusion of false-positive interactions. Library plasmids from positive clones were isolated, transformed into E. coli XL1-blue for amplification of the plasmid, and sequenced. For further verification of the interaction, identified clones were retransformed into the yeast together with the empty pLexA plasmid or the corresponding FHOD3-pLexA constructs and grown on selection plates lacking histidine, tryptophane, and leucine and containing 3-AT in concentrations between 0.5 and 5 mM.
shRNA
Oligonucleotides for shRNA were designed to target 22 nucleotide sequences in the FH1 and FH2 domains of Rattus norvegicus FHOD3 with ends compatible to BglII and XhoI restriction sites. Oligos (FHOD3-shRNA forward, 5′-GATCCCCATAGACCAGTTGGAGAACAATATTCAAGAGATATTGTTCTCCAACTGGTCTATTTTTTGGAAC-3′, and reverse, 5′-TCGAGTTCCAAAAAATAGACCAGTTGGAGAACAATATCTCTTGAATATTGTTCTCCAACTGGTCTATGGG-3′; and control shRNA forward, 5′-GATCCCCAGAAGGTTCCAAAGGTTAAAGATTCAAGAGATCTTTAACCTTTGGAACCTTCTTTTTTGGAAC-3′, and reverse, 5′-TCGAGTTCCAAAAAAGAAGGTTCCAAAGGTTAAAGATCTCTTGAATCTTTAACCTTTGGAACCTTCTGGG-3′) were obtained from Sigma-Aldrich. 2 nmol of forward and reverse oligonucleotides were diluted to 50 µl of 30 mM Hepes, 100 mM potassium acetate, and 2 mM magnesium acetate, pH 7.4, incubated for 4 min at 98°C and 10 min at 70°C, and then slowly cooled down to 4°C. Annealed oligos were ligated into the BglII and XhoI restriction sites of a modified cDNA 3.1 vector, which was constructed by inserting the H1 expression cassette from pSUPER (Brummelkamp et al., 2002; donated by R. Agami, Netherlands Cancer Institute, Amsterdam, Netherlands), as well as an EGFP gene under the control of a cytomegalovirus promoter (Fukuzawa et al., 2008). Successful cloning was confirmed by restriction digest and sequencing. shRNA constructs were used to transiently transfect COS-1 cells and NRCs as described earlier.
Actin depolymerization
NRCs were cultured and transfected as described earlier. Cells were then grown for 5 d in long-term maintenance medium containing 1 µg/ml verapamil and then treated with 20 µM latrunculin B (VWR International) in long-term maintenance medium containing verapamil overnight. Latrunculin was washed out, and cells were incubated for 30 min or 2 h in maintenance medium containing verapamil before fixation and antibody staining as described earlier.
Cycloheximide
COS-1 cells were cultured and transfected as described earlier. 24 h after transfection, 1 µg/µl cycloheximide was added to the cells to inhibit protein synthesis. Cells were harvested at different time points between 0 and 72 h. Lysates were normalized by total protein content and applied to immunoblotting as described earlier.
Quantification of actin polymerization activity
HeLa cells were transfected as described earlier, changed to starvation medium (0.1% fetal calf serum, 1% penicillin–streptomycin, and 4 mM glutamine in DMEM) 24 h after transfection, and fixed as described earlier after culturing for another 24 h. Cells were stained and imaged as described earlier. Activation of actin polymerization was analyzed with CellProfiler Image Analysis software (Carpenter et al., 2006) essentially by measuring the phalloidin pixel intensity and calculating the ratio between transfected and untransfected cells. Presented values are the means between three separate experiments with 9–12 quantified images per construct per experiment.
Antibodies
For production of a polyclonal rabbit (pRb) anti-FHOD3 antibody, the N-terminal 339 amino acids of FHOD3 were amplified from human cDNA with 5′-GGAATTCCGCCACCATGGCCACGCTGGCTTGCC-3′ forward and 5′-CGGGATCCCGCCCGGCACCCACTGGG-3′ reverse primers and cloned into the EcoRI and BamHI restriction sites of a pET-3b (EMD) vector. The His6 fusion protein was expressed as described earlier and purified with 1 ml high performance columns (HisTrap; GE Healthcare) on an HPLC system (Äkta; GE Healthcare).
