Abstract
This study introduces new information fusion algorithms to enhance disease surveillance systems with Bayesian decision support capabilities. A detection system was built and tested using chief complaints from emergency department visits, International Classification of Diseases Revision 9 (ICD-9) codes from records of outpatient visits to civilian and military facilities, and influenza surveillance data from health departments in the National Capital Region (NCR). Data anomalies were identified and distribution of time offsets between events in the multiple data streams were established. The Bayesian Network was built to fuse data from multiple sources and identify influenza-like epidemiologically relevant events. Results showed increased specificity compared with the alerts generated by temporal anomaly detection algorithms currently deployed by NCR health departments. Further research should be done to investigate correlations between data sources for efficient fusion of the collected data.
Introduction
Timely and accurate detection of both naturally occurring and bioterrorism-related disease outbreaks is crucial for executing an efficient public health response to limit mortality and morbidity in the population. Preventing the spread of natural disease also reduces the economic effects of treatment costs and lost productivity. Although traditional methods of surveillance relying on confirmed laboratory tests are specific, they may not be obtained early enough to halt rapidly spreading disease outbreaks.
To address the need for timely response, developers have implemented several electronic syndromic surveillance applications to collect and analyze near-real-time data from different health indicator sources. These applications attempt to provide public health situational awareness to decision makers in their regions. 1–8 Many of these applications were rapidly developed to support surveillance for acts of bioterrorism during special, large public events. 9,10 These applications support the detection of statistical anomalies in health indicators such as Emergency Department (ED) chief complaints, over-the-counter (OTC) drug sales, and military medical facility visits. Beginning in 1999, The Johns Hopkins University Applied Physics Laboratory (JHU/APL) developed the Electronic Surveillance System for Early Notification of Community-based Epidemics (ESSENCE), 8,11 a system that the Department of Defense deploys worldwide and that has multiple civilian installations within the United States. Improved performance has been a goal of ESSENCE since its introduction, and it is currently one of the most widely used disease surveillance systems. Many syndromic surveillance systems have created tools that quickly identify potentially large disease events, 12–18 but to do so specificity is often traded for sensitivity. 19–24 Despite the capability that these projects have provided, there is still a critical need to develop data fusion techniques that will assimilate multiple sources of information to provide situational awareness and automatically differentiate epidemiologically significant events from statistical anomalies unrelated to actual disease. Some studies have been conducted to combine multiple data streams, 25–27 but they have focused on the outbreak detection problem rather than on providing functionality to support decision making. There are many successful examples of clinical decision support systems, 28–30 but few examples of population-based decision support systems. These successes in clinical medicine systems along with the current study suggest that the development of similar systems for disease surveillance will significantly enhance public health practitioners' ability to monitor disease trends.
Currently, available syndromic surveillance systems rely on a trained user to interpret system alerts and determine which are likely to be associated with a true disease event. Understanding the epidemiological significance of the detected anomalies requires a cognitive analysis of multidimensional data components where the availability and quality of some of the components may be uncertain. The sequence of events; time lags between events; the size, nature, and frequency of the event(s); and the demographic characteristics, signs, and symptoms exhibited by, and the geographic location of the affected people are all important elements of the decision-making process. Once a temporal data anomaly, or alert, is detected, it must be evaluated to determine whether it is likely to be associated with a real disease event. This process can be time-consuming, especially if the probability of false alerts is high. Algorithms that are both sensitive and specific and include the criteria users employ to rule out anomalies are needed to streamline the user's review process.
These considerations illustrate a need for intelligent decision support capabilities. This paper introduces new data fusion algorithms and decision support concepts that can satisfy this need. The described system emulates an epidemiologist's cognitive analysis of the mathematical anomalies to seek patterns suggestive of disease scenarios. Core algorithms of the system are based on temporal data anomaly detection and Bayesian Networks (BNs). 31–33 The system performs cascaded multilevel data processing and refines its detections at each level. As a result, it shows high sensitivity and significant improvement in specificity.
Methods
Study Design
Anonymized, aggregated clinical encounter record data collected by the ESSENCE system were used to create a BN designed to detect the onset of influenza epidemics in a population. Influenza prevalence data collected by the Maryland and Virginia Departments of Health influenza surveillance programs were used to identify influenza-like illness (ILI) and non-ILI periods in the National Capital Region (NCR).
To estimate the validity of the new system, the frequency and timing of detection “alerts” generated by the BN were compared with those generated by the standard univariate ESSENCE detection algorithms during ILI and non-ILI time periods.
