Abstract
Cadmium (Cd+2), a known carcinogen mimics the effects of estrogen in the uterus and mammary gland suggesting its possible involvement in the development and progression of breast cancer. This lab showed through analysis of a small set of archival human diagnostic specimens that the third isoform of the classic Cd+2 binding protein metallothionein (MT-3), is not expressed in normal breast tissue, but is expressed in some breast cancers and that expression tends to correlate with a poor disease outcome. The goals of the present study were to verify that overexpression of MT-3 in a large set of archival human diagnostic specimens tends to correlate with poor disease outcome and define the mechanism of MT-3 gene regulation in the normal breast epithelial cell. The results showed that MT-3 was expressed in approximately 90% of all breast cancers and was absent in normal breast epithelium. The lack of MT-3 staining in some cancers correlated with a favorable patient outcome. High frequency of MT-3 staining was also found for in situ breast cancer suggesting that MT-3 might be an early biomarker for breast cancer. The study also demonstrated that the MCF-10A cell line, an immortalized, non-tumorigenic model of human breast epithelial cells, displayed no basal expression of MT-3, nor was it induced by Cd+2. Treatment of the MCF-10A cells with the demethylation agent, 5-Aza-2′-deoxycytidine, or the histone deacetylase inhibitor, MS-275, restored MT-3 mRNA expression. It was also shown that the MT-3 metal regulatory elements are potentially active binders of protein factors following treatment with these inhibitors suggesting that MT-3 expression may be subject to epigenetic regulation.
Introduction
Cadmium is classified by the International Agency for Research on Cancer as a human carcinogen (IARC, 1980, 1993). Precise role of Cd+2 remains undefined in the development and progression of individual types of human cancers. Cadmium, a major environmental pollutant is a contaminant in most human foodstuffs because of high rates of soil-to-plant transfer in conjunction with continuing mobilization of Cd+2 from non-bioavailable geologic matrices into biologically accessible situations (Oskarsson et al, 2004; Clemens, 2006; Franz et al., 2008). This makes diet a primary exposure source among non-smoking, non-occupationally exposed populations. Studies showed that Cd+2 mimics the effects of estrogen in the uterus and mammary gland, suggesting a possible involvement of Cd+2 exposure in the development and progression of breast cancer (Johnson et al., 2003; Darbre, 2006). There is also epidemiological evidence that Cd+2 exposure might increase breast cancer risk (McElroy et al., 2006). This study reinforces an earlier report from Finland that defined a possible association between human breast concentrations of Cd+2 and breast cancer (Antila et al., 1996). Other reports demonstrated that Cd+2 influenced estrogenic responses in breast cancer cells (Stoica et al., 2000; Garcia-Morales et al., 1994; Brama et al., 2007). The metallothionein proteins (MT) are members of a gene family that encode low molecular weight (6 kD), intracellular proteins that have a very high conserved number of cysteine residues that allow the efficient binding of transition metals, including Cd+2 (Hamer, 1986). The MT-1 and MT-2 members of this gene family have been extensively studied and are believed to serve an important role in the homeostasis of essential metals such as zinc (Zn+2) and copper (Cu+2) during growth and development as well as in the detoxification of heavy metals such as Cd+2 and mercury (Hg+2); rendering the MT important mediators and attenuators of heavy metal-induced toxicity (Hamer, 1986; Andrews, 2000; Cousins, 1983; Goering and Klaassen, 1983; Kagi, 1993; Kagi and Hunziker, 1989). Sens et al. (2001) found through analysis of a small set of archival human diagnostic specimens that the third isoform of metallothionein (MT-3) is not expressed in tissues of the normal breast, but is expressed in some breast cancers and that expression tends to correlate with a poor disease outcome.
The MT-3 isoform has seen only limited study in general and studies from this lab apparently are the only studies of MT-3 in human breast cancer. The MT-3 isoform also has unique properties when compared to the extensively studied MT-1 and MT-2 isoforms. MT-3 was originally named growth inhibitory factor (GIF), but was renamed when it was shown to possess all the characteristic features of the traditional MT (Tsuji et al., 1992; Uchida et al., 1991). There are several important differences between MT-3 and other members of the MT gene family. The MT-3 isoform is not ubiquitous in its expression like the MT-1 and MT-2 members of the MT family. MT-3 was found to possess 7 additional amino acids that are not present in any other member of the MT gene family; a 6 amino acid C-terminal sequence and a Thr in the N-terminal region (Tsuji et al., 1992; Uchida et al., 1991; Palmiter et al., 1992). Functionally, MT-3 was demonstrated to possess a neuronal cell growth inhibitory activity which is not duplicated by the other human MT classes (Uchida et al., 1991). The goals of the present study were (1) to extend the preliminary observation of MT-3 overexpression in human breast cancer to a larger set of patient specimens and (2) further define the mechanism of MT-3 gene regulation in the normal breast epithelial cell.
Materials and methods
Archival specimens for immunohistochemical analysis of MT-3 expression
The study utilized 276 formalin-fixed, paraffin-embedded archival specimens removed from 222 patients undergoing treatment for breast cancer. Each specimen was examined on Hematoxylin and Eosin (H&E) stained sections before immunostaining to confirm the diagnosis and then subsequently stained for immunoreactivity to MT-3. The patient blocks that were used for immunohistochemical analysis had patient identifiers stripped off and the relevant clinical data of each patient was extracted from the charts. The patient identifiers were then burned off the appropriate block casements. Since there is no personal confidentiality risk to the patient, the study was granted exempt status by the University Institutional Review Board and no consents from the patients were obtained for the study.
Demographics of the patient specimens
Of the 222 patients, two patients were male and 220 were female. The average age was 60.05 years with a range from 27–93 years of age. Eight patients had only CIS lesions (DCIS, LCIS); one had a comedocarcinoma, the rest had invasive breast carcinoma. Menopausal status for women included 155 post-menopausal; 32 pre-menopausal and 33 clinically described as peri-menopausal or were in the age range 45–55 without menopausal status present in the medical record. Staging information for the patients was Stage 0: 8 patients; Stage 1: 15 patients; Stage 2: 85 patients; Stage 3: 41 patients and Stage 4: 39 patients. In this retrospective study, 34 patients did not have sufficient information to distinguish between stage 1 or 2. For 198 patients, tissue was available from the breast neoplasm; in 15 patients, only nodal or metastatic lesions were available for study; in 9 patients only CIS lesions were available for study. Outcome information was available for 179 patients; the average follow-up time was 7.31 years with a range of 1 – 311 months (25.9 years).
