Abstract
Staphylococcus aureus (S. aureus) has entered the spotlight as a globally pervasive drug-resistant pathogen. While historically associated exclusively with hospital-acquired infections in immunocompromised hosts, the methicillin-resistant form of S. aureus has been spreading throughout communities since the 1990s. Indeed, it has now become a common household term: MRSA. S. aureus has developed numerous mechanisms of virulence and strategies to evade the human immune system, including a host of surface proteins, secreted enzymes, and toxins. In hospital intensive care units, the proportion of MRSA-related S. aureus infections has increased strikingly from just 2 percent in 1974 to 64 percent in 2004. Its presence in the community has been rising similarly, posing a significant public health burden. The growing incidence of MRSA unfortunately has been met with dwindling efforts to develop new, more effective antibiotics. The continued emergence of resistant strains of bacteria such as MRSA demands an urgent revival of the search for new antibiotics.
Keywords: Staphylococcus aureus, methicillin-resistant Staphylococcus aureus, penicillin, antibiotic resistance, virulence factors, penicillin-binding proteins, community-associated MRSA, antibiotic development, 10x‘20 initiative
Introduction
Infectious diseases are the second leading cause of death worldwide and the third leading cause of death in developed countries [1]. The rapid emergence and spread of drug-resistant organisms, such as Staphylococcus aureus (S. aureus), both in the healthcare setting and the community prompts great urgency in the development of and advocacy for prevention and treatment efforts.
S. aureus, a Gram-positive bacterium, is both a commensal organism found as part of the normal human flora in 30 percent of the population as well as a resourceful human pathogen able to cause severe and devastating illness [2] (Figure 1). The precipitous spread of methicillin-resistant strains of S. aureus (MRSA) — a so-called “superbug” — has created new challenges for governments, healthcare systems, and drug development. From skin abscess and cellulitis to invasive bacteremia, endocarditis, and septic arthritis, MRSA is capable of causing significant human disease [3]. Once thought of as a hospital-acquired infection of immunocomprised hosts, MRSA found its way out of the hospitals and into communities, infecting individuals with no known risk factors. The sudden development and spread of antibiotic-resistant bacteria such as MRSA, coupled with a dwindling culture of antibiotic research and development, sets the stage for a bold multidisciplinary campaign called the 10x‘20 initiative to reinvigorate the antibiotic pipeline. In this review, we describe the pathogenesis of S. aureus-related illness, discuss common mechanisms of resistance to methicillin, and highlight the necessity for developing novel antibiotic therapies.
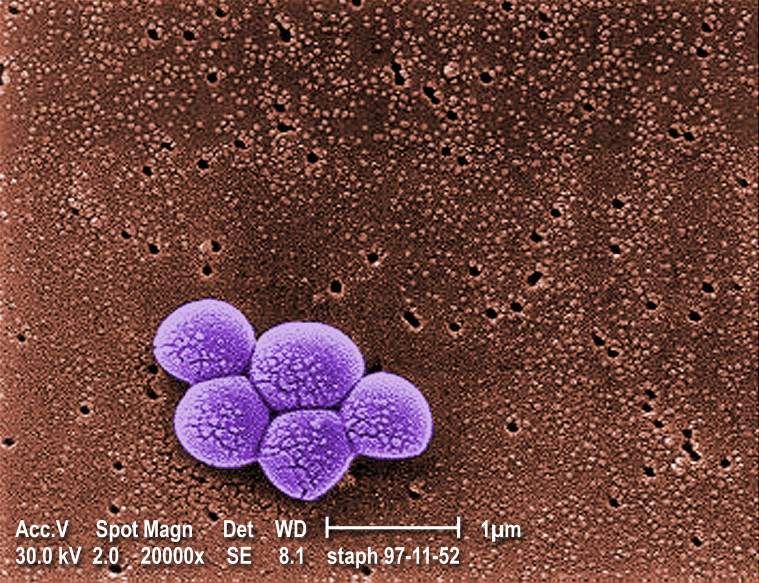
Figure 1.
Scanning electron micrograph of a cluster of Staphylococcus aureus. Image obtained from the Public Health Image Library of the Centers for Disease Control and Prevention (Janice Carr) (http://phil.cdc.gov/Phil/default.asp).
S. aureus: Pathogenesis and Virulence Factors
A critical first step in the pathogenesis of S. aureus infection is colonization. Asymptomatic colonized individuals provide a reservoir for the human-to-human spread of disease. The primary modes of transmission include direct skin-to-skin contact with a colonized source and, to a lesser extent, contact with colonized fomites [4]. Disruption of the normal skin barrier (e.g., abrasion, burn) as well as immunosuppressive conditions (e.g., HIV, steroid use, genetic diseases) predispose colonized hosts to infection [5].
