Abstract
Detection of disease gene interaction effects among the enormous array of single nucleotide polymorphism (SNP) combinations represents the next frontier in genome-wide association (GWA) studies. Here we propose a novel strategy on the basis of the pattern and nature of the interaction, which can be classified as essential (EI) or removable (RI). We provide an analytical framework, including the qualitative conditions for screening EIs/RIs and a RI-to-EI likelihood ratio score to quantitatively measure the effect. In analyzing six GWA data sets, we find that the scores follow an exponential distribution, except in the upper 10−8 tail region in which the scores become irregular and unpredictable. Our approach is conceptually simple, computationally efficient and detects interactions that can be visualized and unequivocally interpreted.
Keywords: Genome-wide association study, gene-gene interaction, removable interaction, essential interaction, likelihood ratio statistics
1. INTRODUCTION
A growing number of successful genome-wide association (GWA) studies have been reported in the past several years [6]. All have initially focused on the main effects of individual single nucleotide polymorphisms (SNPs), which for the most part have been found to be relatively weak. Yet to be explored in this wealth of data are the interaction effects among SNPs, wherein the main effect of one SNP may be weak because that SNP confers risk only in the presence of one or more other SNPs. Taking these gene-gene interactions into account may lead to the discovery of stronger and more interesting effects [2, 8]. However, searching for meaningful interactions is a challenge for even the simplest case of examining all two-way interactions among a million SNPs (~half of 1012 combinations).
To aid in efficiency and interpretability of this formidable computational problem, we present an analytical framework for two types of interactions, removable interactions (RIs) and essential interactions [1, 12]. Interaction is usually defined as departure from additivity of effects on a specific outcome scale. If a monotone transformation exists that induces additivity, the interaction is called removable; otherwise, the interaction is essential.
What makes the mathematics hard is the sheer volume of networks in GWA data, for which no universal law underlying the system can be deduced. Our idea to detect interactions in GWA studies is based on distinct structures which can be visualized and epidemiological interpretations of the effects. We developed a qualitative tool that can rapidly screen specific interactions, essential or removable, and a likelihood score that quantifies the extent of the effect. We have identified empirical regularities in applying the proposed method to six GWA data sets with number of SNPs ranged from 100,000 to 1,000,000.
Under the simplest scenario of two SNPs (SNP1 and SNP2), each having two genotypic variants denoted by 0 and 1, respectively, we derived the necessary and sufficient conditions to use as a screening tool to quickly and reliably identify pair interactions as essential or removable. We then derived the EI-to-RI likelihood ratio (EI-RI score) to quantify the effects. In investigation of six existing GWA studies we find a consistent pattern of the EI-RI score distributions. Extension to scenarios with SNPs having three genotypes is straightforward, albeit involving tedious calculations.
2. METHODS
2.1 Essential/removable interactions
The joint risks of a pair of dichotomous markers can be graphically represented by two lines and are depicted in Fig. 1. We use the OR and log(OR) risk scales; however the results presented can be generalized to any risk scale. Figure 1 illustrates that all two-SNP combinations can be classified into one of three mutually-exclusive categories:
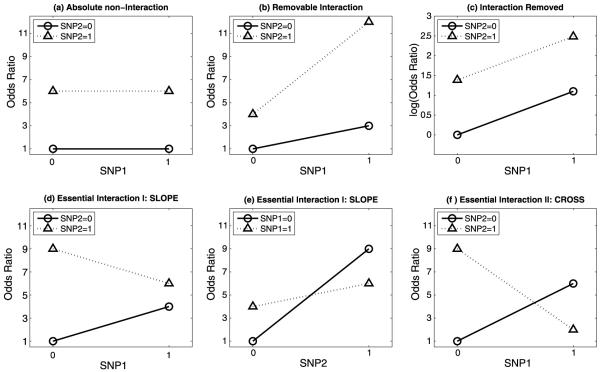
Figure 1.
Graphical representations of absolute non-interaction, removable interaction, and essential interaction. (a) Absolute non-interaction (ANI): in the presence of a given genotypic variant of SNP2, risk does not vary by genotypic variant of SNP1 (two lines are parallel with zero slope). This pattern represents ANI because no monotone transformation can force the two lines to not be parallel. (b) There is an interaction (non-intersecting, non-parallel lines with slopes of the same direction) between two SNPs on the OR risk scale, and (c) the interaction is removed (parallel lines) by transformation to the log (OR) scale. In this instance, a monotone transformation made the two lines parallel. (b) and (c) correspond to the example illustrated in Table 1. (d) and (e) The same essential interaction (EISLOPE) plotted in two different ways, (d) with SNP1 represented on the x-axis, in which the two lines have slopes of opposite directions but do not intersect and (e) with SNP2 represented on the x axis, in which the two lines have slopes of the same direction, and intersect. The patterns shown in (d) and (e) are equivalent. (f) Essential interaction EICROSS, in which the two lines have slopes of opposite direction and intersect. (d), (e), and (f) represent EIs because no monotone transformation can make the two lines parallel under any of these conditions.