Immunohistochemical localization of MT-3
The preparation of the affinity purified antibody against MT-3 and the methods used for immunostaining the tissue have been described previously (Garrett et al., 1999; Zhou et al., 2006). The semi-quantitative assessment of immunohistochemical staining of MT-3 was analyzed using the multiplicative quickscore method (Detre et al., 1995), which accounts for both the intensity and the percentage of cells staining, yielding values from 0 to 18. Intensity of staining was categorized as negative = 0, weak = 1, moderate = 2 or intense = 3. This number was multiplied by a number based on the % of tissue showing positive staining (0–4% = 1, 5–19% = 2, 20–39% = 3, 40–59% = 4, 60–79% = 5, 80–100% = 6).
Cell culture, stable transfection and analysis of MT-3 mRNA and protein
The MCF-10A cell line was obtained from the American Type Culture Collection and were cultured as described previously (Gurel et al., 2005). Preliminary experiments were performed on the MCF-10A cell line to determine the approximate concentrations of cadmium chloride (CdCl2) (Sigma-Aldrich, St. Louis, MO) that would result in variable degree of cell toxicity over a 16-day period of exposure. These 4 concentrations of Cd+2 were found to be 3, 7, 15 and 30 μM. Cell viability, as an indicator of cytotoxicity, was determined by measuring the capacity of the cells to reduce MTT (3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide) to formazan (Rossi et al., 2002). Triplicate cultures were analyzed for each time point and concentration.
The MCF-10A cell line was stably transfected with the MT-3 coding sequence using methods described previously (Dutta et al., 2002; Kim et al., 2002). Clones were selected using cloning rings and the stable transfectants were identified, recloned and preserved in liquid nitrogen storage.
The procedures for the isolation of total RNA and analysis of MT gene expression by RT-PCR from cultured cells and the immuno-blot protocol used for the determination of the levels of MT-1 and 2, and MT-3 protein in cultured cell lysates has been described previously by this lab (Zhou et al., 2006; Somji et al., 2006).
Treatment of MCF-10A cells with 5-Aza-2′-deoxycytidine and histone deacetylase inhibitor MS-275
The MCF-10A cells were seeded at a ratio of 1:10 and the next day they were exposed to 0.5 and 1.5 μM 5-aza-2′-deoxycytidine (AZC) (Sigma-Aldrich) or the histone deacetylase inhibitor MS-275 (ALEXIS Biochemicals, Lausen, Switzerland) at 1, 3, or 10 μM for three days until the cells reached confluency. Cells were then harvested for MT-3 expression which was assessed by Real Time PCR analysis as described previously (Somji et al., 2006) or for EMSA binding reactions.
Electrophoretic Mobility Shift Assay (EMSA)
Cell cultures from T-75 flasks (75 cm2) were trypsinized, pelleted, and stored frozen at -80oC. Whole-cell extracts were prepare by resuspending cell pellets in 3 volumes of harvest buffer consisting of 20 mM HEPES pH 7.6, 0.4 M KCl, 1.5 mM MgCl2, 25% glycerol, 0.1% triton, 0.2 mM PMSF, 0.5 mM DTT, 15 mM NaF, and 0.75 mM Na3VO4. DNA binding reactions consisted of 5 μg of protein extract, 40 fmole of double stranded, 5-prime biotin-labeled oligonucleotides (sequences shown in Table 1), 4 μg poly-dI-dC, 5 μg BSA, in binding buffer (20 mM HEPES pH 7.6, 32 mM KCL, 1 mM DTT, 2 mM MgCl2, and 10% glycerol) in a final volume of 25 μl. The reactions were assembled at room temperature and the labeled oligos were added last after a 10 min preincubation with all other components. Complexes were allowed to form for 30 min at room temperature and then 1/10 volume of loading dye without EDTA (50% glycerol, 100 mM Tris, pH 8, and 0.04 % bromophenol blue) was added and the samples were loaded onto a 5% PAGE gel with 1x Tris-borate buffer (no EDTA). Electrophoresis was performed at 80 V for one hr. The position of the labeled oligos was visualized using the Chemiluminescent Nucleic Acid Detection Model (Pierce, Rockford, IL).
TABLE 1.
Sequences of MRE’s in the MT-3 Promoter
| MRE | Sequences |
|---|---|
| 2Aa | TTTTGCACTCGTCCCGGG |
| MREa | GCATGCGCGCCCCCGCCC |
| MREb | CGGTGCGCAGGGGCGGGC |
| MREc | CAGTGCACACACACGGCA |
| MREd | TGGGGGCGCACGCGCGGG |
| MREe | TCGTGCACGCGGATGCGG |
| MREf | CAGTGGGCGCACAGCGGC |
To identify the MRE elements in the human MT-3 gene, the promoter sequences within the first 435 base pairs upstream of the transcriptional start site were searched for MRE-like sequences using the Transcriptional Element Search System (TESS). Each element is designated a-g based on the proximity to the transcription start site (Figure 1).
Figure 1. Metal regulatory elements in the human MT-3 gene.
To identify the MREs, the promoter sequences within the first 435 base pairs upstream within the transcriptional start site were searched for MRE-like sequences. Each element is designated a–g based on the proximity to the start site. The TATA box and the transcriptional start site have also been indicated.
The MT-3 MRE sequences are shown in Table 1. The consensus MRE sequence is CTNTGCRCNCGGCCC which contains the required core sequence TGCRCNC (R, purine, N, any nucleotide) (Andrews, 2001). Note that MREd is in the reverse orientation. Double stranded oligos of each element was synthesized with the additional sequences, three bases upstream and 8 bases downstream of each element.
Statistics
The statistical association of MT-3 immunostaining in the initial staging of the breast tumor and the progression of breast cancer was performed using the Pearson Chi Square test at P=0.019. Statistical analysis of Kaplan-Meier survival curves were performed using the Mantel-Cox method at a significance of P=0.019. Statistical analysis for all other assays consisted of ANOVA with Tukey post-hoc testing performed by Graphpad PRISM 4. All statistical significance is denoted at P<0.05.
Results
Classification of archival breast cancer specimens
There were 198 patients with breast cancer where diagnostic material from the breast was available for evaluation (Table 2). There were 131 patients with invasive ductal carcinoma, 25 patients with lobular carcinoma, 14 patients with mixed ductal and lobular carcinoma, and 8 patients with mixtures of ductal and another histologic type (mucinous, micropapillary) of breast cancer. Eighteen patients had breast cancers with rarer histologic patterns; 13 of these patients had patterns generally regarded as having a better prognosis (8 mucinous carcinomas; 3 medullary carcinomas, 2 tubular carcinomas) while the remaining histologic patterns included solid papillary (2), micropapillary (1), apocrine (1) and comedocarcinoma (1). Two patients had metaplastic or “inflammatory” carcinoma, generally regarded as having a worst prognosis. Invasive ductal carcinomas (IDC) were graded 1, 2, 3 based on tumour morphology, tubule/acinar/glandular formation, nuclear atypia/pleomorphism and frequency of mitoses (Elston and Ellis, 1991).