Population and Data Sources
Data used in the study were collected in the NCR, which includes Washington, D.C. and the seven surrounding counties in Maryland and Virginia. As expected in a passive data collection system, reporting levels in ESSENCE data vary somewhat by jurisdiction and data source. Some counties supply records from all hospitals in their region, and some from only select hospitals. Similarly, reporting from physician offices is more complete in some jurisdictions. The detection system was built and tested with data from Prince William, Fairfax, and Loudoun counties in Northern Virginia and Montgomery County in Maryland because data from these counties were the most complete in the region. These four jurisdictions are also representative of the NCR. They include 53% of the geographic area, 53% of the total 3.98 million residents in the NCR, and together have a median population density very similar to that of the whole NCR (0.67 vs. 0.66 people/km2, respectively). It is important to note that the proposed method will work for the regions with less complete datasets. One of the strengths of the BN is the capability to function with limited data. While the network is designed to work with a maximum number of data inputs, it can also function and provide results when limited data are available. When some of the inputs for the leaf nodes of the BN are missing, the BN estimates the value for the corresponding nodes based on the known values of the other nodes. BN output is a probabilistic value. When fewer data are available, probability of the true state for the output value will be lower. This is comparable to the human decision making process in situations with uncertainty; the more data available, the more certain we are that the event is happening.
The study used the following data sources: chief complaints from ED visits, International Classification of Diseases Revision 9 (ICD-9) codes from records of outpatient visits to civilian and military facilities, and influenza surveillance data from Maryland and Virginia state health departments. Precise catchment areas for the data sources cannot be determined for privacy reasons, but all eligible EDs and military facilities in the NCR provided data to ESSENCE during the study period. While civilian office visit data were available only from selected doctors' offices, the ILI time series produced from this source mimicked those created from the ESSENCE ED data and from ILI reports from the Maryland and Virginia Departments of Health, suggesting the sources were good candidates for inclusion in the BN.
We evaluated system performance in Montgomery County between June 2003 and May 2006, and in Prince William, Fairfax and Loudoun counties between June 2005 and May 2006.
Evaluation of Data Sources and Epidemiological Methods
Anomalous data trends were identified by an experienced ESSENCE user. This process was observed by the BN developer to identify tasks important to the evaluation process that could be included in the BN design. Information gathered during observation was discussed extensively with users and subject matter experts. An investigation was completed that compared relationships among the different data streams. Anomalies identified in one data stream were sought in other data streams by comparing time series trends and the timing of events. As a result, the distribution of time offsets between events in the data streams was established. The study also examined the degree of dependence between data streams.
The regional, state, and national influenza activity reports were evaluated to estimate the time periods when epidemiologically significant events most likely occurred in each county. Reports included several ILI cases that were reported by Sentinel Physicians in Northern Virginia region and reported by the Virginia Department of Health, cases of the Maryland laboratory-confirmed influenza activity reported by the Maryland Department of Health, and CDC national influenza surveillance data. The time series behaviors among available data sources during these estimated event periods were evaluated for detection consensus and relative timeliness. These time frames were compared with the detection system's output. The findings were discussed with the subject matter experts to verify their relevance to the epidemiologist's objective.
Analytic Methods
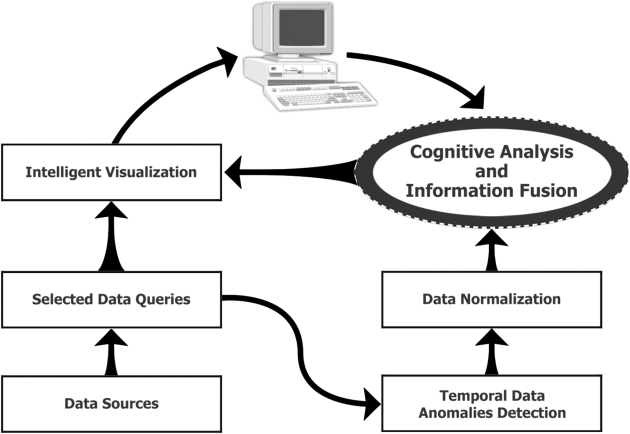
The core algorithm can be described as a multistep information process (▶). As a first step, we reviewed all available hospital patient record data and selected ILI syndrome-specific combinations of words from chief complaint strings. Examples of word combinations included fever and cough or fever and sore throat. The ESSENCE database was queried using these selected words and phrases. For each selected combination, we formed the time series of daily counts of records whose chief complaint string contained that combination. We applied ESSENCE alerting algorithms 8,17 to each of these series. These algorithms are univariate time series detectors based on adaptive regression models and control charts. This step created the sets of daily anomalies found for each combination. Table 1 (available as an online data supplement at http://www.jamia.org) lists the queries selected for the influenza detection model.
Figure 1.
Multilevel information processing diagram.
Step 2 supplies the daily set of anomalies for each query as an input for the Information Fusion Network (IFN). Each bottom level, data-driven node of the IFN is associated with one of these queries. It uses corresponding algorithm results to evaluate the assertion that an anomaly exists within the most recent 7 days for the chief-complaint combination underlying that query.