Absolute non-interaction (ANI) (Fig. 1a): No interaction between SNP1 and SNP2 means that the effect of SNP1+SNP2 is the sum of the individual effects (i.e., effects are additive). ANI is manifest by a two-SNP combination that does not produce an interaction under any risk scale. This phenomenon occurs when in the presence of a given genotypic variant of SNP2, risk does not vary by genotypic variant of SNP1.
Removable interaction (RI), Fig. 1b–c, is manifest by a two-SNP combination that produces an interaction under at least one risk scale (Fig. 1b), and does not produce an interaction under at least one other risk scale (Fig. 1c). If an RI exists, the direction, positive or negative, of the risk difference for one SNP is not affected by the other SNP (and is not zero). However, under a risk scale in which the RI manifests itself as an interaction, the magnitude of the effect of SNP1 varies by SNP2 genotypic stratum, and vice versa. That is, with an RI, the magnitude, but not the direction, of the effect for one SNP may be modified by the other SNP. An example of an RI is given in Table 1.
Table 1.
An example of a removable interaction (RI). The interaction between two SNPs (SNP1 and SNP2) is significant under the odds ratio (OR) scale, but is removed under the log(OR) scale. This example is graphically illustrated in Fig. 1b–c
| SNP1 | SNP2 | # of cases | # of controls | Odds ratio(OR) | Log(OR) |
|---|---|---|---|---|---|
| 0 | 0 | 100 | 100 | 1.0 | 0.0 |
| 0 | 1 | 300 | 100 | 3.0 | 1.099 |
| 1 | 0 | 400 | 100 | 4.0 | 1.386 |
| 1 | 1 | 1200 | 100 | 12.0 | 2.485 |
| Expected risk of “11” with no interaction | 6.0 | 2.485 | |||
| Estimation of interaction | 6.0 | 0.0 | |||
| F-Test of interaction | P = 0.0001 | P = 1.0 |
Essential interaction (EI), Fig. 1d-f, is manifest by a two SNP combination that produces an interaction under all risk scales: In contrast to RI, an EI exists when the direction of the effect of one SNP is dependent on the genotypic variant of the other SNP. That is, the effect of one SNP is reversed by the other SNP. And as in RI, for an EI the magnitude of the effect for one SNP may also be modified by the other SNP. EI can be further classified into two subclasses, EISLOPE and EICROSS. EISLOPE occurs when the reversal of the direction of effect is one way (e.g., SNP1 by SNP2 but not SNP2 by SNP1) (Fig. 1d–e), while EICROSS occurs when the reversal of the direction of effect is reciprocal (SNP1 by SNP2 and SNP2 by SNP1) (Fig. 1f).
From the above reasoning we can derive a set of relationships in terms of magnitudes and directions of the effect change of one SNP by the other:
Condition I (slopes have opposite direction): (OR10 − OR00)(OR11 − OR01) ≤ 0 in which one risk difference in the product can equal zero, but not both.
Condition II (lines intersect): (OR01 − OR00)(OR11 − OR10) ≤ 0 in which one risk difference, but not both, can equal zero.
An interaction is essential if and only if Condition I and/or II holds. If only one of these conditions holds, we have an EISLOPE; if both conditions hold, we have an EICROSS. If both conditions are not satisfied, then we have RI (unless both product terms in Condition I or Condition II are zeros, in which case we have ANI).
2.2 An EI-RI score: a quantitative measure for the effect
Since the Conditions I-II are invariant under any monotone transformation, ORab in two conditions could be replaced by its logarithm, the log odds ratio of genotypic variants combination ab with respect to baseline 00. Based on the Condition I and/or Condition II, the necessary and sufficient(N&S) condition for EIs, a statistical test can be constructed for the null hypothesis that no EI exists. By replacing ORs with their logarithms, the testing hypothesis depends only on log-odds ratios. The standard theory [11, 13, 14] assures that the likelihood ratio test for such kind of hypothesis based on the prospective likelihood function would be valid. The prospective likelihood function is proportional to
| (1) |
where nab and mab respectively are observed numbers of cases and controls for joint genotypic variant ab, pab = eθab /(1 + eθab) is the corresponding affected probability and θab represents the log odd of the joint genotypic variant ab. Let θ = (θ00, θ01, θ10, θ11) be the vector of parameters. The null hypothesis parameter space, the complement of the subset for parameters satisfying the N&S condition, is not closed. We will use instead its closure, i.e.,
| (2) |
as a working null parameter space. By doing so, we can simplify the computation and the test will be slightly conservative.
We took the EI-RI score to be the likelihood ratio statistics [7], i.e.
| (3) |
where ϴ = {θ : −∞ ≤ θ00, θ01, θ10, θ11 < is the uncon-strained parameter space.
Obviously, the unconstrained MLE (θ ∈ ϴ) has a closed form, namely θab = log(nab/mab). To obtain the EI-RI score, one needs to calculate the constrained MLE under the null hypothesis, which can be solved by a non-linear programming (NLP) algorithm. However, the NLP algorithm is time-consuming and could break down especially when the constrained MLE is on the boundary of ϴ0. Therefore, the NLP algorithm is not suitable for GWA study.