Purified His6 fusion proteins were dialyzed against PBS and concentrated to a final concentration of >2 µg/µl. Purity was tested by SDS-PAGE and Coomassie staining. The purified His6 fusion protein was sent for the immunization of rabbits to BioScience. Anti-FHOD3 immunsera were affinity purified with antigen-coupled Sepharose columns.
The monoclonal mouse anti–sarcomeric α-actinin (clone EA-53) antibody, as well as the pRb anti–all actin antibody, was obtained from Sigma-Aldrich. The monoclonal mouse anti-GFP (mixture of clones 7.1 and 13.1), as well as the monoclonal rat anti-HA antibody (clone 3F10), was purchased from Roche. The monoclonal mouse anti–phospho-threonine (clone 4H4; donated by G. Fruhwirth, King’s College London, London, England, UK) and the pRb LC3B antibodies were obtained from Cell Signaling Technology, and the monoclonal mouse anti-p62 antibody was obtained from Abcam. The monoclonal mouse anti–cardiac actin antibody (clone Ac1-20.4.2) was obtained from Progen. The polyclonal goat anti-GST antibody was obtained from GE Healthcare. The monoclonal mouse anti-myomesin (clone B4) was a gift from J.-C. Perriard’s Laboratory (Institute of Cell Biology, Swiss Federal Institute of Technology Zürich Hönggerberg, Zürich, Switzerland). The pRb antibodies anti–myosin-binding protein C and anti-p62 were gifts from M. Gautel. Horseradish peroxidase–conjugated anti–mouse Igs were purchased from Dako. Horseradish peroxidase–conjugated anti–rabbit and anti–goat Igs were purchased from EMD. Cy2-conjugated anti–rabbit Igs, Cy3-conjugated anti–mouse Igs, and Cy5-conjugated anti–rabbit and anti–mouse Igs were all purchased from Jackson ImmunoResearch Laboratories, Inc.
Image processing
Original digital images obtained either from the confocal microscope or by scanning of x-ray films using a scanner (Perfection 4870 Photo; Epson) were assembled to the figures and labeled using Photoshop or Illustrator (Adobe). Only linear contrast adjustments were used and were always applied to the entire image.
Online supplemental material
Fig. S1 shows a modified diagram of the gene structure of human (Homo sapiens), chimpanzee (Pan troglodytes), and mouse (M. musculus) FHOD3 as annotated on Ensembl. Fig. S2 shows that all FHOD3 isoforms colocalize with F-actin and affect actin polymerization to a comparable extent. Fig. S3 shows the characterization of the affinity-purified polyclonal antibody against FHOD3. Fig. S4 depicts that the muscle FHOD3 isoform shows increased stability in COS-1 cells compared with FHOD3 lacking the T(D/E)5XE exon and with mutants that cannot be phosphorylated. Fig. S5 shows that knockdown of FHOD3 in NRCs leads to myofibril disassembly. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201005060/DC1.
Acknowledgments
We are grateful to Prof. Mathias Gautel for pointing out the CK2 phosphorylation sites in the T(D/E)5XE exon, to Dr. Gilbert Fruhwirth and Prof. Jean-Claude Perriard for antibodies, and to Prof. Reuven Agami for providing pSUPER. We would like to thank Dr. Alexander Alexandrovich for help with protein purifications; Dr. Sagair Hussain, Dr. Katja Gehmlich, and Dr. Isabel Dominguez for helpful suggestions and discussions; and Dr. Friederike Cuello for ARCs. The help of Miss Nadine Gose (recipient of a fellowship by the Studienstiftung des Deutschen Volkes) is gratefully acknowledged.
T. Iskratsch is the recipient of a King’s College London Strategic Investment PhD Fellowship that was awarded to the Cardiovascular Division and a British Heart Foundation Research Excellence Centre Pump Priming fellowship. S. Lange is funded by a development grant from the Muscular Dystrophy Association. Work in the laboratory of E. Ehler was supported by a Medical Research Council Career Establishment Grant.