The third step is an IFN process. The IFN is a BN designed to allow multisource information fusion in a manner emulating the domain expert's decision-making process. A BN is a probabilistic model typically visualized as a directed acyclic graph. It has nodes representing variables and directed edges representing probabilistic dependencies, and it offers a compact representation of the relationships among all variables in the graph. Expert knowledge is embedded into the BN at two levels. First, the gross structure of the network captures an expert's view of the interdependencies of the causal factors in the network. The factors are elements of data sources. Second, the conditional probability table attached to each node quantifies the probabilistic relationships between connected variables. The values for probabilistic relationships in this table are calculated based on both the opinions of subject matter experts and anomaly correlation among data sources. The subject matter experts are involved only at the initial formation of the BN structure; subsequent adjustments are done from data.
The following observation and development procedure was used to guide formation of the BN structure and associated conditional probability tables in an attempt to select relevant database queries that are unbiased and supported by the data. Experienced ESSENCE users, masters or doctoral level epidemiologists, review the daily system alerts. Their aim is to eliminate alerts that do not detect events that are a public health threat. To do that, they characterize the alert by the type and severity of illness described, and the geographic proximity of cases and their demographic characteristics, such as age and sex. They also consider whether and how often such an alert is expected considering the season, weather, and time of year, and whether similar alerts have been seen recently and, if so, the number and the temporal sequence of those alerts, along with other issues specific to the suspected disease, locale and population. An analyst observed this process and identified commonly used queries and data selection criteria. The resulting queries, for example ICD-9 code 487 for infants aged 0–4, were run using data from known outbreak periods and from outbreak-free time frames. We calculated the ratio of algorithm alerts found between the outbreak and outbreak-free periods. This analysis also took into account noisiness and timeliness relative to the beginning of flu season as defined by health department laboratory reports.
The resulting findings were used to create both the BN structure and the conditional probability tables. For example, adult visit counts peaked mainly during flu season, while infant visit counts peaked at other times. Thus, there is conditional dependency between influenza and age distribution factor, and the BN structure reflects this dependency. Increases in the data counts were detected using temporal anomaly detection regression algorithms currently deployed by the ESSENCE system. 8,17 Our initial analysis was performed on the data collected in Montgomery County. During flu season, both infant (0–4 age group) and adult (18–64 age group) counts showed timely increases in the number of sick visits. However, in the preinfluenza period (Jun 1, 2005 to Dec 15, 2005), the count of ill infants was anomalously high 6 times, while the number of ill adults was anomalously high 3 times. This difference may be explained by the noninfluenza respiratory outbreaks that usually occur earlier in the season and severely affect infants more than adults, for example, respiratory syncytial virus (RSV). As a result, because our goal in this experiment was to differentiate flu from other respiratory outbreaks, for the BN that specifically targets influenza, the conditional probability tables for infant nodes contributed less and the probability values reflected the degree of influence observed. Our finding was cross-validated in the different regions. In Prince William County for the same preflu time frame (Jun 1, 2005 to Dec 15, 2005), the anomalous number of adults was detected 2 times and for infants 7 times. In Fairfax County, the anomalous number of adults was detected 3 times and for infants 7 times. Similar analysis was performed for all other BN input nodes. The results from one year to another and from one geographic region to another were slightly different. For each node, the generic mean of the range of outputs was calculated and presented to epidemiologists familiar with the ESSENCE system to verify that these values were realistic based on their experience. These generic values are the basis for defining the relative strength of conditional dependency between the nodes. This process is labor-intensive but necessary to create conditional probability tables that are both data dependent and epidemiologically realistic.
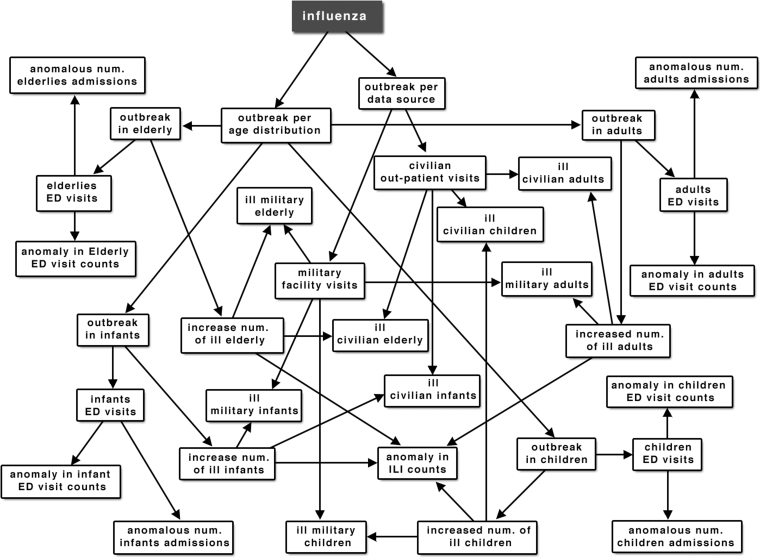
The function of the IFN is to determine the probability of occurrence of an epidemiologically significant event by estimating the likelihood of the detected temporal data anomalies. The network's structure reflects the epidemiologist's decision-making process. It is structured to calculate the probability of outbreak within different age categories as represented in different data sources. ▶ illustrates the structure of the IFN for influenza detection.