By virtue of the concavity of the log likelihood function, we developed an efficient algorithm for calculating the constrained MLE. Details of the algorithm are given in Appendix A.
2.3 EI-RI scores in six GWA data sets
Under the null hypothesis of ANI or RI, the null distribution of the EI-RI score is mathematically intractable as it is a mixture of zero (when the maximum point belongs to the null parameter space) and a positive distribution (when the constrained MLE does not belong to the null parameter space) with the component weights unknown. Estimation of p-values by the permutation test fails because the conventional permutation procedure is not suitable for our composite null hypothesis. In fact, randomly allocating the samples under the assumption of no joint effects or marginal effects as in the conventional permutation approach inevitably introduces false positives. And indeed, serious inflated type I error rates by the permutation test have been observed in our simulation studies (data not shown here).
Re-sampling approaches [4] might be applicable for estimating p-values with small numbers of SNP markers, but it is beyond computational capability for GWA data with large numbers of SNP pairs. We therefore sought to examine the empirical distribution patterns of the scores among six GWA data sets. All empirical distributions of EI-RI scores in six data sets give rise to similar upper tails, which, in turn, may justify the existence of a universal threshold for declaring the significance of an EI.
3. RESULTS FOR EMPIRICAL STUDY
Descriptive statistics of six GWA data sets are given in Table 2. The numbers of SNPs in these studies ranged from 105 to 106. We dichotomized each SNP genotype based on the empirical mode of inheritance [9]. In the six data sets examined, about half (48.83% on average) of the SNP pairs were in the category of EIs (Fig. 1d–f). Of the EI pairs, about two-thirds (65.97% on average) were EISLOPE pairs and one-third (34.03%) were EICROSS pairs.
Table 2.
Descriptive information of six GWA data sets
| GWA1_100K | GWA2_100K | GWA3_300K | GWA4_300K | GWA5_300K | GWA6_1.8m | |
|---|---|---|---|---|---|---|
| Sample information | ||||||
|
| ||||||
| Number of samples | 500 | 441 | 541 | 183 | 268 | 292 |
| Number of cases | 354 | 313 | 270 | 92 | 176 | 175 |
| Number of controls | 146 | 128 | 271 | 91 | 92 | 117 |
|
| ||||||
| SNP information | ||||||
|
| ||||||
| Number of SNPs | 105834 | 109924 | 317503 | 317503 | 317503 | 909622 |
| Number of SNPS on sex chromosome | 0 | 0 | 9173 | 9173 | 9173 | 37380 |
| SNPs with call rate less than 95% | 1584 | 1018 | 3101 | 7235 | 28124 | 131770 |
| SNPs with minor allele frequency (MAF) < 0.01 |
5005 | 10642 | 252 | 30970 | 45173 | 39408 |
| SNPs with no polymorphism observed | 1810 | 2150 | 91 | 20540 | 31852 | 66 |
| SNPs with only heterozygotes observed | 0 | 2 | 0 | 0 | 113 | 2046 |
| Polymorphic SNPs with no heterozygote observed |
25 | 94 | 5 | 139 | 358 | 130957 |
| SNPs with HWE χ2 > 50 in case group | 26 | 205 | 9126 | 396 | 10084 | 6511 |
| SNPs with HWE χ2 > 50 in control group | 330 | 666 | 7741 | 596 | 2117 | 4555 |
| SNPs with HWE χ2 > 50 in combined samples |
752 | 1112 | 9265 | 5443 | 12160 | 10023 |
|
| ||||||
| SNP QC Results Summary: | ||||||
|
| ||||||
| Number of autosomal chromosome Markers | 105834 | 109924 | 308330 | 308330 | 308330 | 872242 |
| Autosomal SNPs Passing Call Rate/MAF | 100610 | 96260 | 304342 | 269795 | 256811 | 606999 |
| Threshold (0.01 ≤ MAF < 0.05 and call rate ≥ 0.99) or (0.05 ≤ MAF < 0.10 and call rate 0.97) or (0.10 ≤ MAF and call rate ≥ 0.95) |
||||||
| Pass above, no polymorphism observed | 0 | 0 | 0 | 0 | 0 | 0 |
| Pass above, only heterozygotes observed | 0 | 2 | 0 | 0 | 0 | 6 |
| Pass above, no heterozygote observed | 0 | 0 | 0 | 2 | 100 | 0 |
| Pass above, fail HWE test (χ2 > 50 in case group or χ2 > 50 in control group) |
76 | 191 | 8 | 28 | 3443 | 1303 |
| Number of usable SNPs | 100534 | 96067 | 304334 | 269765 | 253268 | 605690 |
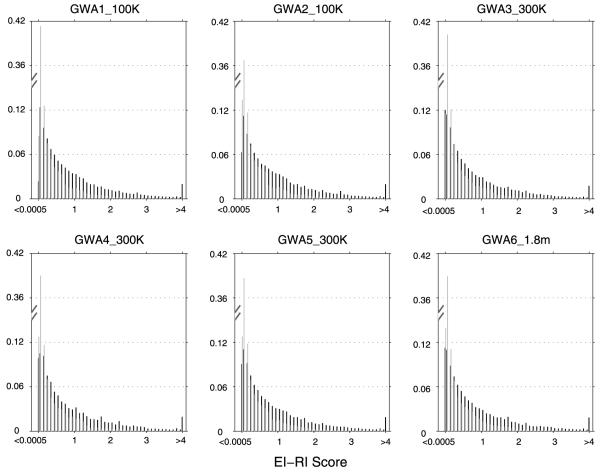
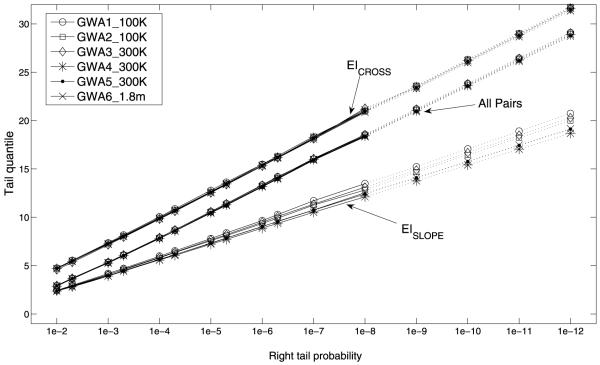