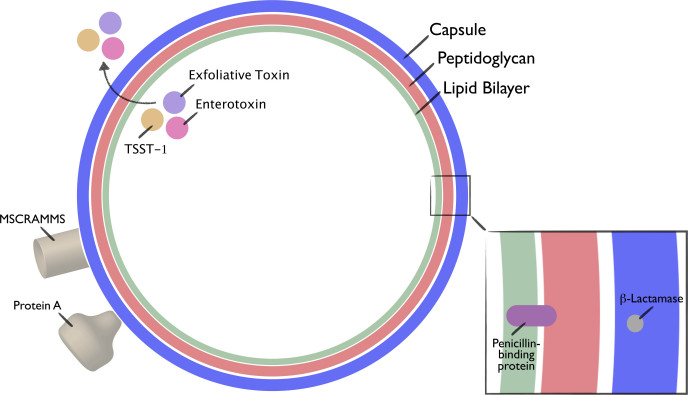
A plethora of surface proteins and secreted virulence factors endow S. aureus with great potential to cause disease (Figure 2). Understanding the detailed mechanisms by which these factors cause S. aureus-related illness provides opportunities to develop targeted therapeutics.
Figure 2.
Schematic diagram of S. aureus, depicting basic structure and a selection of virulence factors. Adapted from Gordon et al., 2008.
Surface proteins
To initiate infection, S. aureus must first adhere to host tissues or prosthetic devices. To accomplish this, S. aureus uses a constellation of surface proteins known as microbial surface components recognizing adhesive matrix molecules (MSCRAMMS) [6]. Each strain of S. aureus has its own genetic repertoire of MSCRAMMS, yielding strain-specific adhesion preferences and concomitant infection patterns. Once affixed to a surface, S. aureus capitalizes on various resources to evade the host immune system in order to yield sufficient time for an infection to take hold. One such intriguing mechanism is the formation of biofilms, surface-associated bacterial collections situated within self-made extracellular polymeric matrices that give microbial communities protection against host defenses and antibiotics [7]. Scientific inquiry into the genetic programs responsible for biofilm formation has yielded insight into the development of adjunctive therapies [8,9].
When circumstances permit invasion past physical host barriers, S. aureus deploys several surface protein-based mechanisms to survive in the midst of the host immune system. Its antiphagocytic capsule provides the primary mechanism against host phagocytic immune cells, namely neutrophils, monocytes, and macrophages [3]. Research also has demonstrated that S. aureus uses bacterial fibronectin-binding proteins (a type of MSCRAMM) to evade host immune cell phagocytosis. Specifically, to find refuge, it creates a fibronectin bridge between the bacterium and host endothelial and epithelial cell β1 integrins, allowing for its internalization and protection against extracellular immune cells [10-12]. To further avoid antibody-mediated immunity, S. aureus utilizes an additional surface protein virulence factor called Protein A. In binding to the universal Fc region of host immunoglobulins, Protein A inhibits opsonization and phagocytosis [3,13].
Secreted proteins
S. aureus employs secreted protein-based mechanisms to defend itself from host immune system phagocytosis. Greater than one-half of S. aureus isolates secrete a substance called chemotaxis inhibitory protein of S. aureus (CHIPS), which impairs neutrophil recruitment [3,14]. Isolates also can produce leukocidins, factors that disrupt leukocyte membranes by creating pores [15]. Many other secreted factors, including lactamases, proteases, lipases, nucleases, hyaluronate lyase, phospholipase C, and metalloproteinases also play a significant role in infectious spread and tissue destruction [16].
Toxins
Numerous S. aureus isolates produce toxins capable of causing specific physiologic disturbances. Toxins classified as superantigens produce a cytokine storm and provoke T cell proliferation. One such superantigen, toxic shock syndrome toxin-1 (TSST-1), results in the clinically devastating toxic shock syndrome [17]. Exfoliative toxins induce erythema as well as skin exfoliation, as is observed in the staphylococcal scalded skin syndrome [18]. Finally, S. aureus enterotoxin results in a self-limited food poisoning [19].
In summary, in response to infection, the host immune system readies for battle, and its phagocytic immune cells are its infantry. Many surface and secreted virulence factors play major roles in both avoiding and inhibiting phagocyte-based destruction. Coupled with virulence factors that aid in the disruption of tissue structure, S. aureus affords itself the opportunity to disseminate by reaching local lymphatics and blood vessels [11]. S. aureus isolates also can produce toxins, which, when present, are capable of causing specific physiologic dysfunction.
S. aureus and Disease
The clinical presentation of S. aureus infection is highly variable. It depends on the clinical isolate’s complement of virulence factors as well as the site and timing of infection [11]. Skin infections are the most frequently encountered S. aureus infections. In the hospital setting, it is most common to find S. aureus skin infections postoperatively [11]. In the community, skin infections include abscesses, bullous impetigo, folliculitis, furunculosis, and necrotizing fasciitis [20]. Clinically, in deciding whether skin infection is the result of S. aureus, important clues are provided by the skin distribution as well as the site of infection (e.g., facial, follicular, or deeper tissue) [11]. If S. aureus penetrates the barrier afforded by the skin and successfully evades the host immune system using the virulence factor-based mechanisms discussed above, it can disseminate, resulting in serious infections, including sepsis, septic arthritis, osteomyelitis, and endocarditis [3].
