Abstract
This study evaluated the performance of an electronic screening (E-screening) method and used it to recruit patients for the NIH sponsored ACCORD trial. Out of the 193 E-screened patients, 125 met the age criterion (“age ≥ 40”). For all of these 125 patients, the performance of E-screening was compared with investigator review. E-screening achieved a negative predictive accuracy of 100% (95% CI: 98–100%), a positive predictive accuracy of 13% (95% CI: 6–13%), a sensitivity of 100% (95% CI: 45–100%), and a specificity of 84% (95% CI: 82–84%). The method maximized the use of a patient database query (i.e., excluded ineligible patients with a 100% accuracy and automatically assembled patient information to facilitate manual review of only patients who were classified as “potentially eligible” by E-screening) and significantly reduced the screening burden associated with the ACCORD trial.
Introduction
According to recent reports, recruitment difficulties caused delays from 1 to 6 months for 86% of clinical trials, with the remaining 14% experiencing even longer delays. 1 Complex inclusion and exclusion criteria contribute to recruitment challenges. Usually done manually, screening for clinical trials is a laborious and inefficient process. Moreover, patient information that satisfies eligibility criteria can be scattered in multiple information systems, databases, and patient documents as either coded data or free text in various linguistic forms. Experienced and knowledgeable clinical research staff often manually assemble and sift through large amounts of patient information to determine patient eligibility. The increased adoption of electronic health records (EHR) in recent years invites E-screening solutions to automatically identify potentially eligible patients. However, few studies have evaluated the performance and value of E-screening for reducing the workload of clinical research staff while improving recruitment. In this paper, we describe an E-screening design that accommodates the data characteristics in EHR systems and maximizes the value of automatic database queries by reducing manual screening. Our method reduced the screening burden for clinical research staff by automatically and accurately excluding ineligible patients and by assembling pertinent clinical information needed for manual review by clinical research staff.
Methods
This study was reviewed by the Columbia University Medical Center Institutional Review Board (IRB) and was granted a waiver of HIPAA authorization from the Institutional Privacy Board. Below we describe our design rationale, system architecture, and evaluation design.
The E-screening Rationale
Screening charts manually is time-consuming for research personnel who must search for information in patient records to determine whether a patient meets the eligibility criteria for a clinical trial. With E-screening, we aimed to exclude ineligible patients and establish a much smaller patient pool for manual chart review. E-Screening will help clinical research personnel transition from random and burdensome browsing of patient records to a focused and facilitated review. Consistent with concerns for patient safety and trial integrity, clinical research personnel should review all patients classified as “potentially eligible” by E-screening to confirm their eligibility. Generally, E-screening systems essentially perform “pre-screening” for clinical research staff and should not fully replace manual review. We hypothesized that the screening burden can be reduced automatically by, (1) searching electronic patient information to exclude ineligible patients, and by (2) extracting and assembling pertinent electronic patient records to facilitate manual review.
As Kahn pointed out, EHR systems configured to support routine care are a good source of demographic and structured laboratory test data, but a poor source of narrative patient reports and questionnaires about patient health behaviors. 2 If we could implement all the eligibility criteria for a clinical trial in E-screening, we would still fail to exclude ineligible patients who have missing data. Some exclusion criteria are rarely available in EHRs, such as “life expectancy greater than 3 months” or “women who are breastfeeding”. To address missing clinical data and subjective eligibility criteria definitions, we took an important step in our query formulation by analyzing which eligibility criteria had corresponding EHR data elements, where and how these data were stored (structured vs. free-text), and which criteria could be addressed automatically and which needed manual review. On this basis, we selected the best negative predictive variables, primarily those that had corresponding EHR data elements and could be queried automatically.