Histogram plots are drawn for EI-RI scores of the six data sets and are shown in Fig. 2a. Comparing histogram plots of the two EI groups, EICROSS pairs tend to have higher EI-RI scores than EISLOPE pairs. Patterns of histogram plots are consistent across six data sets, particularly, when EI-RI scores become large. This consistency implies that the empirical distributions of EI pairs in the six data sets have similar tail behavior. This conclusion was confirmed by the consistency of tail quantiles for EI-RI scores, as shown in Fig. 2c. The consistent tail distribution of EI-RI scores in six data sets may justify the existence of a universal critical value for declaring the significance of an EI.
Figure 2a.
The histogram plots of EI-RI scores in six data sets. Dark black is the histogram for EICROSS pairs and gray is the histogram for EISLOPE pairs. For each type of EI pair, the leftmost column is the frequency for EI-RI scores less than 0.0005 and the rightmost column is the frequency for EI-RI scores larger than 4.
Figure 2c.
The tail quantiles of the EI-RI scores stratified by EICROSS pairs and EISLOPE pairs in the six GWA data sets based on the real data (solid lines) and the extrapolated tail quantiles from an estimated exponential distribution (dotted lines).
In GWA studies, the critical values are chosen as the (1-p)-quantiles of EI-RI scores under the null hypothesis for an extremely small p. One way to estimate the extreme quantiles is to inverse the linear interpolation of the empirical cumulative distribution function [10]. We found that the empirical (1-p)-quantiles are consistent across the six data sets for 10−8 ≤ p ≤ 10−2 (Fig. 2c, Table 3). However, in the upper 10−8 tail region the quantiles became irregular and unpredictable, which is due to random error of the extreme value distribution of p-values. Therefore, the interpolation method is not suitable for estimation of extreme quantiles. To overcome this problem, we approximated extreme quantiles using much lower quantiles which can be estimated by intermediate order statistics.
Table 3.
Right tail quantiles of the EI scores in the six GWA data sets. Quantiles between 10−8 and 10−2 were estimated directly from the data, outside that range were estimated by extrapolation
| Tail probability | GWA1_100K | GWA2_100K | GWA3_300K | GWA4_300K | GWA5_300K | GWA6_1.8m |
|---|---|---|---|---|---|---|
| 1.0E-02 | 2.9360 | 2.9277 | 2.9534 | 2.8668 | 2.9263 | 2.9106 |
| 5.0E-03 | 3.6737 | 3.6716 | 3.7104 | 3.6421 | 3.6652 | 3.6160 |
| 1.0E-03 | 5.3524 | 5.3309 | 5.3555 | 5.2672 | 5.3139 | 5.3129 |
| 5.0E-04 | 6.1056 | 6.0776 | 6.1174 | 6.0195 | 6.0526 | 6.0277 |
| 1.0E-04 | 7.9038 | 7.8264 | 7.9108 | 7.7986 | 7.8368 | 7.8085 |
| 5.0E-05 | 8.7071 | 8.6172 | 8.7007 | 8.5751 | 8.6036 | 8.5792 |
| 1.0E-05 | 10.5998 | 10.5225 | 10.5844 | 10.4760 | 10.4865 | 10.4403 |
| 5.0E-06 | 11.4246 | 11.3137 | 11.4054 | 11.2845 | 11.2738 | 11.2374 |
| 1.0E-06 | 13.3533 | 13.2277 | 13.3188 | 13.1858 | 13.1961 | 13.1545 |
| 5.0E-07 | 14.1986 | 14.0553 | 14.1349 | 14.0286 | 14.0091 | 13.9621 |
| 1.0E-07 | 16.0027 | 16.0606 | 16.0798 | 15.9701 | 15.9223 | 15.9039 |
| 1.0E-08 | 18.5659 | 18.4649 | 18.5488 | 18.4146 | 18.3703 | 18.3299 |
| 1.0E-09 | 21.1983 | 21.0851 | 21.1762 | 21.0338 | 20.9728 | 20.9296 |
| 1.0E-10 | 23.830 | 23.7053 | 23.8035 | 23.6531 | 23.5754 | 23.5293 |
| 1.0E-11 | 26.4631 | 26.3254 | 26.4308 | 26.2723 | 26.1779 | 26.1291 |
| 1.0E-12 | 29.0955 | 28.9456 | 29.0582 | 28.8915 | 28.7804 | 28.7288 |
For each empirical distribution of the six data sets, the relationship between the (1 – p) quantile and log(p) was approximately linear in the range 10−8 ≤ p ≤ 10−2 (Fig. 2c). The linearity implies that extreme value indices [5] are zero for all six empirical distributions. The conclusion was confirmed by constructing 95% confidence intervals of the extreme value index with different numbers of upper orderstatistics (for details see Appendix B). The value zero belongs to almost all these confidence intervals, which does not contradict the hypothesis that the extreme value index is zero.