The clinical management of localized S. aureus skin infections is two-fold. First, the correct type and form (intravenous, topical, or oral) of antimicrobial therapy must be selected. Second, infected abscesses must be drained [11]. Severe and advanced infections require more significant and intensive management in order to address limb-threatening and/or life-threatening sequelae [21]. Several main principles govern the management of such complicated cases [11,21]: 1) diagnose and begin intravenous antimicrobial treatment early in the progression of the disease; 2) differentiate between necrotizing and non-necrotizing fasciitis; 3) identify resistant profiles as well as virulence factors; and 4) rapid surgical drainage and/or removal of necrotic tissue. Toxin-mediated S. aureus-related illnesses (e.g., toxic shock syndrome and gastroenteritis) require symptom-directed management. For example, treatment of toxic shock syndrome involves fluid replacement and blood pressure surveillance. In combating S. aureus toxins, such as TSST-1, some studies have demonstrated the utility of intravenous immunoglobulin therapy [22,23].
Antimicrobial therapy: from Penicillin to Methicillin
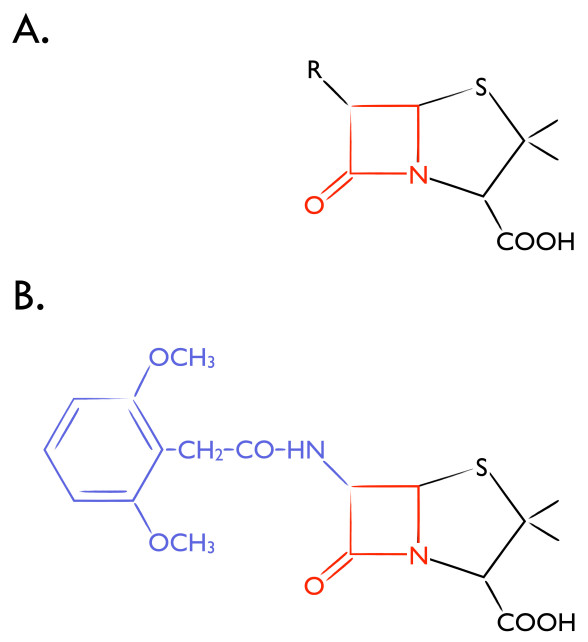
In 1928, in his laboratory in London, Scottish biologist and pharmacologist Alexander Fleming discovered that he had left open the cover of a Petri dish containing Staphylococcus, which had become contained by a blue-green mold. To his amazement, the growth of the Staphylococcus adjacent to the mold was inhibited. Fleming posited that the mold was secreting a substance that lysed its bacterial neighbors. It was this discovery of a blue-green mold, Penicillium notatum, and the resulting unearthing of the inhibiting substance, penicillin, for which Fleming shared the 1945 Nobel Prize in Physiology or Medicine [24,25] (Figure 3).
Figure 3.
β-lactam Antibiotics. Chemical structure of (A) Penicillin and (B) Methicillin. Purple indicates modification of the carbonyl group side chain of the penicillin core that facilitates β-lactamase resistance. β-lactam ring in red.
The bacterial cell wall is important for maintenance of cell shape and protection against osmotic lysis. Therefore, pharmacologic agents that disrupt this critical bacterial component would likely be bactericidal. Penicillin, a β-lactam antibiotic, covalently binds to and inhibits a collection of bacterial proteins called penicillin binding proteins (PBPs), which are responsible for the construction, maintenance, and regulation of the peptidoglycan portion of the cell wall [26]. S. aureus normally has four PBPs (PBP1-4); PBP1, 2, and 3 are essential and exhibit high affinity for β-lactam antibiotics while PBP4 carries out secondary cross-linking of the peptidoglycan [27,28].
Shortly after the introduction of penicillin as a clinical therapeutic in the 1940s, a strain of Staphylococcus aureus rapidly emerged that secreted an enzyme called penicillinase, which hydrolyzes penicillin into inactive penicilloic acid. In the 1950s, scientists at a United Kingdom-based pharmaceutical company, Beecham, discovered that placing bulky substituents on the penicillin side chain would protect penicillins from penicillinase-mediated destruction due to steric hindrance. Subsequently, in 1959, Beecham introduced methicillin, a class of penicillinase resistant β-lactams [29] (Figure 3). Unfortunately, however, the first cases of methicillin-resistant S. aureus were reported in Europe just a few years later [30]. Then, less than a decade later, MRSA was identified in the United States at Boston City Hospital in almost 20 patients with evidence of patient-to-patient spread [31].
Mechanisms of S. aureus resistance to Methicillin
In order to evade the bactericidal properties of methicillin, S. aureus has developed several modes of resistance. Such mechanisms include the expression of a methicillin-hydrolyzing β lactamase as well as the expression of an altered form of PBP2 that binds to methicillin with lower affinity and with higher rates of methicillin release.
The most prevalent mode of methicillin resistance in S. aureus, however, involves the chromosomal presence of a large 40-60 kilobase stretch of foreign DNA called the mec element. The mecA gene, nested within the mec element, encodes for the 76 kDa protein called PBP2a, a unique and newly acquired PBP that exhibits a low affinity for β-lactam antibiotics. Therefore, when the normal staphylococcal PBPs are inhibited by β-lactams, PBP2a can resume cell wall assembly, enabling viability in the presence of methicillin [24].