Figure 2.
Influenza detection BN structure.
Table 2 (available as an online data supplement at http://www.jamia.org) illustrates how queries from Table 1 are mapped to Influenza Detection BN nodes. Each node has a true value if the corresponding mapped query returned temporal anomaly detection within the past 7 days.
Probability tables for the intermediate nodes are based on the joint probability distribution of child node tables. For example, ED_child node probabilities are calculated using tables for nodes spike_in_child and child_discharge_factor. Node child_discharge_factor is true when the number of children admitted to the hospital within the past 7 days is greater than expected one. The output of the BN is the probability of a true influenza outbreak, which is represented by the Influenza node (▶). The probability of the influenza outbreak I, given that the age distribution in the ill population is consistent with the age distribution expected during influenza, a condition represented as A, is
where
The probability of the influenza outbreak given that the civilian and military populations are affected by influenza (condition denoted S) is
where
The probability of the influenza outbreak given an anomalous increase in self-care activities (condition SC), such as increase in OTC medication sales, is:
where
The probability of influenza given A, S, and SC is then
The intermediate nodes can also provide outputs, for example, the population-specific probability of outbreak as indicated by military (military_factor node) or civilian (civilian_factor node) data sources.
Our BN structure is constructed by a data-driven but user-centric principle, where correlations between different nodes are supported by available data and confirmed as relevant by the subject domain expert. For example, the data might show that age distribution in an ill population is a significant factor, and the epidemiologist looks at it to verify the presence of an outbreak. These findings are relevant to the objective of the responsible health agency, so the BN is structured to incorporate sick visit counts from each of the data sources by different age groups, and the probability of the outbreak within each age group will be calculated. Another important factor is outbreak within a population represented by a particular data source. For example, if the outbreak occurs in a Military Academy, most of the patients will go to the military facility; in that case, we will see the spike of visits within one source rather than across multiple sources. Therefore, the BN calculates probability of anomaly within military and civilian sources with consideration of the ill population's age distribution in each source. Patient disposition—whether the patient is admitted, discharged, transferred, or deceased—is also available in the ED visit records, and disposition anomalies were added to the BN structure. What percent of people visiting hospital emergency rooms are admitted to the hospital? The final value for the probability of influenza is calculated based on the joint probability of all of these factors (▶).
The final step is an intelligent data visualization process, where the reasons for an alert are presented to the user and drill-down capabilities are implemented to allow the user to see actual records that might be causing a high outbreak probability.
Results
To measure specificity of the IFN model, we identified a known 4.5 month long flu-free time period from Jun 1, 2005 to Oct 15, 2005, and evaluated the model's performance within the one county in Maryland and the three counties in Virginia. The BN output of the parent node should be reviewed as a relative measure and not an absolute probability that the statement underlying the node is true. Therefore, operational thresholds are required. A parent node output probability threshold of 0.35 was chosen because it was the lowest multiple of 0.05 that gave perfect sensitivity for the identified events. Use of the lowest multiple of 0.05 ensures that sensitivity will remain as high as possible.
This threshold also gave 100% specificity during the flu-free period. The influenza probability never exceeded 0.05 in data from Montgomery County in Maryland and Loudoun and Prince William counties in Virginia. Although the influenza probability from Fairfax County, Virginia exceeded 0.05 two times during the flu-free period, 0.32 on Jun 1 to Jun 5, and 0.11 on Jul 3 to Jul 4—both times it was below the 0.35 threshold.
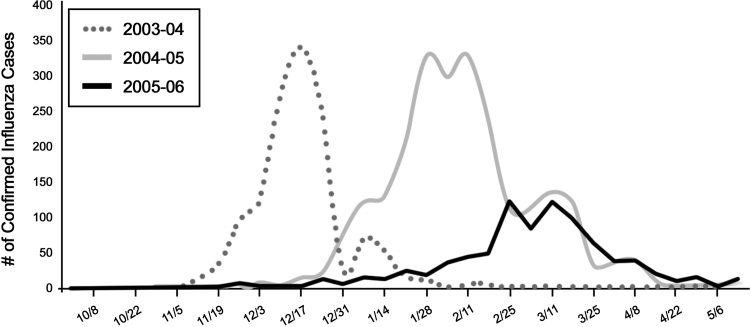
To measure sensitivity of the IFN model, influenza outbreaks were identified from the publicly available health reports from the Maryland and Virgínia Health Departments. Based on Maryland's confirmed flu laboratory tests (▶) and Virginia's Sentinel Physicians' reports (▶), six influenza outbreaks were identified, counting separate county-based events. Three outbreaks were in Montgomery County for influenza seasons 2003–04, 2004–05, and 2005–06; and one for the 2005–06 season in each of the Virginia counties–Prince William, Loudoun, and Fairfax. All six outbreaks were detected by the model at the threshold of 0.35. For detection at the beginning of the influenza seasons, the sensitivity was 100%. Given the small number of events, we cannot claim perfect sensitivity in general, but this evidence is very promising. The model detected all six outbreaks at least as soon as the onset dates in the health department reports.