With a zero extreme value index, the (1 − p)-quantile for small p can be approximated by
| (4) |
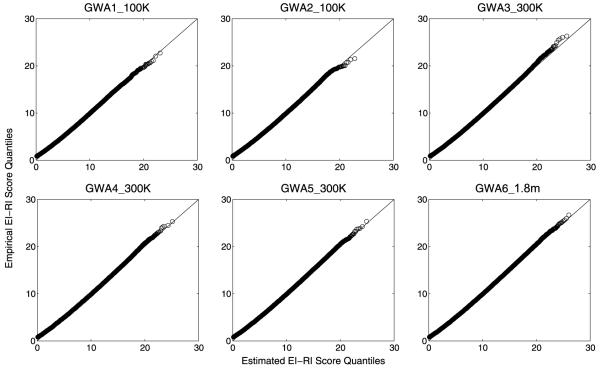
for some μ and σ(σ > 0). That is, the upper tail of the EI-RI score approximately follows an exponential distribution with location parameter σ and scale parameter σ.The two parameters were estimated by fitting the regression line defined in (4) for p in the range [10−8, 10−2]. As shown in Table 4, all R-square statistics are close to 1, which indicates that regression lines defined in (4) almost perfectly fit the data (also see quantile-quantile plots in Fig. 2b).
Table 4.
Estimation for intercept μ and slope σ by fitting (1 – p)-empirical quantile vs −log(p) for 10−8 ≤ p ≤ 10−2
| GWA1_100K | GWA2_100K | GWA3_300K | GWA4_300K | GWA5_300K | GWA6_1.8m | |
|---|---|---|---|---|---|---|
| μ | −2.493 | −2.492 | −2.475 | −2.519 | −2.450 | −2.468 |
| σ | 2.632 | 2.620 | 2.627 | 2.619 | 2.603 | 2.600 |
| R-square | 0.9994 | 0.9991 | 0.9993 | 0.9992 | 0.9992 | 0.9992 |
Figure 2b.
Quantile-quantile plots of EI-RI scores for six GWA data sets. Each point corresponds to a probability p: the x-coordinate represents the EI-RI score corresponding to the p-th quantile of the exponential distribution (details in text) and the y-coordinate represents the EI-RI score corresponding to the p-th sample quantile from the empirical data.
Furthermore, estimators of the two parameters are consistent across all six data sets. This, again, suggests that distributions of EI-RI scores have similar tail behavior.
Based on these data and a reasonable underlying assumption of an exponential tail distribution for EI-RI scores under the null hypothesis, we can deduce a simple proposition: the EI-RI score corresponding to the (1 − p)-quantile is approximately equal to (−2.617) × log(p) – 2.483, for small p, and equivalently, the p-value is approximately equal to exp{−(EI-RI score + 2.483)/2.617}, for large EI-RI scores.
It will be interesting to see whether a similar trend of the scores will hold and the above estimated p-values can be generalized for any GWA study. If so, this in turn may provide a simple guideline of choosing significance levels for SNP pairs worthy of further investigation such as replication studies. For example, here we postulate an EI-RI score less than 18 indicates an inconclusive interaction; a score between 18 and 26 suggests a possibility of a true EI; and a score greater than 26 indicates a statistically significant EI. These speculations warrant further investigation.
4. DISCUSSION
Detection of interaction effects among SNPs represents the next frontier in GWA studies. However, screening the enormous array of SNP combinations represents a daunting task. To begin to address this challenge, we classified all two-SNP combinations into three mutually-exclusive categories, absolute non-interactions, removable interactions, and essential interactions, and we characterized the conditions for each of these categories. In essence, an interaction is removable when the magnitude, but not the direction, of the effect of one SNP is modified by the other SNP, and an interaction is essential when the direction of the effect for at least one of the SNPs is altered in the presence of the other SNP (and the magnitude may be modified as well).