Data suggests that mecA originated from Staphylococcus sciuri, but the mechanism by which this genomic transfer occurred remains mysterious, perhaps involving recombinase proteins capable of excising DNA from one place and integrating it into another [25,26]. Investigating the clonality of 472 MRSA isolates using DNA hybridization technology, Kreiswirth et al. determined that this genetic relocation occurred once and that all MRSA isolates are descendants of this single clone [27].
Research on the basic properties of PBP2a has revealed novel targets for therapeutic intervention. Like other PBPs, PBP2a catalyzes the formation of peptide crosslinks (transpeptidation) between the gylcan chains of the cell wall. However, PBP2a requires atypical cell wall precursor molecules: a pentaglycine-decorated side chain attached to the position 3-L-lysine of the stem peptide, as well as an amidated D-glutamine in position 2. Construction of these peculiar substrate molecules requires the assistance of many accessory genes, including femABC and fmhV, which are responsible for adding these critical residues. Therefore, any therapeutic intervention that perturbs the function of these accessory genes would diminish methicillin resistance despite the continued presence of PBP2a.
β-lactam antibiotics effectively inhibit the normal PBPs produced by S. aureus. Therefore, therapeutics specifically targeted against PBP2a would be of great utility against MRSA. In 2002, Lim et al. solved the crystal structure of a soluble derivative of PBP2a, providing a detailed molecular map of the active site cavity [32]. This crystallographic analysis provides important clues for understanding how some β-lactams target PBP2a and for the rational design of novel molecular therapies [33].
The Spread of MRSA
MRSA is spreading and causing disease at a rapid rate. Outbreaks have frequently been reported in neonatal and surgical intensive care units, burn units, inpatient wards, and operating rooms [34]. The patient-to-patient nosocomial transmission responsible for such outbreaks predominantly occurs through the hands of healthcare workers [35]. The proportion of MRSA-related S. aureus infections in hospital intensive care units has increased steadily from 2 percent in 1974 to 22 percent in 1995 and 64 percent in 2004 [36]. In 2005, 58 percent (278,000) of hospitalizations that included a diagnosis of S. aureus were caused by MRSA (including those admitted to the hospital for community-acquired infections) [37]. According to a 2005 estimate, nearly 19,000 deaths were caused by MRSA [38]. Moreover, MRSA is now considered one of the leading causes of death by any single infectious pathogen [39]. MRSA, therefore, has expeditiously become a significant health burden to society.
Fortunately, major hospital efforts and campaigns to reduce hospital-acquired infections have helped diminish the incidence of MRSA-related illness [40]. These efforts include: 1) reduction in antibiotic use; 2) healthcare worker education; 3) hand washing protocols and monitoring; 4) surveillance cultures; and 5) isolation of patients colonized with MRSA or perceived as a high risk for infection [41]. Active surveillance cultures and isolation protocols remain controversial as some studies have demonstrated the lack of utility in employing widespread screening efforts outside of the intensive care units [41].
Hospital-based movements to reduce the incidence of MRSA have undoubtedly demonstrated success. Between 2001 and 2007, MRSA central line-associated bloodstream infections within intensive care units have decreased by nearly 50 percent [42]. Similarly, a separate study that investigated hospitalized patients between 2005 and 2008 demonstrated a 34 percent reduction in the incidence of MRSA-related bloodstream infections [40]. Continued efforts must be exercised to prevent the spread of MRSA in the hospital setting. New strategies may include attempts to minimize the length of hospital stay and improved surveillance techniques, as well as more strict hand hygiene regulations [35].
From the Hospital to the Community
While MRSA was once just a hospital-acquired infection observed in immunocompromised hosts, the rapid and persistent emergence of community-associated MRSA (CA-MRSA) has caused considerable concern. Illustrative of its means to quickly disseminate through populations is its pervasive international presence and its ability to spread between continents [39,43]. The medical community recognized the first cases of CA-MRSA in the 1990s, first in remote Western Australia [44] and then with a group of otherwise healthy children in the Midwestern United States [45]. None of the affected individuals had contact with the healthcare setting or exhibited any identified risk factors prior to infection. The ability of CA-MRSA to cause disease in immunocompetent healthy hosts suggests that CA-MRSA harbors novel mechanisms of virulence [39]. Genetic analyses of CA-MRSA isolates support this assertion, as several isolates harbor a unique mec element as well as express the Panton-Valentine leukocidin (PVL), a factor linked to severe skin infections in the community [3,46]. Given that nearly 90 percent of CA-MRSA cases are skin and soft tissue infections, 90 percent of which are abscesses or cellulitis [39], it is likely that PVL plays a significant role in the pathogenesis of CA-MRSA. Future concern and attention is certainly warranted. The presence of severe community-acquired infections such as purpura fulminans, myositis, osteomyolitis, and necrotizing fasciitis suggests the potential for increased virulence [39].
The emergence of CA-MRSA as a growing threat has posed new preventive and therapeutic challenges. Akin to hospital-acquired MRSA, the community variant demonstrates extensive resistance to β-lactam antibiotics. Currently, little clinical evidence exists regarding the efficacy of alternative agents. Thus, most non-invasive CA-MRSA skin infections are not treated with antibiotics. Instead, abscesses are simply drained [39]. If antibiotics are indicated as the result of more significant signs of infection, clindamycin, tetracyclines (including doxycyline), trimethoprim-sulfamethoxazole, and linezolid are the drugs of choice [47]. More severe cases of CA-MRSA are treated with parenteral vancomycin, daptomycin, teicoplanin, or linezolid [48].