TABLE 2.
Immunostaining of MT-3 in all diagnostic categories.
| Type | |||||
|---|---|---|---|---|---|
| Invasive Carcinomas | N | % Positive | Quick Score Average | SEM +/− | |
| IDC | Invasive Ductal Carcinoma | 131 | 87.8% | 11.626 | 0.548 |
| ILC | Lobular Carcinoma | 25 | 76.0% | 10.273 | 1.143 |
| MixD/L | Mixed Ductal and Lobular Carcinoma | 14 | 92.9% | 12.500 | 1.493 |
| MixD/O | Mixed Ductal/Other type | 8 | 100.0% | 13.250 | 2.144 |
| Other | Other types (SPC, Papillary, Comedo, Apocrine) | 5 | 100.0% | 14.000 | 1.673 |
| GoodST | Other types (good prognosis – medullary, mucinous, tubular) | 13 | 69.2% | 5.143 | 1.410 |
| BadST | Other types “Inflammatory” (poor prognosis) | 2 |
100.0% | 14.000 | 4.000 |
| 198 | |||||
| Carcinoma in Situ | N | % Positive | Quick Score Average | SEM +/− | |
| DCIS | Ductal Carcinoma in Situ | 84 | 92.9% | 12.404 | 0.602 |
| LCIS | Lobular Carcinoma in Situ | 13 | 84.6% | 12.938 | 1.510 |
| D/L CIS | Mixed DCIS/LCIS | 5 |
100.0% | 12.000 | 3.146 |
| 102 | |||||
| Invasive Ductal Grading | N | % Positive | Quick Score Average | SEM +/− | |
| ICD-3 | 78 | 91.0% | 12.052 | 0.805 | |
| IDC-2 | 58 | 89.7% | 10.526 | 1.655 | |
| IDC-1 | 19 | 78.9% | 10.273 | 1.143 | |
MT-3 staining as a function of histologic tumor type, tumor staging and established tumor markers
The typical profiles of archival specimens of breast cancer stained with the MT-3 antibody have been described previously (Sens et al., 2001). When MT-3 staining was evaluated using the criteria that a tumor was positive if any malignant cells stained for MT-3, then a high % of breast cancers expressed the MT-3 protein across all diagnostic categories (Table 2). Using this criteria, 93% of DCIS, 88% of IDC, 85% of LCIS, 76% of ILC, and 93% of tumor composed of mixed ductal/lobular components were positive for expression of the MT-3 protein. Although sample numbers were small, several trends could be noted using this criteria for staining. Those carcinoma types with a favorable prognosis had the lowest MT-3 staining, with 69% demonstrating some MT-3 staining. When MT-3 staining was evaluated by a three-tiered system of “Strong”, “Weak” and “Negative” similar results were obtained (data not shown). Strong MT-3 staining was seen in the majority of unfavorable cancers. The “good” prognostic classifications were the only group with a low % (31%) of carcinomas demonstrating strong MT-3 staining. The degree of the MT-3 staining in each specimen was also evaluated using the quick score method, which takes into account both the intensity of the staining and the % of cells within the tumor that stain positive for MT-3. Quick score showed similar trends to those noted above. For invasive ductal carcinoma, grading had minimal effects on MT-3 staining. Positive staining for MT-3 was seen in 79% of Grade 1 IDC tumors (well differentiated) which was somewhat lower but not statistically different from the Grade 2 IDC with 90% positive for MT-3 or Grade 3 (poorly differentiated) IDC with 91% positive MT-3 staining. Again, Quickscore measurements were similar in all grades.
There was also no statistically significant association between initial staging of the breast tumor and MT-3 staining, although there was a trend that stage 1 tumors had a lower incidence of positive MT-3 staining (62%) than higher stage tumors (Table 3). A similar pattern was seen if only IDC was examined, thus eliminating influences from the histologic types which tend to have lower stages of tumor burden. An analysis of staining by Quickscore did not alter the above findings.
TABLE 3.
Correlation of MT-3 immunostaining with staging of breast cancers.
| All histologic types | Ductal Only | |||||||
|---|---|---|---|---|---|---|---|---|
| MT-3 | MT-3 | |||||||
| Stage | N | Positive | Mean* | SEM +/− | N | Positive | Mean* | SEM +/− |
| 1 | 13 | 61.5% | 8.769231 | 2.315372 | 7 | 57.1% | 10.28571 | 3.636549 |
| NS(1–2) | 33 | 72.7% | 9.515152 | 1.288881 | 22 | 68.2% | 8.727273 | 1.60651 |
| 2 | 77 | 89.6% | 11.93506 | 0.711044 | 49 | 93.9% | 13.02041 | 0.8312 |
| 3 | 38 | 94.7% | 11.78947 | 0.902941 | 26 | 92.3% | 12.15385 | 1.09922 |
| 4 | 36 |
91.7% | 11.47222 | 0.932215 | 27 |
96.3% | 11.2963 | 0.969352 |
| 198 | 131 | |||||||
Mean of the Quickscore
There was no association between MT-3 staining and other common markers of breast cancer, such as estrogen receptors (ER), progesterone receptors (PR) or Her2neu receptors in either incidence of MT-3 staining or intensity of staining as measured by Quickscore. Within an individual patient, MT-3 staining was generally consistent at all sites (CIS lesions, invasive carcinoma, nodal metastasis and non-nodal metastasis) in both incidence of MT-3 staining, intensity and Quickscore. Previously Sens et al. (2001) showed and confirmed in the current study, that normal breast tissue did not stain for MT-3 protein (data not shown). In the present study, approximately 40% of the tumors contained clearly defined areas of normal breast which were well removed from the primary tumor, and these normal areas showed no staining for MT-3.