We developed an efficient computer program to screen for EIs and to calculate the EI-RI scores (likelihood ratio statistics) for each SNP pair in GWA data of any number of SNPs. With this algorithm we examined the distributions of EI-RI scores in six GWA data sets with 105 to 106 SNPs. A consistent pattern emerged, which allowed us to derive the tail quantiles by extrapolation from the stable portion of the empirical distributions. Finally, we suggested an EI-RI score threshold for identifying prime candidate EIs.
The EI-RI score presented here emphasizes EIs because they are not dependent on risk scale and represent the most extreme form of interaction (reversal of direction of risk difference). Furthermore, while one may reasonably screen for RIs solely among SNPs that exhibit significant main effects, this approach may not work for EIs, especially for EICROSS. This is because in the presence of an RI, both SNPs have non-zero marginal effects (of course, the magnitude of the effects depends on the joint effects and MAFs). With EI, however, the marginal effect for at least one SNP can be close to zero even when the EI effect is substantial. With EICROSS the marginal effect for both SNPs can be close to zero. Thus, many EIs may be missed if only SNPs with main effects are screened. This conjecture requires further investigation.
Even though RIs have the undesirable mathematical property of dependence on the risk scale, it is likely that many RIs in GWA data sets will prove to be potentially interesting, as they have proved to be in traditional case-control studies. It will be straightforward to apply the methods presented here, which are independent of risk scale, to screen for RIs as well as EIs.
Taken together, the qualitative and the quantitative methods we proposed to detect interaction effects in GWA data by way of EIs and RIs is conceptually simple, computationally manageable and, most of all, the resulting interactions can be readily visualized and unequivocally interpreted. We plan to determine the practical utility of our current methodology to address real disease problems.
All algorithms have been implemented in C++ and the program is available upon request from authors.
ACKNOWLEDGEMENTS
The research was partially supported by NIH/NEI grants (R01 EY015771, R21 EY018127), NIH/NHGRI grant (K25 HG000060), NIH grant (5-R37-GM047845-18), NSF grant (DMS-05-04871), NSFC grants (10701067, 10671189), and grants provided by Verto Institute, American Health Alliance Foundation, Macular Vision Research Foundation, and Ellison Medical Researching Foundation. This work was carried out while Zhang was a postdoctoral associate and Yang was a visiting scholar in Hoh’s Lab at Yale. We gratefully acknowledge Yale University Biomedical High Performance Computing Center and NIH grant: RR19895, which funded the instrumentation. Our thanks also extend to the database of Genotypes and Phenotypes (dbGaP) for the GWA data sets. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors would like to thank the editor for his comments which improved this article.
APPENDIX A. ALGORITHM FOR CONSTRAINED MLE
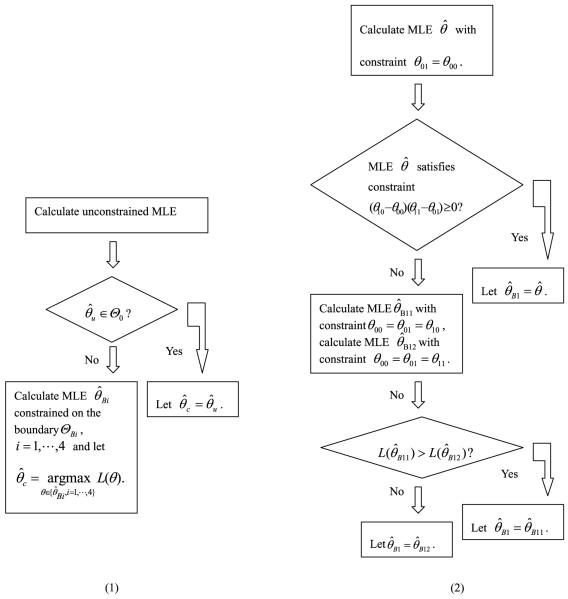
If the unconstrained MLE belongs to ϴ0, then it is of course the constrained MLE. If the constrained MLE is an interior point of ϴ0, it must be the unconstrained MLE since the log likelihood function is concave so that there are no local maxima other than the global maxima. Therefore, if the unconstrained MLE does not belong to ϴ0, the constrained MLE must be on its boundary ϴB. This leads to the algorithm for obtaining the constrained MLE, which is illustrated in Fig. A1.
Figure A.
Algorithm for calculating MLE constrained on the null hypothesis. (1) Calculation of constrained MLE . (2) Calculation of constrained MLE on boundary ϴB1.
The boundary ϴB is the union of four subboundaries: , , and . We can calculate the MLEs on four sub-boundaries separately and the one with largest likelihood function value is the constrained MLE on boundary ϴB. We also develop an algorithm for obtaining the MLE on sub-boundary ϴB1(the other three constrained MLEs can be similarly obtained). The algorithm is graphically displayed in Fig. A2.