The Antibiotic Pipeline
The Centers for Disease Control (CDC) estimates that roughly two million people in the United States will develop bacterial infections while in the hospital and that nearly 90,000 will die from associated complications. Close to 70 percent of the bacteria responsible for these infections will be resistant to at least one commonly used antibiotic. In recent decades, antibiotic resistant organisms have spread at alarming rates, causing the Institute of Medicine, CDC, National Institutes of Health, and the Food and Drug Administration (FDA) to caution that drug-resistant organisms pose a serious public health concern [49].
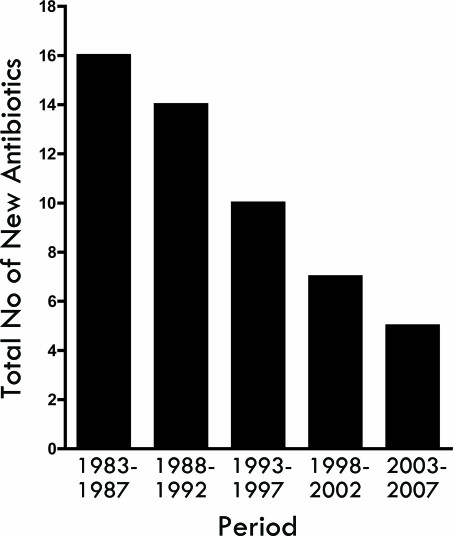
Given the rapid spread of drug-resistant bacteria, one might suspect that the pharmaceutical and biotechnology industries would be countering with an equally impressive course of antimicrobial development. However, the spread of drug-resistant organisms has largely outpaced antibiotic research and development. In fact, in comparing the periods of 1998 to 2002 and 1983 to 1987, FDA approval of new antibiotics decreased 56 percent, with no new antibacterial agents approved in 2002. A startling downward trend emerges when examining the number of new systemic antibiotics approved by the FDA over the last 25 years [50] (Figure 4).
Figure 4.
Number of new antimicrobials approved by the U.S. Food and Drug Administration, per five-year period. Adapted from Spellberg et al., 2008.
Many reasons could explain the paucity of research and development in antimicrobial therapeutics, most of which point to financial considerations. Two independent studies from 2001 and 2003 estimate that the cost of discovering and then developing a new drug in the United States exceeds $800 million [51,52]. This figure presents a significant investment challenge for the development of drugs targeted against short course therapies that cure their target disease, leaving most of drug discovery focused on chronic lifelong non-curative diseases. A further impediment to ongoing development is that antibiotics are the only class of drugs whose effectiveness decreases the more they are employed, which causes leaders to advocate for restrictions on and judicial use of new antimicrobial therapies. Lastly, the pharmaceutical industry cites the lack of clear guidance from regulatory agencies as a deterrent for ongoing antibiotic development. They express uncertainty about what types of safety and efficacy data will be deemed appropriate at the time of formal drug application [53].
In response to the pervasive problem of drug-resistant organisms coupled with a limited developmental pipeline for drug discovery, the Infectious Diseases Society of America (IDSA), with the broad support of leading medical societies and organizations, has launched the 10x‘20 initiative. The bold aim of this collaboration between scientific, industry, political, economic, policy, medical, intellectual property, and philanthropic leaders is the development of 10 new, safe, and effective antibiotics by 2020. To do this, the 10x‘20 initiative advocates for the development of new systemic antibacterial therapeutics not only through the discovery of new drug classes, but also through the evolution of more effective drugs from existing classes of antibiotics. Global stakeholders recognize the imminent need for antimicrobial development. With the IDSA’s support, in 2009, U.S. President Barack Obama and Swedish Prime Minister Fredrik Reinfeldt, acting on behalf of the European Union, created a transatlantic task force to focus on solutions to the dwindling antibiotic pipeline and find ways to fortify infection control interventions and practices [54].
The Search for Novel Antibiotics
While the impending crisis has worsened over the past decade, academic and biotechnology companies have made significant and noteworthy progress toward development of effective antimicrobial therapeutics. One such example is the research of Dr. Andrew G. Myers of the Department of Chemistry and Chemical Biology at Harvard University, who revealed a novel route and robust platform for the synthesis of new tetracycline antibiotics [55,56]. Tetraphase Pharmaceuticals, a company based on this research, already has developed a cadre of drug candidates with the potential to treat a wide range of infectious diseases. Moreover, a lead drug candidate called TP-434 has shown great preclinical promise as a potent antibacterial against a broad spectrum of susceptible and multidrug-resistant organisms [57].