The System Infrastructure
To query for potentially eligible patients, we used the Columbia University Medical Center's clinical data warehouse, 3 which is an electronic repository of patient information gathered from over 20 ancillary and departmental systems, optimized for cross-patient analytic queries. The clinical data warehouse, created in 1989, contains laboratory test results, imaging reports, and clinical notes such as operative notes, pathology reports, and discharge summaries. It provides a comprehensive collection of information to facilitate patient care, administration, and clinical research. 4 These data have been used to support many levels of organizational decision-making and research. A key advantage of the clinical data warehouse is that data can be aggregated across millions of electronic patient records quickly without impacting clinical information systems used for daily patient care. Integration of semantically related clinical data collected by various hospital information systems using different data standards is enabled by a standards-based, controlled clinical terminology called the medical entities dictionary (MED) 5 (shared by all our hospital information systems). The MED includes comprehensive terminologies for drugs, diseases, clinical findings, and procedures, bridging the translations among these terminologies through a shared language, thus allowing for concept-based data integration and retrieval. Each entity has a unique identifier called the MED code, which helps achieve standardization by relating synonymous terms from different terminologies that share the same MED code. The MED contains more than 100,000 concepts from controlled terminologies that are used by local applications and some national standard terminologies (such as ICD-9-cm) and contains their mappings to other national standards, such as LOINC, CPT and UMLS. 6 The clinical data warehouse is updated daily at midnight by input from the Columbia University Medical Center's EHR system, a Web-based Clinical Information System (WebCIS). 7 WebCIS uses the MED codes to automatically assemble semantically related patient laboratory tests, diagnoses, ancillary reports, and medication histories and enables easy web-based browsing of EHR data one patient at a time.
Our E-screening method used a data query (automatically assisted by MED) that interrogated the clinical data warehouse to search for potentially eligible patients, and then only these patients were reviewed by clinical research staff using WebCIS and/or paper charts as needed.
The Evaluation Design
Our evaluation goals were to assess the accuracy of E-screening to exclude ineligible patients and determine the sensitivity of E-screening to include eligible patients. We chose to use the Action to Control Cardiovascular Risk in Diabetes (ACCORD) Trial 8 as a source for our experimental data because it was actively enrolling at the Columbia University Medical Center. To determine the accuracy of E-screening, a clinical investigator (acting as our gold standard) independently reviewed the electronic and paper medical records to classify patients as eligible or ineligible. The investigator's results were cross classified with the E-screening categories “potentially eligible” and “ineligible” as shown in ▶. The ACCORD is a challenging case for E-screening because it has 6 major inclusion and 19 major exclusion criteria as well as 5 eligibility criteria associated with substudies. 8 We believed that an E-screening method that worked well in a complex trial like ACCORD would likely be applicable to many other clinical research studies. In this study, we relied exclusively on structured data, such as ICD-9 codes and laboratory values. The subset of inclusion and exclusion criteria of ACCORD selected for E-screening is shown below:
- Inclusion Criteria:
- 1 Age ≥ 40
- 2 Type 2 diabetes mellitus determined by at least one of the following:
- • fasting plasma glucose ≥ 126 mg/dL
- • random plasma glucose ≥ 200 mg/dL
- • ICD-9 code for type 2 diabetes mellitus
- • hemoglobin A1c (HbA1c) between 7.5 and 11.0%
- • use of antidiabetic medications
- Exclusion Criteria:
- 1 A diagnosis of any malignancy (except nonmelanoma skin cancer) first made in the last 2 years
- 2 History of organ transplant except skin and cornea transplants
- 3 Diagnosis of dementia (including Alzheimer's disease)
- 4 History of alcohol or substance abuse
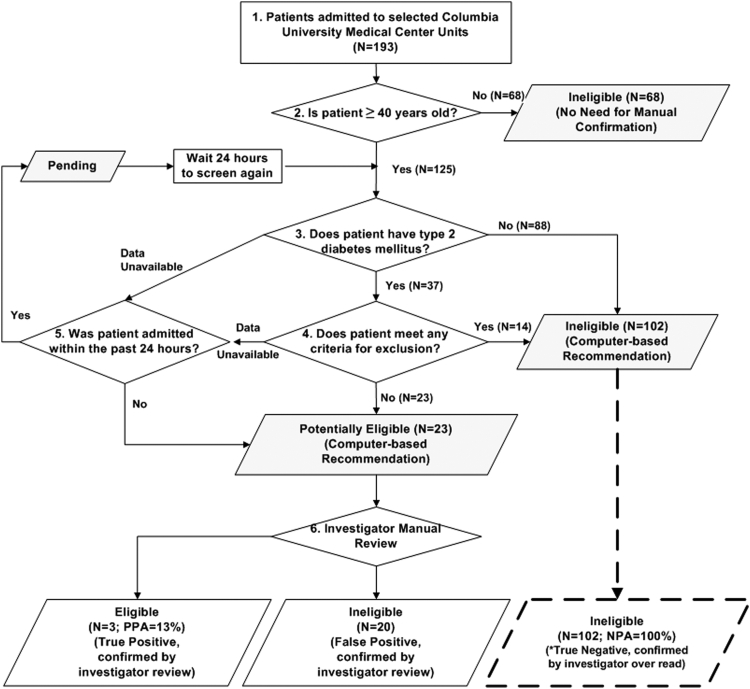