The algorithm is very easy to implement and is time efficient since there are closed forms for these constrained MLEs that need to be calculated in the algorithm. For example, under constraint θ01 = θ00, the resulting MLE of θ is (log[(n00 + n01)/(m00 + m01)], log[(n00 + n01)/(m00 + m01)], log[n10/m10], log[n11/m11]). Under constraint θ00 = θ01 = θ10, MLEs of θijs are θ00 = θ01 = θ10 = log[(n00 + n01 + n10)/(m00 + m01 + m10)] and θ11 = log[n11/m11]. Similarly, under constraint θ00 = θ01 = θ11, the MLE of θ00 = θ01 = θ11 is log[(n00 + n01 + n11)/(m00 + m01 + m11)] and the MLE of θ10 is log[n10/m10].
APPENDIX B. MOMENT ESTIMATION AND CONFIDENCE INTERVAL FOR EXTREME VALUE INDEX
The key issue in extreme value theory is the estimation of the extreme value index c, which governs the tail behavior of a distribution function. The Hill estimator is the most popular estimator which is restricted to the case c > 0. The moment estimator , an adaptation of the Hill estimator, is a consistent estimator for all real value c’s. Let X1,n ≤ X2, n ≤ ⋯ ≤ Xn,n be the order statistics for sample X1, … , Xn, the moment estimator of c is given by
with for j = 1,2.
Under some regular conditions, asymptotically follows a zero mean normal distribution with variance [3]
An approximate (1 – α)100% confidence interval is then given by
where is the asymptotic variance with c being replaced by its moment estimator, and Zα/2 is the (1 − α/2)-quantile of the standard normal distribution.
Table A gives the 95% asymptotic confidence intervals for various values of k. The value zero belongs to almost all these confidence intervals, which does not contradict the hypothesis that the extreme value index is zero.
Table A.
Moment estimator and the 95% asymptotic confidence interval for extreme value index c with different k’s, where k is the number of upper order statistics used in the moment estimator
| k | Moment Estimation |
95% Confidence Interval |
Moment Estimation |
95% Confidence Interval |
Moment Estimation |
95% Confidence Interval |
|||
|---|---|---|---|---|---|---|---|---|---|
| Lower | Upper | Lower | Upper | Lower | Upper | ||||
| GWA1_100K | GWA2_100K | GWA3_300K | |||||||
| 50 | −0.0592 | −0.3266 | 0.2082 | −0.035 | −0.3050 | 0.2349 | 0.0789 | −0.1991 | 0.3570 |
| 100 | −0.0570 | −0.2462 | 0.1322 | −0.2421 | −0.4453 | −0.0390 | −0.0275 | −0.2192 | 0.1642 |
| 200 | −0.0503 | −0.1843 | 0.0838 | −0.2524 | −0.3972 | −0.1075 | −0.0150 | −0.1517 | 0.1218 |
| 300 | −0.0420 | −0.1519 | 0.0678 | −0.2184 | −0.3335 | −0.1033 | −0.0825 | −0.1913 | 0.0263 |
| 400 | 0.0186 | −0.0794 | 0.1166 | −0.1098 | −0.2042 | −0.0154 | 0.0072 | −0.0908 | 0.1052 |
| 500 | 0.0542 | −0.0335 | 0.1420 | −0.096 | −0.1803 | −0.0117 | 0.0001 | −0.0875 | 0.0878 |
| 1000 | −0.0044 | −0.0662 | 0.0573 | −0.0296 | −0.0902 | 0.0309 | 0.0129 | −0.0491 | 0.0749 |
| 2000 | 0.0066 | −0.0372 | 0.0504 | −0.0199 | −0.0629 | 0.0232 | 0.0528 | 0.0089 | 0.0967 |
| 3000 | −0.0194 | −0.0546 | 0.0158 | −0.0021 | −0.0378 | 0.0336 | 0.0113 | −0.0245 | 0.0471 |
| 4000 | −0.0391 | −0.0692 | −0.0090 | 0.0341 | 0.0031 | 0.0651 | 0.0396 | 0.0086 | 0.0706 |
| 5000 | −0.0131 | −0.0405 | 0.0143 | −0.0021 | −0.0297 | 0.0256 | 0.0347 | 0.007 | 0.0624 |
| 10000 | −0.0345 | −0.0536 | −0.0155 | 0.0283 | 0.0087 | 0.0479 | 0.0472 | 0.0276 | 0.0668 |
|
| |||||||||
| GWA4_300K | GWA5_300K | GWA6_1.8m | |||||||
|
| |||||||||
| 50 | −0.0833 | 0.1949 | 0.3614 | −0.0858 | 0.1924 | 0.3640 | 0.0349 | −0.2424 | 0.3123 |
| 100 | 0.013 | −0.1830 | 0.2091 | −0.0255 | −0.2174 | 0.1665 | −0.0255 | −0.2175 | 0.1664 |
| 200 | −0.0827 | −0.2159 | 0.0506 | −0.0939 | −0.2271 | 0.0394 | −0.1262 | −0.2602 | 0.0079 |
| 300 | −0.0561 | −0.1654 | 0.0531 | −0.0531 | −0.1625 | 0.0562 | −0.0848 | −0.1935 | 0.0240 |