The Nobel Prize for Chemistry in 2009 highlights a seminal scientific discovery holding immense relevance to antimicrobial therapeutic development. Drs. Ada Yonath (Israel), Thomas Steitz (United States), and Venki Ramakrishnan (United Kingdom) were awarded the Nobel Prize for elucidating the atomic three-dimensional structure of the ribosome using X-ray crystallography, the machine within cells responsible for making proteins and the target of many of today’s antimicrobial agents. In pioneering work, Dr. Steitz, a Sterling Professor of Molecular Biophysics and Biochemistry and Howard Hughes Medical Institute Investigator at Yale University, and colleagues demonstrated the structural basis of bacterial resistance to clinically important antibiotics, revealing important insights into novel antibiotic development [58]. Based on these important observations, Dr. Steitz and his colleagues have founded Rib-X Pharmaceuticals [59]. Rib-X is developing a novel class of broad-spectrum antibitiocs called RX-04. In addition, delafloxaxin, Rib-X’s novel fluroquinolone antibiotic, has successfully completed three Phase 2 clinical studies and demonstrated utility in the fight against MRSA [60].
Conclusion and Outlook
Without a doubt, the medical community is now astutely aware of the global burden of disease imposed by drug-resistant S. aureus infections. The ubiquitous knowledge of MRSA in communities is evident by its common use as a new household term. The consequences of the MRSA pandemic are profound and worrisome. MRSA itself is only a symptom of a broader phenomenon, a harbinger of a growing pool of resistant pathogens found both in hospitals and in the community. While researchers, both academic and pharmaceutical, are working to develop therapeutic responses and have already have made significant advances, considerable work remains. The continued emergence of resistant strains of bacteria such as MRSA demands an urgent response. An efficient, comprehensive, multidisciplinary search for new antibiotics must commence to prepare us to squelch strains of resistant pathogens as they inevitably strike.
Abbreviations
- MSCRAMMS
microbial surface components recognizing adhesive matrix molecules
- CHIPS
chemotaxis inhibitory protein of S. aureus
- TSST-1
toxic shock syndrome toxin-1
- PBPs
penicillin binding proteins
- CA-MRSA
community-associated MRSA
- PVL
Panton-Valentine leukocidin
- CDC
Centers for Disease Control
- FDA
Food and Drug Administration
- IDSA
Infectious Diseases Society of America
References
- Fauci AS. Infectious diseases: considerations for the 21st century. Clin Infect Dis. 2001;32:675–685. doi: 10.1086/319235. [DOI] [PubMed] [Google Scholar]
- Gorwitz RJ. et al. Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001-2004. J Infect Dis. 2008;197:1226–1234. doi: 10.1086/533494. [DOI] [PubMed] [Google Scholar]
- Foster TJ. Immune evasion by staphylococci. Nat Rev Microbiol. 2005;3:948–958. doi: 10.1038/nrmicro1289. [DOI] [PubMed] [Google Scholar]
- Miller LG, Diep BA. Clinical practice: colonization, fomites, and virulence: rethinking the pathogenesis of community-associated methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46:752–760. doi: 10.1086/526773. [DOI] [PubMed] [Google Scholar]
- Chambers HF, Deleo FR. Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat Rev Microbiol. 2009;7:629–641. doi: 10.1038/nrmicro2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti JM. et al. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- Sritharan M, Sritharan V. Emerging problems in the management of infectious diseases: the biofilms. Indian J Med Microbiol. 2004;22:140–142. [PubMed] [Google Scholar]
- Geoghegan JA. et al. The Role of Surface Protein SasG in Biofilm Formation by Staphylococcus aureus. J Bacteriol. 2010;192(21):5663–5673. doi: 10.1128/JB.00628-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirioni O. et al. Daptomycin and Rifampin Alone and in Combination Prevent Vascular Graft Biofilm Formation and Emergence of Antibiotic Resistance in a Subcutaneous Rat Pouch Model of Staphylococcal Infection. Eur J Vasc Endovasc Surg. doi: 10.1016/j.ejvs.2010.08.009. Forthcoming 2010. [DOI] [PubMed] [Google Scholar]
- Schwarz-Linek U. et al. The molecular basis of fibronectin-mediated bacterial adherence to host cells. Mol Microbiol. 2004;52:631–641. doi: 10.1111/j.1365-2958.2004.04027.x. [DOI] [PubMed] [Google Scholar]
- Tang YW, Stratton CW. Staphylococcus aureus: An old pathogen with new weapons. Clin Lab Med. 2010;30:179–208. doi: 10.1016/j.cll.2010.01.005. [DOI] [PubMed] [Google Scholar]
- Fowler T. et al. Cellular invasion by Staphylococcus aureus involves a fibronectin bridge between the bacterial fibronectin-binding MSCRAMMs and host cell beta1 integrins. Eur J Cell Biol. 2000;79:672–679. doi: 10.1078/0171-9335-00104. [DOI] [PubMed] [Google Scholar]
- Deisenhofer J. Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-A resolution. Biochemistry. 1981;20:2361–2370. [PubMed] [Google Scholar]