▶ illustrates our E-screening workflow. As shown in ▶, “Age ≥ 40” was the first criterion we evaluated in E-screening because it unambiguously excludes many patients. “Presence of type 2 diabetes mellitus” was evaluated next. All patients who satisfied these two inclusion criteria were then evaluated for exclusion criteria. If any exclusion criterion was met, the patient was excluded and the evaluation stopped. All patients who were at least 40 years old were categorized by the E-screening algorithm as “ineligible”, “pending”, or “potentially eligible”, based on the selected inclusion and exclusion criteria. The “pending” status was assigned to potentially eligible patients who had missing data and were first screened < 24 hours of admission. These patients were screened again 24 hours later. If they met any exclusion criteria, the status changed to “ineligible”; otherwise, the status changed to “potentially eligible”, which could mean either (a) patients met all inclusion criteria and no exclusion criteria; or (b) patients had incomplete data for E-screening. To avoid duplicate matches of previously enrolled patients, a list of enrolled patients was maintained and checked each time the query was run.
Table 1.
Table 1 E-screening Results Compared with Investigator Review
| E-screening Results | Investigator Review Results |
Total | |
|---|---|---|---|
| Eligible | Ineligible | ||
| Eligible | 3 | 20 | 23 |
| Ineligible | 0 | 102 | 102 |
| Total | 3 | 122 | 125 |
CI = confidence interval; NPA = negative predictive accuracy; PPA = positive predictive accuracy.
Sensitivity = 100% (95% CI: 45–100%).
Specificity = 84% (95% CI: 82–84%).
PPA = 13% (95% CI: 6–13%).
NPA = 100% (95% CI: 98–100%).
Figure 1.
The E-screening workflow.
The shaded boxes indicate computer-generated recommendations (i.e., “ineligible”, “pending”, or “potentially eligible”). E-screening reduced the cases that need investigator review from 193 to 23. All cases were over read by an investigator to determine the accuracy of E-screening to identify ineligible patients (NPA was 100%). Dashed lines depict investigator over read of patients who were categorized as ineligible by E-screening. Patients with incomplete data were classified as pending if E-screening was done < 24 hours after admission.
We compared E-screening's performance to an independent, standard manual evaluation of the 125 patients' complete electronic and paper-based health records using the full set of ACCORD eligibility criteria. 8 Investigator review determined true positives for ACCORD. The investigator conducting the manual evaluation received a list of patients admitted to the medical service whose age was ≥ 40 years and was blinded to the categorization of “ineligible” or “potentially eligible” automatically made by E-screening. Patients on this list were first screened by investigator review of their available EHR data in WebCIS. Patients who were not excluded in the initial review had their paper health records reviewed by the same, blinded, investigator.
Results
Our evaluation lasted for 2 weeks, during which time we prospectively screened the inpatient medical service at the Columbia University Medical Center. During the 2-week evaluation period (Dec 3, 2002 to Dec 17, 2002), out of the 193 screened inpatients, 125 met the “age ≥40” criterion and were further classified by the E-screening algorithm. The 125 patients' records also were independently evaluated by one of the investigators using WebCIS and, when necessary, paper health records.
The comparison of E-screening and manual screening was summarized by calculating the sensitivity, specificity, positive predictive accuracy (PPA), and negative predictive accuracy (NPA) of the E-screening method (▶). E-screening classified 23 patients as “potentially eligible” and 102 patients as “ineligible”. All 102 patients classified by E-screening as “ineligible” were confirmed by investigator review, yielding a NPA of 100% with a 95% confidence interval (CI) between 98 and 100%. Of the 23 patients classified as “potentially eligible” by E-screening, the investigator confirmed 3 patients (“True Positives”) and reclassified the remaining 20 as “ineligible” (“False Positives”). Thus, the PPA for E-screening was 13% (95% CI: 6–13%). The investigator classified 3 patients “potentially eligible” and 122 patients “ineligible”. The sensitivity was 100% (95% CI: 45–100%) and the specificity was 84% (95% CI: 82–84%). Table 2 (available as an online data supplement at http://www.jamia.org) lists the reasons patients were excluded during the investigator chart review. Table 3 (available as an online data supplement at http://www.jamia.org) lists the reasons the investigator review excluded the 20 false positive patients who were classified as “potentially eligible” by E-screening algorithm.