| 400 | −0.0568 | −0.1514 | 0.0378 | −0.0383 | −0.1336 | 0.057 | −0.0559 | −0.1505 | 0.0387 |
| 500 | −0.0962 | −0.1805 | −0.0119 | −0.0179 | −0.1042 | 0.0684 | −0.0656 | −0.1500 | 0.0188 |
| 1000 | −0.0352 | −0.0956 | 0.0252 | −0.0218 | −0.0827 | 0.0390 | −0.0107 | −0.0721 | 0.0507 |
| 2000 | −0.002 | −0.0457 | 0.0418 | −0.0061 | −0.0497 | 0.0375 | −0.0217 | −0.0647 | 0.0214 |
| 3000 | 0.0107 | −0.0251 | 0.0465 | −0.0227 | −0.0578 | 0.0124 | 0.0067 | −0.0291 | 0.0425 |
| 4000 | 0.0237 | −0.0073 | 0.0547 | 0.0037 | −0.0273 | 0.0347 | 0.0039 | −0.0271 | 0.0349 |
| 5000 | 0.0208 | −0.0069 | 0.0485 | 0.0208 | −0.0069 | 0.0485 | −0.0008 | −0.0285 | 0.0269 |
| 10000 | 0.0126 | −0.0070 | 0.0322 | 0.0018 | −0.0178 | 0.0214 | 0.0014 | −0.0182 | 0.0210 |
Contributor Information
Chengqing Wu, Yale School of Public Health, New Haven, CT, chengqing.wu@yale.edu.
Hong Zhang, Department of Statistics and Finance, University of Science and Technology of China, Hefei, Anhui, P. R. China, zhangh@ustc.edu.cn.
Xiangtao Liu, Department of Applied Mathematics, Yale University, New Haven, CT, xiangtao.liu@yale.edu.
Andrew DeWan, Yale School of Public Health, New Haven, CT, andrew.dewan@yale.edu.
Robert Dubrow, Yale School of Public Health, New Haven, CT, robert.dubrow@yale.edu.
Zhiliang Ying, Department of Statistics, Columbia University, New York, NY, zying@stat.columbia.edu.
Yaning Yang, Department of Statistics and Finance, University of Science and Technology of China, Hefei, Anhui, P. R. China, ynyang@ustc.edu.cn.
Josephine Hoh, Yale School of Public Health, New Haven, CT, josephine.hoh@yale.edu.
REFERENCES
- [1].Breslow NE, Day NE. Statistical Methods in Cancer Research. Volume I - The Analysis of Case-Control Studies. IARC Sci. Publ.; Lyon, France: 1980. [PubMed] [Google Scholar]
- [2].Chen X, Liu CT, Zhang M, Zhang H. A forest-based approach to identifying gene and gene gene interactions. Proc Natl Acad Sci. 2007;104:19199–19203. doi: 10.1073/pnas.0709868104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Dekkers ALM, Einmahl JHJ, Haan LD. A moment estimator for the index of an extreme-value distribution. The Annals of Statistics. 1989;17:1833–1855. MR1026315. [Google Scholar]
- [4].Efron B. The Jackknife, the Bootstrap and Other Resampling Plans. SIAM; Philadelphia: 1982. MR0659849. [Google Scholar]
- [5].Haan LD, Ferreira A. Extreme Value Theory: An Introduction. Springer; New York, London: 2006. MR2234156. [Google Scholar]
- [6].Hindorff L, Junkins H, Manolio T. A Catalog of Published Genome-Wide Association Studies. 2009 Available at: www.genome.gov/26525384.
- [7].Lehmann EL, Romano JP. Testing Statistical Hypotheses. Springer; New York: 2005. MR2135927. [Google Scholar]
- [8].Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37:413–417. doi: 10.1038/ng1537. [DOI] [PubMed] [Google Scholar]
- [9].Matthews AG, Haynes C, Liu C, Ott J. Collapsing SNP genotypes in case-control genome-wide association studies increases the type I error rate and power. Stat Appl Genet Mol Biol. 2008;7 doi: 10.2202/1544-6115.1325. Article23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Parzen E. Nonparametric statistical data modeling. Journal of the American Statistical Association. 1979;74:105–121. MR0529528. [Google Scholar]
- [11].Prentice RL, Pyke R. Logistic disease incidence models and case-control studies. Biometrika. 1979;66:403–411. MR0556730. [Google Scholar]
- [12].Scheffe H. The Analysis of Variance. John Wiley & Sons; New York: 1959. MR0116429. [Google Scholar]
- [13].Scott AJ, Wild CJ. Hypothesis testing in case-control studies. Biometrika. 1989;76:806–808. MR1041428. [Google Scholar]
- [14].Seaman SR, Richardson S. Equivalence of prospective and retrospective models in the Bayesian analysis of case-control studies. Biometrika. 2004;91:15–25. MR2050457. [Google Scholar]