- de Haas CJ. et al. Chemotaxis inhibitory protein of Staphylococcus aureus, a bacterial antiinflammatory agent. J Exp Med. 2004;199:687–695. doi: 10.1084/jem.20031636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladstone GP, Van Heyningen WE. Staphylococcal leucocidins. Br J Exp Pathol. 1957;38:123–137. [PMC free article] [PubMed] [Google Scholar]
- Gordon RJ, Lowy FD. Pathogenesis of methicillin-resistant Staphylococcus aureus infection. Clin Infect Dis. 2008;46(Suppl 5):S350–S359. doi: 10.1086/533591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd J. et al. Toxic-shock syndrome associated with phage-group-I Staphylococci. Lancet. 1978;2:1116–1118. doi: 10.1016/s0140-6736(78)92274-2. [DOI] [PubMed] [Google Scholar]
- Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- Gilbert RJ. et al. Serological detection of enterotoxin in foods implicated in staphylococcal food poisoning. J Hyg (Lond) 1972;70:755–762. doi: 10.1017/s0022172400022592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden MS. Skin and soft tissue infection: microbiology and epidemiology. Int J Antimicrob Agents. 2009;34(Suppl 1):S2–S7. doi: 10.1016/S0924-8579(09)70541-2. [DOI] [PubMed] [Google Scholar]
- Napolitano LM. Severe soft tissue infections. Infect Dis Clin North Am. 2009;23:571–591. doi: 10.1016/j.idc.2009.04.006. [DOI] [PubMed] [Google Scholar]
- Gauduchon V. et al. Neutralization of Staphylococcus aureus Panton Valentine leukocidin by intravenous immunoglobulin in vitro. J Infect Dis. 2004;189:346–353. doi: 10.1086/380909. [DOI] [PubMed] [Google Scholar]
- Yanagisawa C. et al. Neutralization of staphylococcal exotoxins in vitro by human-origin intravenous immunoglobulin. J Infect Chemother. 2007;13:368–372. doi: 10.1007/s10156-007-0551-6. [DOI] [PubMed] [Google Scholar]
- Stapleton PD, Taylor PW. Methicillin resistance in Staphylococcus aureus: mechanisms and modulation. Sci Prog. 2002;85:57–72. doi: 10.3184/003685002783238870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SW. et al. Recruitment of the mecA gene homologue of Staphylococcus sciuri into a resistance determinant and expression of the resistant phenotype in Staphylococcus aureus. J Bacteriol. 2001;183:2417–2424. doi: 10.1128/JB.183.8.2417-2424.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama Y. et al. A new class of genetic element, staphylococcus cassette chromosome mec, encodes methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 2000;44:1549–1555. doi: 10.1128/aac.44.6.1549-1555.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreiswirth B. et al. Evidence for a clonal origin of methicillin resistance in Staphylococcus aureus. Science. 1993;259:227–230. doi: 10.1126/science.8093647. [DOI] [PubMed] [Google Scholar]
- Georgopapadakou NH. Penicillin-binding proteins and bacterial resistance to beta-lactams. Antimicrob Agents Chemother. 1993;37:2045–2053. doi: 10.1128/aac.37.10.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau R, Pharmaceutical innovation : revolutionizing human health. Philadelphia: Chemical Heritage Press; 1999. [Google Scholar]
- Barber M. Methicillin-resistant staphylococci. J Clin Pathol. 1961;14:385–393. doi: 10.1136/jcp.14.4.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett FF. et al. Methicillin-resistant Staphylococcus aureus at Boston City Hospital. Bacteriologic and epidemiologic observations. N Engl J Med. 1968;279:441–448. doi: 10.1056/NEJM196808292790901. [DOI] [PubMed] [Google Scholar]
- Lim D, Strynadka NC. Structural basis for the beta lactam resistance of PBP2a from methicillin-resistant Staphylococcus aureus. Nat Struct Biol. 2002;9:870–876. doi: 10.1038/nsb858. [DOI] [PubMed] [Google Scholar]
- Guignard B. et al. Beta-lactams against methicillin-resistant Staphylococcus aureus. Curr Opin Pharmacol. 2005;5:479–489. doi: 10.1016/j.coph.2005.06.002. [DOI] [PubMed] [Google Scholar]
- Sheretz RJ. et al. A cloud adult: the Staphylococcus aureus-virus interaction revisited. Ann Intern Med. 1996;124:539–547. doi: 10.7326/0003-4819-124-6-199603150-00001. [DOI] [PubMed] [Google Scholar]
- Henderson DK. Managing methicillin-resistant staphylococci: a paradigm for preventing nosocomial transmission of resistant organisms. Am J Med. 2006;119:S45-52, discussion S62-70. doi: 10.1016/j.amjmed.2006.04.002. [DOI] [PubMed] [Google Scholar]
- Klevens RM. et al. Changes in the epidemiology of methicillin-resistant Staphylococcus aureus in intensive care units in US hospitals, 1992-2003. Clin Infect Dis. 2006;42:389–391. doi: 10.1086/499367. [DOI] [PubMed] [Google Scholar]
- Klein E. et al. Hospitalizations and deaths caused by methicillin-resistant Staphylococcus aureus, United States, 1999-2005. Emerg Infect Dis. 2007;13:1840–1846. doi: 10.3201/eid1312.070629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klevens RM. et al. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA. 2007;298:1763–1771. doi: 10.1001/jama.298.15.1763. [DOI] [PubMed] [Google Scholar]