MT-3 staining correlates to disease outcome
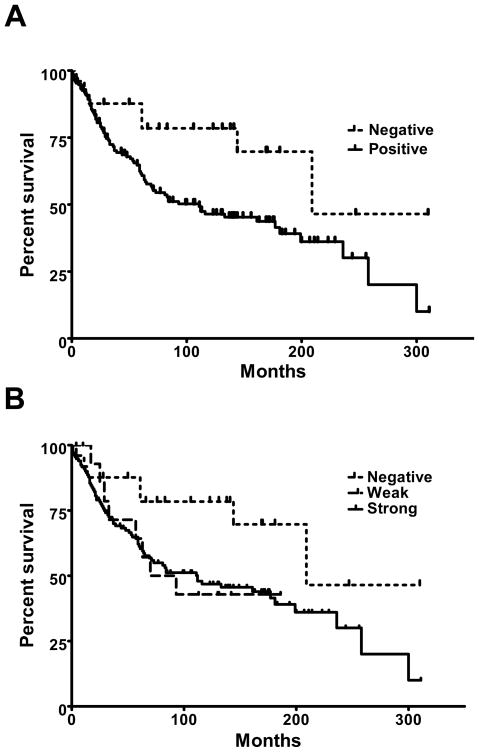
Patients whose tumors showed no staining of MT-3 were more likely to have outcomes without the progression of breast cancer. Patients were classified as a “No progression” if they were alive without evidence of breast cancer or died without evidence of breast cancer. Patients that died of breast cancer or those with active lesions were classified as “Progression of Breast Cancer”. As demonstrated in Table 4, Pearson Chi Square testing demonstrated a significance difference in these two groups based on MT-3 staining. This was further confirmed by Kaplan-Meier Survival Curves. Patients with MT-3 negative breast cancer had prolonged survival curves (Figure 2A). The mean survival time for MT-3 negative patients was 212 months compared to 139 months for MT-3 positive patients. The probability of differences with MT-3 staining by Log Rank (Mantel Cox) Chi Square was 0.019. A similar analysis as shown was conducted comparing tumors with negative, weak MT-3 staining (Quickscores 3–6) and strong MT-3 staining (Quickscore >6). The weak staining tumors had nearly identical survival curves as the strong staining samples and both weak and strong staining was statistically different from the survival curves from patients with negative MT-3 staining (Figure 2B). Analysis was also attempted using MT-3 staining and deaths from breast cancer. Of the 72 patients that died from breast cancer, only 2 had cancers that were MT-3 negative. Although this data was significant, the small numbers are not optimal for firm conclusions from statistical analysis. Thus, while MT-3 negative tumors were rare, they were associated with patients who showed no progression of their disease.
TABLE 4.
Correlation of MT-3 immunostaining with disease outcome.
| MT-3 Negative | MT-3 Positive | Total | |
|---|---|---|---|
| No progression (Alive, NED* or Died, NED) | 15 | 75 | 90 |
| Progression of Breast Cancer (Died, Breast Ca or Alive with Disease) | 5 | 84 | 89 |
| 20 | 159 | 179 | |
No Evidence of Disease
Figure 2. Kaplan Meier plot demonstrating prolonged survival of MT-3 negative breast cancer.
A) The survival distributions between MT-3 positive (solid line) and MT-3 negative (dotted line) breast cancer patients were different by Mantel-Cox Log Rand at a significance of 0.019. B) Kaplan Meier plot of MT-3 staining in breast cancer demonstrates those cancers with weak staining (long dash) had a similar survival to patients with strong staining (solid) line.
Basal and vector-induced expression of MT-3 in MCF-10A cells
The MCF-10A cell line was used as a model to begin to determine the mechanism underlying the differential expression of MT-3 found in human breast cancer. The first set of experiments were to determine the expression, metal inducibility, and permissiveness of MT-3 protein expression. For this purpose, total RNA and protein from confluent cultures of MCF-10A cells were analyzed using RT-PCR and immuno-blot analysis for the expression of MT-3 mRNA and protein, respectively. At a total RNA input of 1 μg and 40 reaction cycles it was shown that there was no basal expression of MT-3 mRNA in the MCF-10A cells (Figure 3A). The absence of MT-3 mRNA expression in the MCF-10A cell line was also confirmed using real time RT-PCR utilizing a previously described MT-3 isoform-specific primer (Somji et al., 2006). There was also no basal expression of the MT-3 protein in the MCF-10A cell lysates (Figure 4).
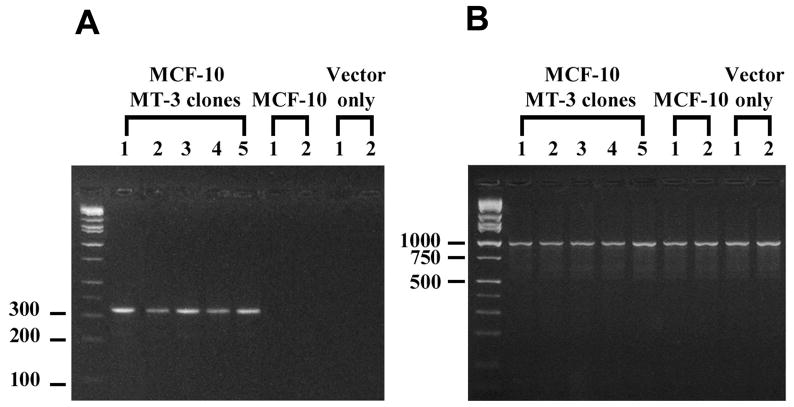
Figure 3. MT-3 and g3pdh gene expression in MCF-10A cells stably transfected with the MT-3 coding sequence.
The expression of MT-3 and g3pdh mRNA was determined using RT-PCR. The reactions were sampled at 25, 30, 35 and 40 cycles of the PCR and the reaction products visualized on ethidium bromide stained 2% agarose gels. Panel (A) shows the MT-3 mRNA at 30 cycles of PCR for the 5 independent MT-3 clones, and at 40 cycles for the 2 clones containing blank vector and 2 samples from parental MCF-10A cells. Panel (B) shows the corresponding g3pdh mRNA expression at 30 cycles of PCR for all the above samples.
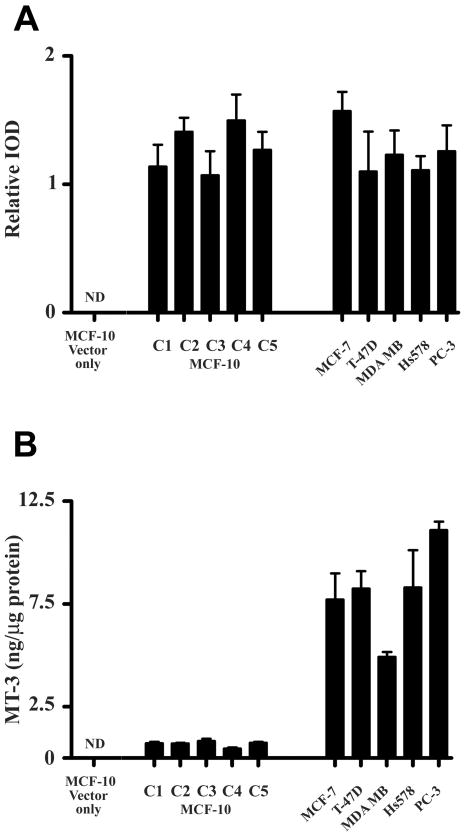
Figure 4. Relative expression of MT-3 in MCF-10A cells compared to breast and prostate cancer cell lines.