Footnotes
Abbreviations used in this paper:
- ARC
- adult rat cardiomyocyte
- DAD
- diaphanous autoregulation domain
- MLP
- muscle LIM protein
- NRC
- neonatal rat cardiomyocyte
- pRb
- polyclonal rabbit
- shRNA
- short hairpin RNA
- UBA
- ubiquitin associated
References
- Agarkova I., Auerbach D., Ehler E., Perriard J.-C. 2000. A novel marker for vertebrate embryonic heart, the EH-myomesin isoform. J. Biol. Chem. 275:10256–10264 10.1074/jbc.275.14.10256 [DOI] [PubMed] [Google Scholar]
- Ahuja P., Perriard E., Pedrazzini T., Satoh S., Perriard J.C., Ehler E. 2007. Re-expression of proteins involved in cytokinesis during cardiac hypertrophy. Exp. Cell Res. 313:1270–1283 10.1016/j.yexcr.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Arber S., Hunter J.J., Ross J.J., Jr, Hongo M., Sansig G., Borg J., Perriard J.-C., Chien K.R., Caroni P. 1997. MLP-deficient mice exhibit a disruption of cardiac cytoarchitectural organization, dilated cardiomyopathy, and heart failure. Cell. 88:393–403 10.1016/S0092-8674(00)81878-4 [DOI] [PubMed] [Google Scholar]
- Brummelkamp T.R., Bernards R., Agami R. 2002. A system for stable expression of short interfering RNAs in mammalian cells. Science. 296:550–553 10.1126/science.1068999 [DOI] [PubMed] [Google Scholar]
- Campellone K.G., Welch M.D. 2010. A nucleator arms race: cellular control of actin assembly. Nat. Rev. Mol. Cell Biol. 11:237–251 10.1038/nrm2867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter A.E., Jones T.R., Lamprecht M.R., Clarke C., Kang I.H., Friman O., Guertin D.A., Chang J.H., Lindquist R.A., Moffat J., et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7:R100 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chereau D., Boczkowska M., Skwarek-Maruszewska A., Fujiwara I., Hayes D.B., Rebowski G., Lappalainen P., Pollard T.D., Dominguez R. 2008. Leiomodin is an actin filament nucleator in muscle cells. Science. 320:239–243 10.1126/science.1155313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesarone M.A., Goode B.L. 2009. Actin nucleation and elongation factors: mechanisms and interplay. Curr. Opin. Cell Biol. 21:28–37 10.1016/j.ceb.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chhabra E.S., Higgs H.N. 2007. The many faces of actin: matching assembly factors with cellular structures. Nat. Cell Biol. 9:1110–1121 10.1038/ncb1007-1110 [DOI] [PubMed] [Google Scholar]
- Cooper T.A. 2005. Alternative splicing regulation impacts heart development. Cell. 120:1–2 10.1016/j.cell.2004.12.030 [DOI] [PubMed] [Google Scholar]
- Cox D.M., Du M., Marback M., Yang E.C., Chan J., Siu K.W., McDermott J.C. 2003. Phosphorylation motifs regulating the stability and function of myocyte enhancer factor 2A. J. Biol. Chem. 278:15297–15303 10.1074/jbc.M211312200 [DOI] [PubMed] [Google Scholar]
- Ehler E., Gautel M. 2008. The sarcomere and sarcomerogenesis. Adv. Exp. Med. Biol. 642:1–14 10.1007/978-0-387-84847-1_1 [DOI] [PubMed] [Google Scholar]
- Ehler E., Horowits R., Zuppinger C., Price R.L., Perriard E., Leu M., Caroni P., Sussman M., Eppenberger H.M., Perriard J.C. 2001. Alterations at the intercalated disk associated with the absence of muscle LIM protein. J. Cell Biol. 153:763–772 10.1083/jcb.153.4.763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehler E., Fowler V.M., Perriard J.C. 2004. Myofibrillogenesis in the developing chicken heart: role of actin isoforms and of the pointed end actin capping protein tropomodulin during thin filament assembly. Dev. Dyn. 229:745–755 10.1002/dvdy.10482 [DOI] [PubMed] [Google Scholar]