Discussion
The E-screening method performed very well for excluding ineligible patients compared with investigator review for the ACCORD trial. The age criterion (“age ≥40”) decreased the study population by 68 patients, i.e., from 193 to 125 patients who comprised the evaluation group. The 100% NPA with a confidence interval between 98 and 100% indicates that it is very unlikely that an investigator review of patients who are categorized as ineligible by E-screening would actually be eligible. In our evaluation study, an investigator served as the gold standard and reviewed all patients (n = 125) regardless of their E-screening eligibility results. In real uses of this E-screening method, only those patients who are classified as “potentially eligible” (n = 23) need manual review, which translates into 81% effort saving for clinical research personnel, who otherwise would manually review records from all admitted patients (n = 193).
Some may think that the E-screening method's PPA of 13% is too low. However, this number should be considered in the appropriate context of our screening strategy. Out of 125 patients who met the age criterion, only three (2.4%) met the eligibility criteria by investigator review. The low prevalence of eligible patients in the examined population caused a low PPA. In addition, the “cost” of losing an eligible patient by E-screening is much higher than that of including an ineligible patient; therefore, our E-screening method was tuned to achieve high NPA at the expense of low PPA. We also identified reasons for the high false positive rate of this study; some of which are open research questions. For example, in EHR systems, a patient usually has multiple laboratory test values; some of them may meet the inclusion criteria and some do not. However, clinical trial protocols often do not provide specific instructions for selecting and/or aggregating multiple values for a variable. In Table 3, 8 patients were included by E-screening but later excluded by investigator review because they had multiple inconsistent HbA1c values. Our E-screening included the patients based on the first value evaluated by the algorithm that satisfied the inclusion criteria, but our investigator review revealed the inconsistency in multiple values and excluded these patients. A solution to this problem requires complete clinical trial protocol specifications that address how to handle multiple inconsistent values for a variable.
We did not include exclusion criteria such as “no evidence of type 2 diabetes mellitus; did not have cardiovascular disease or risk factors; organ transplant; taking prednisone or protease inhibitors; increased transaminases or hepatic disease” in our E-screening query for multiple reasons. A patient may have a disease but the EHR may not have a corresponding diagnosis due to common problems such as missing clinical data, heterogeneous representations, and inconsistent information (the multiple-value problem) in EHR systems. We used ICD-9 codes in our e-screening query to identify patients with certain diseases. The ICD-9 codes tend to be used to support billing or document primary medical problems; therefore, they are not an exhaustive index of patient problems. The coded items may not include those that are critical to screening for a research study. A patient may have “transplant” documented in his or her medical history notes but was coded only for more acute problems other than “transplant” in the EHR. Therefore, our E-screening query did not exclude a few patients with a transplant history, as indicated in Table 3. Our strategy was to include only straightforward exclusion criteria in our E-screening query and require manual review of complicated criteria to maximize the negative predictive accuracy of E-screening.
There is no widely accepted gold standard for evaluating an E-screening system. We believe a key feature is its negative predictive accuracy. A high negative predictive accuracy directly translates into effort savings without loss of eligible patients. A reliably high negative predictive accuracy makes it safe for clinical research staff to exclude patients, who are automatically labeled ineligible by E-screening, without manual review.
Our E-screening method differed from some earlier work, particularly that of Embi, et al, 9,10 that e-mailed alerts of potentially eligible patients to clinicians. In contrast, our semiautomatic E-screening method provides assistance to clinical research personnel, who will confirm each eligible patient before contacting primary care physicians. Embi's method serves and impacts nonresearch clinicians, while ours assists research personnel. Since there is a tradeoff between positive predictive accuracy and negative predictive accuracy, it may be important to achieve a high positive predictive accuracy for methods that interact with nonresearch clinicians, but is not very important for our method. As we stated previously (Section 2.2), our goal is to reduce the screening burden and avoid unnecessary manual review of ineligible patients, measured by the negative predictive accuracy. We consider the negative predictive accuracy as a key measurement for E-screening systems designed to reduce the screening burden for clinical research personnel.