Panel (A) shows the relative expression of MT-3 mRNA in the blank vector controls and clones of the MT-3 transfected MCF-10A cells compared to the relative expressions of MT-3 found previously for MT-3 transfected MCF-7, T-47D, MDA-MB-231, Hs578T and PC-3 cell lines (Dutta et al., 2002; Gurel et al., 2003). Relative IODs were calculated from the IOD of product bands obtained at 500 ng total RNA inputs and 30 PCR cycles for each cell line. Panel (B) shows the corresponding expression of MT-3 protein for the above cell lines. All determinations were performed in triplicate. ND indicates a protein level below the limit of detection.
The MCF-10A cell line was transfected with the MT-3 plasmid construct in the sense direction or with the blank vector using the Effectene protocol. The MT-3 vector and methodology has been utilized previously to stably transfect the MT-3 coding sequence under the control of the CMV promoter into the PC-3 prostate cancer cell line (Dutta et al., 2002), the MCF-7, T-47D, Hs578T or MDA-MB-231 breast cancer cell lines (Gurel et al., 2003), and the immortalized HK-2 human proximal tubule cell line (Kim et al., 2002) with resultant accumulation of MT-3 mRNA and protein. Following stable transfection and selection, 5 clones of the MCF-10A cell line were selected for further characterization of MT-3 expression. It was demonstrated that each of the 5 independent clones selected for characterization overexpressed MT-3 mRNA when compared to wild type cells or cells containing the blank pcDNA3.1 vector (Figure 3A). The MT-3 mRNA was first detected from all of the overexpressing clones between 20 and 25 cycles of PCR using 500 ng total RNA inputs (data not shown). The level of MT-3 mRNA expression was similar to that of the glyceraldehyde 3-phosphate dehydrogenase (g3pdh) housekeeping gene (Figure 3A). It was also demonstrated that the transfection protocol had no marked effect on the expression of the g3pdh housekeeping gene (Figure 3B). The relative level of MT-3 mRNA expression in the MT-3 transfected MCF-10A cells was similar to that obtained in previous studies where the MT-3 sequence was transfected into the MT-3 null background of the PC-3 prostate cancer cell line and the MCF-7, T-47D, Hs578T and MDA-MB-231 breast cancer cell lines (Figure 4A). In contrast to MT-3 mRNA expression, a corresponding analysis of MT-3 protein expression in each of the 5 MT-3 expressing clones of the MCF-10A cell line demonstrated that they expressed low levels of MT-3 protein (Figure 4B). For all 5 MT-3 stable transfectants, the level of MT-3 expression was at or near the detection limit of the assay (between 0.1 – 0.5 ng MT-3/μg total cell protein) and similar to that shown in the parental MCF-10A line, which as detailed above, express no MT-3 mRNA. This finding was unexpected since all 5 clones were found to express elevated levels of MT-3 mRNA (Figure 4A). As an additional control, it was shown that the identical MT-3 sequence, when transfected using identical methodology into MCF-7, T-47D, Hs578T or MDA-MB-231 breast cancer cell lines, or the PC-3 prostate cancer cell line, yielded similar levels of MT-3 mRNA expression, but elevated levels of MT-3 protein expression (Figure 4B). Lastly, to determine if MT-3 protein expression might be an artifact generated by which clones were chosen for analysis, an additional 14 randomly chosen clones of the MT-3 transfected MCF-10A cells were assessed for the expression of MT-3 mRNA and protein. It was demonstrated that all 14 clones expressed similar relative amounts of MT-3 mRNA when assessed against the expression of the g3pdh gene and that all 14 clones failed to express appreciable levels of MT-3 protein (data not shown).
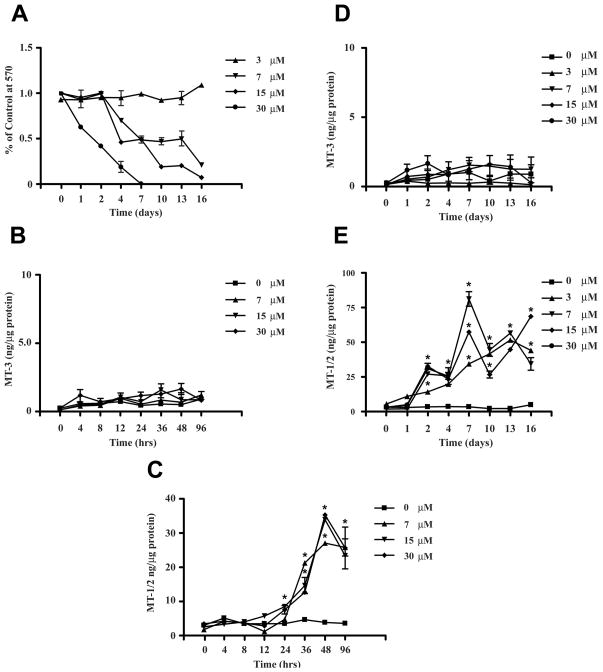
There is evidence that exposure of cells to Cd+2 and Cu+2 can increase the level of the MT-1 and MT-2 protein at the post-transcriptional level by stabilizing the protein against degradation (Vasconcelos et al., 1996, 2002). For this reason, expression of the MT-1/2 and MT-3 proteins were determined in the MCF-10A cell line stably transfected with the MT-3 gene and a blank vector control following exposure of the cells to lethal and sub-lethal levels of Cd+2. The expression of the MT-3 and MT-1/2 proteins were determined on confluent cultures of cells exposed to Cd+2 over a 96 hr and 16 day time course. The concentrations of Cd+2 employed produced both lethal (15 and 30 μM) and sub-lethal (3 and 7 μM) responses over a 16 day time course of exposure to Cd+2 (Figure 5A). Following 96 hr exposure to Cd+2, it was shown that the MT-3 transfected MCF-10A cells displayed no change in MT-3 protein expression, but the cells did increase the accumulation of the MT-1/2 protein under identical conditions of exposure to Cd+2 (Figure 5B,C). To control for the possibility that the time course was not long enough for MT-3 protein to rise in these cell lines, the MT-3 transfected MCF-10A cells were also assessed for MT-3 and MT-1/2 protein accumulation when exposed to Cd+2 for 16 days. It was found that there was no significant change in MT-3 protein accumulation at any concentration of Cd+2 over the 16 day time course; whereas, MT-1/2 protein displayed significant increases at identical concentrations and times of exposure to Cd+2 (Figure 5D,E). There was no difference in toxicity to Cd+2 among the MCF-10A parental cell line, the MCF-10A cells transfected with the blank vector control, or the MCF-10A cells stably transfected with the MT-3 sequence (data not shown). Similarly, there was no difference in Cd+2 toxicity among 5 characterized MT-3 transfected MCF-10A cell clones (data not shown).