Figure 3.
Laboratory-confirmed cases of influenza in the State of Maryland.
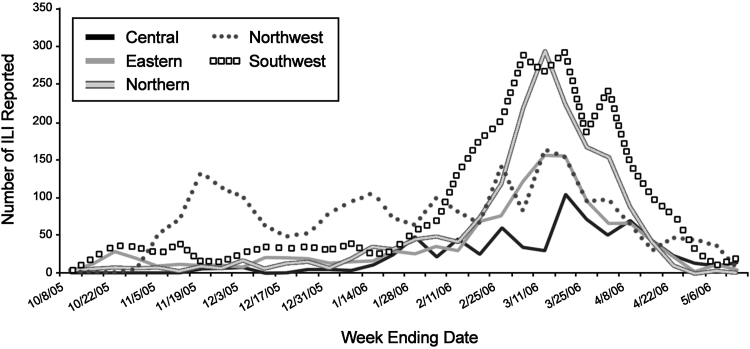
Figure 4.
Influenza-like illnesses (ILI) reported by Sentinel Physicians in Virginia by region, during 2005-06 influenza season (from Virginia Health Department Influenza Surveillance Annual Report).
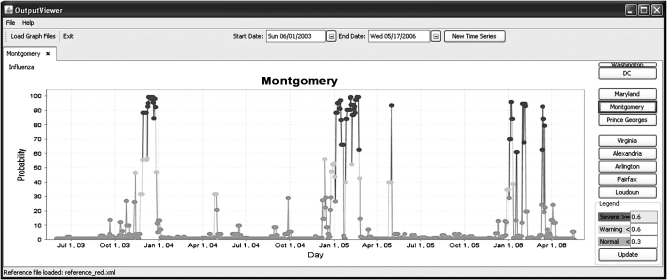
▶ shows the Maryland laboratory-confirmed influenza chart for the 2003–04, 2004–05, and 2005–06 seasons. ▶ shows the model's output indicating probabilities of influenza in Montgomery County between June 2003 and May 2006.
Figure 5.
Output from system indicating probabilities of influenza in Montgomery County between June 2003 and May 2006.
For the 2003–04 season, influenza cases first appear during Week 45–46; the system calculates the outbreak probability as 46% on Nov 12, 2003 (Week 45) and above 80% at the end of Nov (Week 47–48).
For the 2004–05 season, influenza cases first appear during Week 49–50 with an increase in cases on Week 52–1, then the peak at the end of Jan and a second wave in the beginning of Feb. The system calculates the outbreak probability as 56% on Dec 12, 2004 (Week 50) and above 80% at the beginning of January 2006 (Week 1), then another increase of probability at the end of Jan. The system continues to estimate a high probability of outbreak until the end of Feb.
The 2005–06 influenza season came in the spring, later than its customary onset in the early winter. The first few cases appeared on Week 52 then again on Week 2 with the slight increase starting on Week 4–5. The peaks were on Weeks 8 and 11. The system first detects probability of an outbreak on Week 1, then on Week 3 and Week 5, and the last detection is on Week 10 (starting Mar 12).
For the Northern Virginia counties, data were available from civilian and military outpatient office visits for 3 years; however, ED data were consistently available since 2005. Therefore, we calculated probabilities based on available data for 3 years but presented results only for the 2005–06 influenza season, when data were available from all data sources.
The Virginia Health Department Influenza surveillance report shows that the reported cases of influenza in the Northern Virginia region (▶, gray line with black outline) increased in the beginning of Jan, slightly increased in mid-Jan, and consistently increased from the middle of Feb until the middle of Mar. The northern region includes Fairfax, Prince William, Arlington, Alexandria, and Loudoun Counties. For Fairfax County, our system shows an increase in the probability of influenza in the end of Oct, last week of Dec, second week of Jan, second week of Feb, and starting in the end of Feb until the third week of Mar (Figure 6, available as an online data supplement at http://www.jamia.org). Although there is no increase in reported cases at the end of Oct in Northern Virginia, there is a significant increase of the reported cases in the Northwest region (▶, dotted line). For Prince William County, two groups of alerts for the 2005–06 influenza season were indicated; the first starts on Dec 28, and the second starts on Feb 21 (Figure 7, available as an online data supplement at http://www.jamia.org).