- Fukuzawa A., Lange S., Holt M., Vihola A., Carmignac V., Ferreiro A., Udd B., Gautel M. 2008. Interactions with titin and myomesin target obscurin and obscurin-like 1 to the M-band: implications for hereditary myopathies. J. Cell Sci. 121:1841–1851 10.1242/jcs.028019 [DOI] [PubMed] [Google Scholar]
- Gautel M. 2008. The sarcomere and the nucleus: functional links to hypertrophy, atrophy and sarcopenia. Adv. Exp. Med. Biol. 642:176–191 10.1007/978-0-387-84847-1_13 [DOI] [PubMed] [Google Scholar]
- Gauvin T.J., Fukui J., Peterson J.R., Higgs H.N. 2009. Isoform-selective chemical inhibition of mDia-mediated actin assembly. Biochemistry. 48:9327–9329 10.1021/bi901354z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha T., Wooten M.W. 2002. Structure and functional properties of the ubiquitin binding protein p62. FEBS Lett. 512:19–24 10.1016/S0014-5793(02)02286-X [DOI] [PubMed] [Google Scholar]
- Goode B.L., Eck M.J. 2007. Mechanism and function of formins in the control of actin assembly. Annu. Rev. Biochem. 76:593–627 10.1146/annurev.biochem.75.103004.142647 [DOI] [PubMed] [Google Scholar]
- Gregorio C.C., Weber A., Bondad M., Pennise C.R., Fowler V.M. 1995. Requirement of pointed-end capping by tropomodulin to maintain actin filament length in embryonic chick cardiac myocytes. Nature. 377:83–86 10.1038/377083a0 [DOI] [PubMed] [Google Scholar]
- Higgs H.N. 2005. Formin proteins: a domain-based approach. Trends Biochem. Sci. 30:342–353 10.1016/j.tibs.2005.04.014 [DOI] [PubMed] [Google Scholar]
- Huett A., Ng A., Cao Z., Kuballa P., Komatsu M., Daly M.J., Podolsky D.K., Xavier R.J. 2009. A novel hybrid yeast-human network analysis reveals an essential role for FNBP1L in antibacterial autophagy. J. Immunol. 182:4917–4930 10.4049/jimmunol.0803050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsotra A., Xiao X., Ward A.J., Castle J.C., Johnson J.M., Burge C.B., Cooper T.A. 2008. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl. Acad. Sci. USA. 105:20333–20338 10.1073/pnas.0809045105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaya H., Takeya R., Takeuchi K., Watanabe N., Jing N., Sumimoto H. 2005. Fhos2, a novel formin-related actin-organizing protein, probably associates with the nestin intermediate filament. Genes Cells. 10:665–678 10.1111/j.1365-2443.2005.00867.x [DOI] [PubMed] [Google Scholar]
- Katoh M., Katoh M. 2004. Identification and characterization of human FHOD3 gene in silico. Int. J. Mol. Med. 13:615–620 [PubMed] [Google Scholar]
- Ladd A.N., Stenberg M.G., Swanson M.S., Cooper T.A. 2005. Dynamic balance between activation and repression regulates pre-mRNA alternative splicing during heart development. Dev. Dyn. 233:783–793 10.1002/dvdy.20382 [DOI] [PubMed] [Google Scholar]
- Lammers M., Meyer S., Kühlmann D., Wittinghofer A. 2008. Specificity of interactions between mDia isoforms and Rho proteins. J. Biol. Chem. 283:35236–35246 10.1074/jbc.M805634200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange S., Auerbach D., McLoughlin P., Perriard E., Schäfer B.W., Perriard J.C., Ehler E. 2002. Subcellular targeting of metabolic enzymes to titin in heart muscle may be mediated by DRAL/FHL-2. J. Cell Sci. 115:4925–4936 10.1242/jcs.00181 [DOI] [PubMed] [Google Scholar]
- Littlefield R.S., Fowler V.M. 2008. Thin filament length regulation in striated muscle sarcomeres: pointed-end dynamics go beyond a nebulin ruler. Semin. Cell Dev. Biol. 19:511–519 10.1016/j.semcdb.2008.08.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlefield R., Almenar-Queralt A., Fowler V.M. 2001. Actin dynamics at pointed ends regulates thin filament length in striated muscle. Nat. Cell Biol. 3:544–551 10.1038/35078517 [DOI] [PubMed] [Google Scholar]