Figure 5. Effect of Cd+2 on MT-3 protein expression in MCF-10A cells stably transfected with MT-3 gene.
Panel (A) shows cell viability when confluent MCF-10A cells stably transfected with the MT-3 coding sequence are exposed to 3 (▲), 7 (▼;), 15 (◆) or 30 (●) μM Cd+2 for 16 days. Cell viability was determined by the MTT assay and all determinations were in triplicate. Values shown are the % of the mean cell numbers at each time point divided by the mean cell numbers of the control cells for each triplicate determination. Panel (B) shows the expression of the MT-3 protein when cells are exposed for 96 hrs to 0 (■), 7 (▲), 15 (▼;) or 30 (◆) μM Cd+2. The determinations were performed in triplicate and the results shown are the mean ± SEM. Panel (C) shows the expression of the MT-1/2 protein when cells are exposed for 96 hrs to 0 (■), 7 (▲), 15 (▼;) or 30 (◆) μM Cd+2. The determinations were performed in triplicate and the results shown are the mean ± SEM. * denote a significant difference from control (p < 0.05). Panel (D) shows the expression of the MT-3 protein when cells are exposed for 16 days to 0 (■), 3 (▲), 7 (▼;), 15 (◆) or 30 (●) μM Cd+2. Panel (E) shows the expression of the MT-1/2 protein when cells are exposed for 16 days to 0 (■), 3 (▲), 7 (▼;), 15 (◆) or 30 (●) μM Cd+2. The determinations were performed in triplicate and the results shown are the mean ± SEM. * denote a significant difference from control (p < 0.05). A single asterisk above overlapping symbols indicates significance for all overlapping points.
MT-3 mRNA expression in MCF-10A cells following treatment with inhibitors of DNA methylation and histone deacetylation
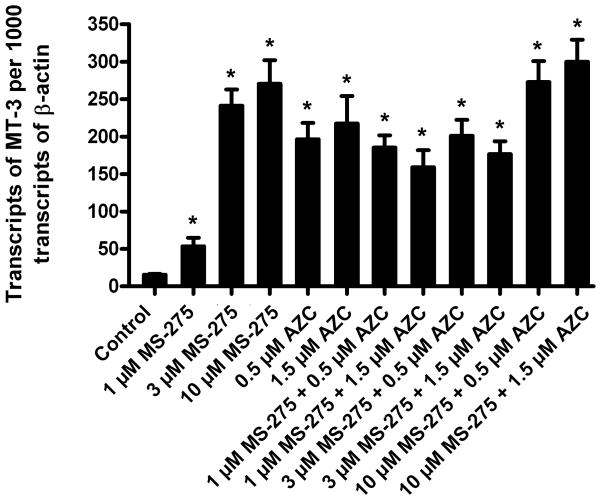
To determine the possible role of histone modifications on MT-3 mRNA expression, the MCF-10A cells were treated with the histone deacetylase inhibitor, MS-275, and the demethylating agent, AZC.. The cells were seeded at a subconfluent density and the next day the subconfluent cells were treated with each drug, alone or in combination, for 3 days. The drug was then removed and the cells were fed fresh growth media. Twenty-four hr later total RNA was prepared from the cells and analyzed for the expression of MT-3 mRNA. This analysis demonstrated that MCF-10A cells treated with MS-275 expressed increased levels of MT-3 mRNA compared to control cells (Figure 6). There was a dose response relationship with a peak in MT-3 mRNA expression between 3 and 10 μM concentrations of MS-275. MS-275 is dissolved in DMSO and it was demonstrated that DMSO exerted no effect on MT-3 mRNA expression in MCF-10A cells (data not shown). The demethylating agent, AZC, also elicited a rise in MT-3 mRNA expression in the MCF-10A cells when compared to untreated control cells (Figure 6). The dose response curve for AZC showed a peak effect on MT-3 mRNA expression between 0.5 and 1.5 μM concentrations of the drug. The elevated expression of MT-3 mRNA was similar in magnitude for each of the drugs at the concentration producing peak increases in MT-3 mRNA. The treatment of the MCF-10A cells with a combination of the two drugs did not lead to a further increase in MT-3 mRNA when compared to than that noted by treatment of either of the two drugs alone (Figure 6).
Figure 6. Expression of MT-3 in MCF-10A cells treated with AZC and/or MS-275.
MCF-10A cells were cultured at a 1:10 ratio in the presence of MS-275 and/or AZC until they reached confluency, following which the cells were harvested and the RNA was extracted. Real time PCR analysis was performed to assess the expression level of MT-3 mRNA. The level of MT-3 expression was normalized to that of β-actin. The determinations were performed in triplicate and the results shown are the mean ± SEM. * denote a significant difference from control (p < 0.05).
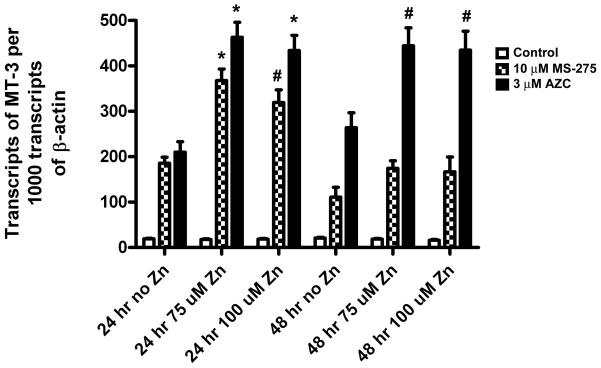
It was also determined if subsequent exposure to Zn+2 would increase MT-3 mRNA expression in the MCF-10A cells that had been treated with AZC or MS-275. In this protocol, the MCF-10A cells were treated with the respective inhibitor for 3 days, followed by recovery of the cells for 24 hr, and then treated with 75 or 100 μM Zn+2 for an additional 24 or 48 hr before preparation of total RNA for analysis of MT-3 mRNA expression. The results of this analysis showed that Zn+2 exposure increased MT-3 mRNA expression in MCF-10A cells that had been exposed to either AZC or MS-275 (Figure 7). The Zn+2-induced rise in MT-3 mRNA appeared to be greater in MCF-10A cells exposed to AZC than to MS-275, but increases were significant for both drugs.
Figure 7. Effect of zinc on the expression levels of MT-3 in the MCF-10A cell line.