Our E-screening method is flexible and strategic in its selection of inclusion and exclusion criteria. E-screening accommodates the data characteristics in a clinical data warehouse, includes eligibility criteria based on data completeness and protocol characteristics and is cognizant of patient status in the E-screening workflow. This E-screening workflow shown in ▶ was effective for separating “ineligible” from “pending” and “potentially eligible.” Anyone who is neither “ineligible” nor “pending” is classified as “potentially eligible”. As research staff uses the E-screening system, they can refine the workflow to improve the performance. The database query results can be communicated to clinical research personnel automatically and regularly (e.g., every 24 h).
We learned that eligibility criteria to be included in E-screening should have corresponding data elements that can be automatically queried. Therefore, it is imperative to conduct an analysis of the information gap between the eligibility criteria and the clinical data before selecting criteria to include in E-screening. During manual review after E-screening, the clinical research staff can focus their clinical judgment on handling incomplete, missing, or difficult to access data and subjective inclusion/exclusion criteria. In the most challenging cases, patients can be interviewed by clinical research personnel to assess their eligibility.
Conclusions
We evaluated a simple E-screening method that substantially reduced the screening burden for the ACCORD clinical trial. The method's high (100%) negative predictive accuracy greatly increased the efficiency of screening for clinical trial recruitment, saving time and effort. The excellent performance was partly due to using a clinical data warehouse which was empowered by a shared semantic dictionary MED for data integration and linked to a web-based EHR system for facilitated manual review. We demonstrated the value of using a carefully designed database query (i.e., selecting a subset of eligibility criteria to maximize the negative predictive accuracy of the query) together with only minimal chart review thereby reducing the trial screening burden. We predict this E-screening method will generalize to other research sites with EHR systems. Future designers of E-screening systems should achieve results similar to ours provided they carefully accommodate the data characteristics in their EHR systems; select eligibility criteria to maximize the negative predictive accuracy of their database queries; and, use a tool similar to MED to integrate heterogeneous clinical data representations. Furthermore, our E-screening method may be enhanced by adding natural language processing of free-text clinical data.
Footnotes
This research was funded under NIH/NHLBI Contracts HHSN268200455208C and N01-HC-95184, NLM grant 1R01 LM009886, and CTSA award UL1 RR024156. Its contents are solely the responsibility of the authors and do not necessarily represent the official view of NIH.
References
- 1.Sullivan J. Subject recruitment and retention: Barriers to success Appl Clin Trials April2004.
- 2.Kahn MG. Integrating electronic health records and clinical trialshttp://www.esi-bethesda.com/ncrrworkshops/clinicalResearch/pdf/MichaelKahnPaper.pdf April2004. Accessed Dec 15, 2008.
- 3. Clinical data warehouse at CPMChttp://ctcc.cpmc.columbia.edu/rdb/ April2004. Accessed Dec 15, 2008.
- 4.Jenders R, Sherman E, Hripcsak G, Fulmer T, Clayton P. Use of a computer-based decision support system for clinical research. Paper presented at: Proceedings of the 1997 AMIA Spring Congress; San Jose, CA, May 28–31, 1997:100.
- 5. The medical entities dictionary (MED)http://med.dmi.columbia.edu/ April2004. Accessed Dec 15, 2008.
- 6. The unified medical language systems (UMLS)http://www.nlm.nih.gov/research/umls/ April2004. Accessed Dec 17, 2008.
- 7.Hripcsak G, Cimino J, Sengupta SW. CIS: Large scale deployment of a web-based clinical information systemPaper presented at: Proc AMIA Symp 1999:804-808. [PMC free article] [PubMed]
- 8.Buse JB, Bigger JT, Byington RP, et al. Action to Control Cardiovascular Risk in Diabetes (ACCORD) Trial: Design and methods Am J Cardiol 2007;99:12-A21i–33i. [DOI] [PubMed] [Google Scholar]
- 9.Embi P, Jain A, Harris CM. Physicians' perceptions of an electronic health record-based clinical trial alert approach to subject recruitment: A survey BMC Med Inform Decis Mak 2008;8(13). [DOI] [PMC free article] [PubMed]
- 10.Embi PJ, Jain A, Clark J, et al. Effect of a Clinical Trial Alert system on physician participation in trial recruitment Arch Intern Med 2005;165:2272-2277. [DOI] [PMC free article] [PubMed] [Google Scholar]