- MacLellan W.R., Schneider M.D. 2000. Genetic dissection of cardiac growth control pathways. Annu. Rev. Physiol. 62:289–319 10.1146/annurev.physiol.62.1.289 [DOI] [PubMed] [Google Scholar]
- Martin A.F. 1981. Turnover of cardiac troponin subunits. Kinetic evidence for a precursor pool of troponin-I. J. Biol. Chem. 256:964–968 [PubMed] [Google Scholar]
- Moscat J., Diaz-Meco M.T., Albert A., Campuzano S. 2006. Cell signaling and function organized by PB1 domain interactions. Mol. Cell. 23:631–640 10.1016/j.molcel.2006.08.002 [DOI] [PubMed] [Google Scholar]
- Moscat J., Diaz-Meco M.T., Wooten M.W. 2007. Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 32:95–100 10.1016/j.tibs.2006.12.002 [DOI] [PubMed] [Google Scholar]
- Paul A.S., Pollard T.D. 2009. Review of the mechanism of processive actin filament elongation by formins. Cell Motil. Cytoskeleton. 66:606–617 10.1002/cm.20379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razi M., Chan E.Y., Tooze S.A. 2009. Early endosomes and endosomal coatomer are required for autophagy. J. Cell Biol. 185:305–321 10.1083/jcb.200810098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten T.E., Stenmark H. 2010. p62, an autophagy hero or culprit? Nat. Cell Biol. 12:207–209 10.1038/ncb0310-207 [DOI] [PubMed] [Google Scholar]
- Seibenhener M.L., Geetha T., Wooten M.W. 2007. Sequestosome 1/p62—more than just a scaffold. FEBS Lett. 581:175–179 10.1016/j.febslet.2006.12.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada A., Nyitrai M., Vetter I.R., Kühlmann D., Bugyi B., Narumiya S., Geeves M.A., Wittinghofer A. 2004. The core FH2 domain of diaphanous-related formins is an elongated actin binding protein that inhibits polymerization. Mol. Cell. 13:511–522 10.1016/S1097-2765(04)00059-0 [DOI] [PubMed] [Google Scholar]
- Sparrow J.C., Schöck F. 2009. The initial steps of myofibril assembly: integrins pave the way. Nat. Rev. Mol. Cell Biol. 10:293–298 10.1038/nrm2634 [DOI] [PubMed] [Google Scholar]
- Taniguchi K., Takeya R., Suetsugu S., Kan-O M., Narusawa M., Shiose A., Tominaga R., Sumimoto H. 2009. Mammalian formin fhod3 regulates actin assembly and sarcomere organization in striated muscles. J. Biol. Chem. 284:29873–29881 10.1074/jbc.M109.059303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallar B.J., Alberts A.S. 2003. The formins: active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 13:435–446 10.1016/S0962-8924(03)00153-3 [DOI] [PubMed] [Google Scholar]
- Wen K.K., Rubenstein P.A. 2009. Differential regulation of actin polymerization and structure by yeast formin isoforms. J. Biol. Chem. 284:16776–16783 10.1074/jbc.M109.006981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Yang D., Ding J.H., Wang W., Chu P.H., Dalton N.D., Wang H.Y., Bermingham J.R., Jr, Ye Z., Liu F., et al. 2005. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 120:59–72 10.1016/j.cell.2004.11.036 [DOI] [PubMed] [Google Scholar]
- Xu Y., Moseley J.B., Sagot I., Poy F., Pellman D., Goode B.L., Eck M.J. 2004. Crystal structures of a Formin Homology-2 domain reveal a tethered dimer architecture. Cell. 116:711–723 10.1016/S0092-8674(04)00210-7 [DOI] [PubMed] [Google Scholar]
- Young K.G., Copeland J.W. 2010. Formins in cell signaling. Biochim. Biophys. Acta. 1803:183–190 10.1016/j.bbamcr.2008.09.017 [DOI] [PubMed] [Google Scholar]