MCF-10A cells were cultured at a 1:10 ratio in the presence of MS-275 or AZC till they reached confluency, following which the cells were allowed to recover for 24 hr without the drug. The cells were then exposed to 100 μM Zinc for 24 or 48 hr. Total RNA was extracted from the cells and the expression of MT-3 was determined by real time PCR. The level of MT-3 expression was normalized to that of β-actin. The determinations were performed in triplicate and the results shown are the mean ± SEM. All treatment groups were statistically significant over untreated controls (p < 0.05). * denotes statistically significant from no zinc treated cells. # denotes statistically significant from no zinc treated cells.
Metal regulatory element activity in MCF-10A cells
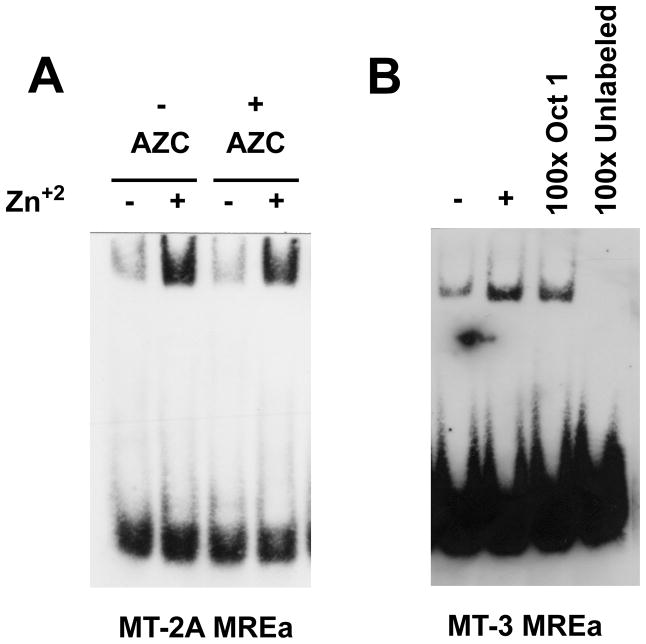
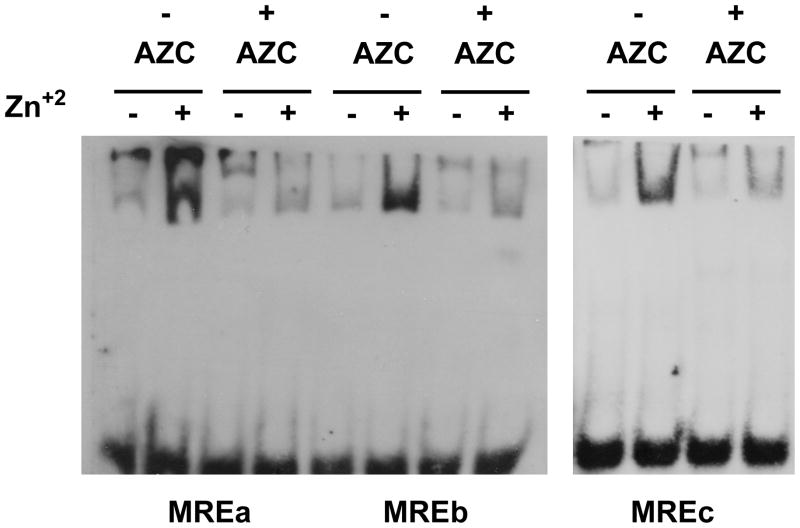
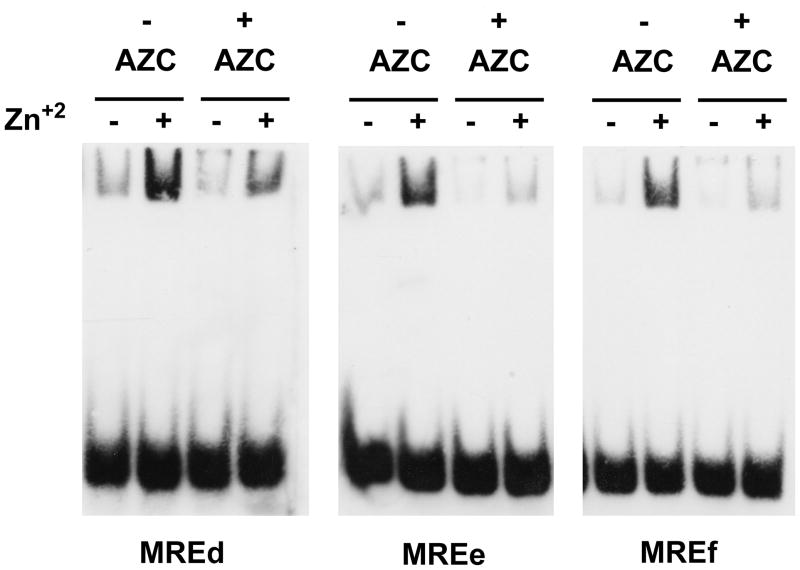
The finding that the MCF-10A cells treated with AZC increased MT-3 mRNA expression when exposed to Zn+2 suggested that the Metal Regulatory Elements (MREs) in the human MT-3 gene are functional, but silenced in the MCF-10A cell under normal conditions. To confirm this, the ability of the MT-3 MRE to bind protein in a Zn+2-dependent fashion was tested in whole-cell extracts from normal and AZC-treated MCF-10A cells. As detailed in the Materials and Methods, 6 MRE were identified and designated MREa for the most proximal to MREf for the most distal MRE element. The MREa element of the MT-2A gene was used as a positive control since this MRE was found to mediate the induction of MT-2A by heavy metals, and bind MTF-1 in a Zn+2 dependent fashion (Koizumi et al., 1999). As expected from the published report, it was demonstrated that the MT-2A MREa oligo bound protein in a Zn+2-dependent fashion (Figure 8A). This was evidenced by oligo gel-retardation when incubation was in whole-cell extracts prepared from Zn+2-treated MCF-10A cells in contrast to little gel-retardation when incubation was performed in extracts prepared from untreated cells. Zinc-dependent interactions with the MREa were also found with extracts from AZC-treated cells (Figure 8A). It was also demonstrated that the most proximal element, MREa, of the MT-3 promoter also formed shifted, Zn+2-dependent complexes in extracts prepared from MCF-10A cells (Figure 8B). It was also demonstrated that the protein-DNA complex of the MREa of MT-3 was effectively competed with 100x of unlabeled oligo in the binding reaction, but not by a nonspecific oligo (Oct 1 binding element) indicating that the Zn+2-dependent complex formation consisted of specific protein-DNA interactions (Figure 8B). Extracts prepared from MCF-10A cells treated with AZC were also able to form complexes with the MT-3 MREa in a Zn+2-dependent fashion, although the amount was less than that of nontreated MCF-10A cells (Figure 9). It was also noted that MREb through MREf were also able to form shifted bands, indicating the dependence of Zn+2 in the formation of protein-DNA complexes with these sequence elements (Figure 9, 10). When these binding reactions were performed using extracts from AZC treated cells, the dependence of Zn+2 treatment was still evident, although to a lesser degree than without AZC treatment (Figure 9, 10). These results indicate that the MREs within the MT-3 promoter are capable of binding protein factor(s) and that the amount of complex formed is greatly enhanced by exposure of the MCF-10A cells to Zn+2.