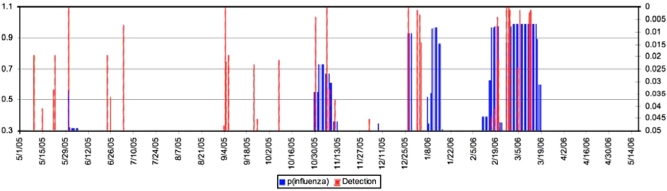
We also compared results with alerts and warnings generated by the ESSENCE detectors. ▶ illustrates a comparison generated by new BN model probabilities of influenza (black, left axis) and the p-values for ESSENCE alerts and warnings (gray, right axis) for ILI (fever and either sore throat or cough). The new model shows significantly higher specificity and also time frames when increasing probabilities of influenza are consistent with the increase of the reported influenza by Sentinel Physicians (▶).
Figure 8.
Probabilities (blue) and ESSENCE Warnings and Alerts (red) for Fairfax County, Virginia.
An obstacle to quantifying the value-added of the BN is that the actual specificity of the traditional algorithms cannot be precisely measured. Because the traditional approach is not disease specific but syndrome specific, one cannot prove in general that a background alert is a false alert. ▶ does compare the number of alerting events obtained with algorithms alone and with the BN, though we would not claim precise specificity measures from this table. The corroboration implicitly required by the BN probabilistic structure removes many of the pure algorithmic alerts—a human monitor should not need to go through this rule-out process, certainly not with alerts from many data streams. The BN structure and conditional probability tables can emulate the weighting process that the human would employ. For interpretation of this table, note that the BN combines algorithm outputs from the most recent 7 days to estimate the outbreak probability for any given day. The BN does not alert for each algorithm anomaly but responds to anomaly patterns. For example, if anomalies are seen only for infant data, then the BN most likely will not show a high outbreak probability. If an anomaly is detected in adult data the next day, the outbreak probability will increase.
Table 3.
Table 3 ESSENCE Alerts Compared with BN Probabilities
| Time Period | Uninterrupted ESSENCE Alerts p-Value < 0.01 | Uninterrupted ESSENCE Alerts p-Value < 0.05 | Uninterrupted Intervals BN Probability > 35% |
|---|---|---|---|
|
3 | 10 | 1 |
|
2 | 5 | 1 |
|
6 | 8 | 5 |
|
0 | 0 | 0 |
| 5/1/05–5/1/06 | 11 | 23 | 7 |
Discussion
This study is one of a few to incorporate epidemiologist decision-making logic in a decision support system. In our experience, public health subject matter experts can effectively evaluate a syndromic surveillance system's detections and recognize patterns suggesting real public health events from statistical anomalies in the data stream. The design includes the epidemiologist's decision-making logic applied to the dynamic environments where events and their resulting data effects do not occur simultaneously but rather in a disease- or syndrome-specific sequence. The benefit of our approach is that it reduces the burden on the user by providing integration of many algorithmic results focused toward specific public health threat hypotheses—the hypothesis of an influenza outbreak in this experiment. This integration has been done only heuristically in common practice. Automated emulation of this process makes the system more intuitive for the user and obviates the decision of whether to investigate individual alerts. Moreover, the graphical BN structure makes the logic transparent so the user may understand the automated logic underlying top-level threat probabilities.
Using age distribution as an outbreak indicator is an important component of the design. The BN is designed so the intermediate nodes show the outbreak probability in different age groups and data sources; this feature allows the system to be sensitive to relatively small outbreaks that are specific to age groups or subpopulations, such as the military. When statistical analyses are performed on visits counts including all ages, increases in a relatively small age category, such as “Children”, may not appear significant. The data fusion approach is sensitive to anomalies within each of the age groups. Furthermore, a data fusion-based system has increased specificity because it estimates the probability of the events across multiple data streams. Another benefit of the transparency of the intermediate nodes is the potential to provide evidence of the spread of disease among subpopulations while an outbreak is in progress. For example, the nodes described in ▶ represent outbreak probabilities based on information from military and civilian data sources and from age-specific categories within them.
We also found that time frames for increase in the probability of influenza in neighboring counties look very similar. Figures 6 and 7 illustrate the probabilities of the outbreak in Fairfax and Prince William counties. The agreement of the multiple-peak increases in probabilities within the same time frames in both regions suggests that the system can detect both the beginning of each season, as well as correctly differentiate multiple waves within the same season.
More research needs to be done to investigate correlations between data sources for efficient fusion of the collected data. A number of surveillance systems collect information from various data sources. Some of these sources, such as ED chief complaint data, have been intensively analyzed for outbreak investigation. Other data sources are less commonly used and need to be analyzed for their appropriate usage for population health monitoring. Furthermore, research is needed to quantify temporal correlations of the effects of health events among different data sources.
Additional research and development is needed to build the network of decision support models for different categories of users. For our prototype system, we developed local and state level decision support models. Additional regional and national models need to be developed. An automated distributed decision support system can be a good vehicle for information exchange between different health departments and will not require the sharing of actual data.