- Deleo FR. et al. Community-associated methicillin-resistant Staphylococcus aureus. Lancet. 2010;375:1557–1568. doi: 10.1016/S0140-6736(09)61999-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallen AJ. et al. Health care-associated invasive MRSA infections, 2005-2008. JAMA. 2010;304:641–648. doi: 10.1001/jama.2010.1115. [DOI] [PubMed] [Google Scholar]
- Tacconelli E. Methicillin-resistant Staphylococcus aureus: source control and surveillance organization. Clin Microbiol Infect. 2009;15(Suppl 7):31–38. doi: 10.1111/j.1469-0691.2009.03096.x. [DOI] [PubMed] [Google Scholar]
- Burton DC. et al. Methicillin-resistant Staphylococcus aureus central line-associated bloodstream infections in US intensive care units, 1997-2007. JAMA. 2009;301:727–736. doi: 10.1001/jama.2009.153. [DOI] [PubMed] [Google Scholar]
- Harris SR. et al. Evolution of MRSA during hospital transmission and intercontinental spread. Science. 2010;327:469–474. doi: 10.1126/science.1182395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udo EE. et al. Genetic analysis of community isolates of methicillin-resistant Staphylococcus aureus in Western Australia. J Hosp Infect. 1993;25:97–108. doi: 10.1016/0195-6701(93)90100-e. [DOI] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention. Four pediatric deaths from community-acquired methicillin-resistant Staphylococcus aureus—Minnesota and North Dakota, 1997-1999. JAMA. 1999;282:1123–1125. [PubMed] [Google Scholar]
- Prevost G. et al. Panton-Valentine leucocidin and gamma-hemolysin from Staphylococcus aureus ATCC 49775 are encoded by distinct genetic loci and have different biological activities. Infect Immun. 1995;63:4121–4129. doi: 10.1128/iai.63.10.4121-4129.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorwitz RJ, Strategies for clinical management of MRSA in the community: Summary of an experts’ meeting convened by the Centers for Disease Control and Prevention. Centers for Disease Control [Internet] 2006. Available at: http://www.cdc.gov/ncidod/dhqp/pdf/ar/CAMRSA_ExpMtgStrategies.pdf .
- Nathwani D. et al. Guidelines for UK practice for the diagnosis and management of methicillin-resistant Staphylococcus aureus (MRSA) infections presenting in the community. J Antimicrob Chemother. 2008;61:976–994. doi: 10.1093/jac/dkn096. [DOI] [PubMed] [Google Scholar]
- Bad Bugs, No Drugs: As Antibiotic Discovery Stagnates ... A Public Health Crisis Brews. Infectious Diseases Society of America [Internet] 2004. Available at: http://www.idsociety.org/10x20.htm .
- Spellberg B. et al. Trends in antimicrobial drug development: implications for the future. Clin Infect Dis. 2004;38:1279–1286. doi: 10.1086/420937. [DOI] [PubMed] [Google Scholar]
- DiMasi JA. et al. The price of innovation: new estimates of drug development costs. J Health Econ. 2003;22:151–185. doi: 10.1016/S0167-6296(02)00126-1. [DOI] [PubMed] [Google Scholar]
- Tollman P, A Revolution in R&D: How Genomics and Genetics are Transforming the Biopharmaceutical Industry. Boston: Boston Consulting Group; 2001. [Google Scholar]
- Spellberg B. et al. The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis. 2008;46:155–164. doi: 10.1086/524891. [DOI] [PubMed] [Google Scholar]
- Infectious Diseases Society of America. The 10 x ‘20 Initiative: pursuing a global commitment to develop 10 new antibacterial drugs by 2020. Clin Infect Dis. 2010;50:1081–1083. doi: 10.1086/652237. [DOI] [PubMed] [Google Scholar]
- Charest MG. et al. A convergent enantioselective route to structurally diverse 6-deoxytetracycline antibiotics. Science. 2005;308:395–398. doi: 10.1126/science.1109755. [DOI] [PubMed] [Google Scholar]
- Sun C. et al. A robust platform for the synthesis of new tetracycline antibiotics. J Am Chem Soc. 2008;130:17913–17927. doi: 10.1021/ja806629e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tetraphase to Present Extensive Data on Novel Broad Spectrum Antibiotic TP-434 at ICAAC. Drugs.com [Internet] 2010. Available at: http://www.drugs.com/clinical_trials/tetraphase-present-extensive-data-novel-broad-spectrum-antibiotic-tp-434-icaac-10049.html .
- Tu D. et al. Structures of MLSBK antibiotics bound to mutated large ribosomal subunits provide a structural explanation for resistance. Cell. 2005;121:257–270. doi: 10.1016/j.cell.2005.02.005. [DOI] [PubMed] [Google Scholar]
- The Nobel Prize in Chemistry 2009 ― Press Release. Nobelprize.org [Internet] 2010. [cited 2010 Oct 24]. Available at: http://nobelprize.org/nobel_prizes/chemistry/laureates/2009/press.html .
- Rib-X Pharmaceuticals Presents Data Supporting Delafloxacin as a Potential Best-in-class Fluoroquinolone at ICAAC ― Delafloxacin's Unique Chemical Structure Leads to Increased Potency and Efficacy at the Site of Infection. Rib-X.com [Internet] 2010. [cited 2010 Oct 24]. Available at: http://www.rib-x.com/news_and_events/press_releases .