The study presented here was conducted with data only from the Washington, D.C. metropolitan area. Although the data covered several counties, one should consider this study as a single-region experiment. This restriction is a limitation of the study; data from multiple locations should be evaluated to confirm these findings and to evaluate system performance in a variety of urban, suburban, and rural surveillance regions. Additional experiments in more congested areas, such as New York City, and low-density rural areas will enable the design of models in varied regional environments.
Several points about operational thresholds should be clarified: methods such as testing with historical data and likely outbreak intervals, bootstrapping, and cross-validation, preferably with the guidance of local epidemiologist users, should be used to determine operational thresholds. It would be misleading to specify a statistical formula for the number of days of historical data required for a desired power to detect. Such formulas from the literature are based on fixed data distributions, and surveillance data streams satisfying these distributions are rare. In some situations, data streams may be modeled well enough that the algorithm results input to a BN may have a normal distribution, but such results are generally not portable across neighboring subregions, as seen in the multiple region analyses in Craigmile. 34 Furthermore, a steady-state data environment is the exception, not the rule. Therefore, a certain amount of data analysis is necessary at the local level for robust BN detection performance. Based on this study, we suggest that for a BN system intended to replicate this study, at least 1 year of quality training data at the time resolution of the BN be required for all data streams included. By “quality data” we mean that the data steams are available with fairly continual provider participation with no dropouts or other problems lasting more than a couple of weeks. Based on the experience of the current and previous studies, the BN paradigm can offer substantial practical decision support guidance in an evolving, nonstationary data environment. However, any operational system should be evaluated for recalibration, possibly annually.
Finally, competing hypotheses can be readily incorporated in BN structures as multiple parent nodes. The availability of truth data for other illnesses such as RSV or adenovirus would have allowed evaluation of multiple hypothesis nodes. Such nodes can be easily added to the current structure, but truth data would be needed for credible evaluation. When dealing with multivariate, highly correlated hypotheses, unbiased simulation is very difficult, and with the best of intentions, it is easy to obtain deceptive results.
Conclusions
The results show that the hybrid statistical probabilistic data fusion model for influenza detection exhibits both high specificity and sensitivity. The discussion surrounding ▶ demonstrates that detections are timely compared with both laboratory-confirmed influenza and reported sentinel Physicians' cases. ▶ demonstrates that the IFN can improve specificity of the ESSENCE system for event detection and can differentiate epidemiologically significant events from mathematical data anomalies and possibly detect different waves of influenza. Given the subjective, expert-dependent portion of the development process described above, more implementation and validation experiences are needed to standardize this fusion capability and make it robust across various data environments and health threat types.
Footnotes
This article was supported in part by a Stuart S. Janney Fellowship award from The Johns Hopkins University Applied Physics Laboratory and Grant Number P01 HK000028-02 from the Centers for Disease Control and Prevention (CDC). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of CDC.
References
- 1.Lober WB, Karras BT, Wagner MM, et al. Roundtable on bioterrorism detection: Information system-based surveillance J Am Med Inform Assoc 2002;9(2):105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zelicoff AP, Brillman J, Forslund DW, et al. The rapid syndrome validation project In: Proceedings AMIA Symposium 2001:771-775. [PMC free article] [PubMed]
- 3.Wagner MM, Tsui FC, Espino JU, et al. The emerging science of very early detection of disease outbreaks J Public Health Manag Pract 2001;7:51-59. [DOI] [PubMed] [Google Scholar]
- 4. Bioterrorism Preparedness and Response: Use of Information Technologies and Decision Support Systems Summary, Evidence Report/Technology Assessment No. 59. Rockville, MD: Agency for Healthcare Research and Quality; July2002. http://ahrq.gov/clinic/epcsums/bioitsum.htm July2002. Accessed September 1, 2007.
- 5.Mandl KD, Overhage JM, Wagner MM, et al. Implementing syndromic surveillance: A practical guide informed by the early experience J Am Med Inform Assoc 2004;11:141-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bradley CA, Rolka H, Walker D, Loonsk J. Biosense: Implementation of a national early event detection and situational awareness system Morb Mortal Wkly Rep 2004;54(SU1):11-19. [PubMed] [Google Scholar]
- 7.Tsui FC, Espino JU, Dato VM, et al. Technical description of RODS: A real-time public health surveillance system J Am Med Inform Assoc 2003;10:399-408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lombardo JS, Buckeridge DL. )Disease Surveillance: A Public Health Informatics Approach, Hoboken (NJ): John Wiley & Sons; 2007.
- 9.Mundorff MB, Gesteland PH, Rolfs RT. Syndromic surveillance using chief complaints from urgent care facilities during the Salt Lake 2002 Olympic Winter Games Morb Mortal Wkly Rep 2004;53(SU1):254. [Google Scholar]
- 10.Dafni UG, Tsiodras S, Panagiotakos D, et al. Algorithm for statistical detection of peaks-Syndromic surveillance system for the Athens 2004 Olympic Games Morb Mortal Wkly Rep 2004;53(SU1):86-94. [PubMed] [Google Scholar]
- 11.Lombardo J, Burkom HS, Elbert E, et al. A systems overview of the Electronic Surveillance System for the Early Notification of Community-based Epidemics (ESSENCE II) J Urban Health 2003;80(2)S1:i32–42. [DOI] [PMC free article] [PubMed]
- 12.Buckeridge DL, Burkom H, Campbell M, et al. Algorithms for rapid outbreak detection: A research synthesis J Biomed Inform 2005;38:99-113. [DOI] [PubMed] [Google Scholar]
- 13.Kulldorff M. A spatial scan statistic Commun Stat Theory Methods 1997;26:1481-1496. [Google Scholar]
- 14.Kulldorff M, Heffernan R, Hartman J, et al. A space-time permutation scan statistic for disease outbreak detection PLoS Med 2005;2:e59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wong W-K, Moore A, Cooper G, Wagner M. Rule-based anomaly pattern detection for detecting disease outbreaks In: AAAI-02; Edmonton, Alberta: AAAI Press/The MIT Press 2002:217-223.
- 16.Reis BY, Pagano M, Mandl KD. Using temporal context to improve biosurveillance Proc Natl Acad Sci USA 2003;100:1961-1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Burkom H. Development, adaptation, and assessment of alerting algorithms for biosurveillance Johns Hopkins APL Tech Digest 2003;24:335-342. [Google Scholar]
- 18.Lawson A. Large scale: SurveillanceIn: Lawson A, editor. Statistical Methods in Spatial Epidemiology. New York: Wiley; 2001. pp. 197-206.
- 19.Marsden-Haug N, Foster V, Hakre S, et al. Evaluation of joint services installation pilot project and BioNet syndromic surveillance systems—United States Morb Mortal Wkly Rep 2004;54(Suppl):194. [Google Scholar]
- 20.Elbert E, Burkom HS, Nelson K. Performance evaluation of temporal alerting algorithms for syndromic surveillance dataProceedings of the Eighth US Army Conference on Applied Statistics 2002. Raleigh, NC.
- 21.Bravata DM, McDonald KM, Smith WM, et al. Systematic review: Surveillance systems for early detection of bioterrorism-related diseases Ann Intern Med 2004;140:910-922. [DOI] [PubMed] [Google Scholar]
- 22.Buckeridge DL, Burkom H, Moore AW, et al. Evaluation of syndromic surveillance systems: Development of an epidemic simulation model. In: Syndromic Surveillance: Reports from a National Conference, 2003. MMWR Morb Mortal Wkly Rep 2004;53(Suppl):137-143. [PubMed] [Google Scholar]
- 23.Buckeridge DL, Switzer P, Owens D, et al. An evaluation model for syndromic surveillance: Assessing the performance of a temporal algorithm Morb Mortal Wkly Rep 2005;54(Suppl):109-115. [PubMed] [Google Scholar]
- 24.Jackson ML, Baer A, Painter I, Duchin J. A simulation study comparing aberration detection algorithms for syndromic surveillance BMC Med Inform Decis Mak 2007;7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burkom HS. Biosurveillance applying scan statistics with multiple, disparate data sources J Urban Health 2003;80(2):157-165(Suppl 1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weng K-W, Cooper G, Dash D, et al. Use of multiple data streams to conduct Bayesian biologic surveillance Morb Mortal Wkly Rep 2005;54(Suppl):63-69. [PubMed] [Google Scholar]
- 27.Kulldorff M, Mostashari F, Duczmal L, et al. Multivariate scan statistics for disease surveillance Stat Med 2007;26(8):1824-1833. [DOI] [PubMed] [Google Scholar]
- 28.Garg AX, Adhikari NK, McDonald H, et al. Effects of computerized clinical decision support systems on practitioner performance and patient outcomes: A systematic review J Am Med Assoc 2005;293:1223-1238. [DOI] [PubMed] [Google Scholar]
- 29.Payne TH. Computer decision support systems Chest 2000;118(2):47S-52S. [DOI] [PubMed] [Google Scholar]
- 30.Sim IP, Gorman P, Greenes RA, et al. Clinical decision support for the practice of evidence-based medicine J Am Med Inform Assoc 2001;8(6):527-534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Castillo E, Gutiérrez JM, Hadi AS. Expert Systems and Probabilistic Network ModelsNew York: Springer-Verlag; 1997.
- 32.Pearl J. Fusion, propagation, and structuring in belief networks Artif Intell 1986;29(3):241-288. [Google Scholar]
- 33.Kahneman D, Slovic P, Tversky A. )Judgment Under Uncertainty: Heuristics and Biases, Cambridge: Cambridge University Press; 1982.
- 34.Craigmile PF, Kim N, Fernanadez B, Bonsu B. Modeling and detection of respiratory-related outbreak signatures BMC Inform Decis Mak 2008;28:7. [DOI] [PMC free article] [PubMed] [Google Scholar]