Figure 8. EMSA to assess protein binding to the most proximal MRE in the MT-2A and MT-3 gene promoter.
Whole cell protein extracts prepared from MCF-10A cells treated with 100 μM zinc for 24 hr were gel-shifted with a double-stranded oligo containing either the MT-2A MREa or the MT-3 MREa. A) MCF-10A cells were previously treated with or without 1.5 μM 5-azacytidine (AZC) for three days followed by another 24 hr period with or without 100 μM zinc in the absence of AZC. Extracts were prepared and were shifted with the MT-2A MREa oligo. B) Extracts prepared from zinc treated MCF-10A cells where shifted with the MT-3 MREa oligo. Specificity of binding is shown by the addition of an oligo for the Oct 1 transcription factor binding site or unlabeled MT-3 MREa at 100 times the amount of the labeled oligo in extracts from zinc treated MCF-10A cells.
Figure 9. EMSA to determine protein binding to the MREa, MREb and MREc in the MT-3 promoter.
Whole cell extracts were prepared from MCF-10A cells pretreated with or without 1.5 AZC for 24 hr followed another 24 hr period with or without 100 μM zinc in the absence of AZC.
Figure 10. EMSA to determine protein binding to the MREd, MREe and MREf in the MT-3 promoter.
Whole cell extracts were prepared from MCF-10A cells pretreated with or without 1.5 AZC for 24 hr followed another 24 hr period with or without 100 μM zinc in the absence of AZC.
Discussion
Sens et al. (2001) reported through analysis of a small set of archival human diagnostic specimens that MT-3 is not expressed in tissues of the normal breast, but is expressed in some breast cancers and that expression tends to correlate with a poor disease outcome. The first goal of the present study was to confirm and extend these findings to a larger set of archival specimens from patients with breast cancer. The results of this larger study showed several important differences from the results reported earlier. A major difference was in the number of patient specimens that contained breast cancer cells that were positive for the expression of MT-3. In the present study, approximately 90% of all breast cancers showed some cells within each tumor that were immunoreactive for the MT-3 protein, a value higher than the previous study.
The fact that the confined lesion, DCIS, also showed similar high % levels of expression provides evidence that the gain in MT-3 expression probably occurs early in the development of the cancer. From this perspective, the expression of MT-3 may be looked upon as a potential biomarker due to its very high occurrence rate for breast cancer. In addition, impacting on its potential applicability as a biomarker is the confirmation of the earlier study that breast cancers that have no expression of MT-3 are associated with a favorable disease outcome. Although an absence of MT-3 expression was rare in patients with breast cancer, its absence in archival specimens did correlate with a favorable disease outcome, making it a potential prognostic marker of disease progression. The fact that MT-3 expression did not correlate with established markers such as ER, PR and Her2neu indicates MT-3 might be a valuable adjunct to these established prognostic markers of disease progression.
Based on the above findings, one might expect that differences in the expression of MT-3 should have been identified in the literature as a result of the numerous past genomic and proteomic screens that have been conducted on specimens of normal breast and breast cancer. There are probably several reasons why this has not been the case, all of which are likely related to the unique properties of the MT gene family. The first is the organization of the genes comprising the MT gene family. The MT gene family is encoded by 10 functional and several nonfunctional isoforms located at 16q13 that display extremely high degrees of sequence homology (Palmiter et al., 1992; West et al., 1990; Stennard et al., 1994; Quaife et al., 1994). This high degree of homology makes it likely that MT are being identified as an overall family, rather than single members, in the majority of microarray and genomic analyses. The degree of homology among human MT family members is such that it is not possible, using coding sequences of the individual MT genes, to construct primers for mRNA analysis of the specific members of the MT gene family (Mididoddi et al., 1996). There is only sufficient diversity in the 5′ and 3′ untranslated regions of the individual MT genes to develop primers for analysis of specific MT gene transcripts (Mididoddi et al., 1996). It was also shown that the human MT genes exhibit unique expression profiles and examples are demonstrated for inducer-specific and tissue-specific (Richards et al., 1984; Jahroudi et al., 1990; Pauwels et al., 1994; Soumillion et al., 1992; Sadhu and Gedamu, 1988) and developmental-specific (Schmidt and Hamer, 1986) regulation. At the level of MT protein expression, the only MT family member that possesses sufficient sequence diversity for production of a specific antibody is the MT-3 isoform, due to a unique 6 amino acid C-terminal sequence (Garrett et al., 1999). The most common and widely used antibody to analyze the expression of MT in tissues is based on the monoclonal antibody raised against the E-9 epitope of the MT protein and this antibody recognizes all MT family members except MT-3 (Jasani and Schmid, 1997). This antibody has been used extensively to characterize MT staining in archival specimens of breast cancer (Jasani and Schmid, 1997). The small size (6 kD) of the MT proteins and only slight differences in charge among the isoforms also create challenges for proteomic screens. Many analytical preparations discard such low molecular weight proteins before analysis and, even when present, would not be searching for charge separation at such low molecular weights. A further complication in the analysis of MT expression in tissue extracts is the expression pattern of the MT-1 and MT-2 isoforms in normal breast and breast cancer tissue. While the normal breast epithelial cells are unreactive with the MT E-9 antibody, the myoepithelial cells lining the breast duct are immunoreactive for the MT-1/2 proteins (Bier et al., 1994; Jin et al., 2001). Thus, analysis of extracts of normal breast tissue would show expression of MT. Thus, there are multiple reasons why previously performed genomic and proteomic analysis would fail to identify alterations in MT-3 expression in breast cancer.
This lab used the MCF-10A cell line as a model to explore the mechanisms that might regulate the absence of MT-3 expression in the normal breast epithelial cell. Similar to normal breast tissue, it was demonstrated that the MCF-10A cells do not have basal expression of MT-3 mRNA. It was also shown that MT-3 expression could not be induced by exposure of MCF-10A cells to the heavy metal, Cd+2. That the absence of basal and induced expression might be due to an epigenetic mechanism of gene regulation was indicated through treatment of the cells with the inhibitor of histone deacetlyation or the demethylation agents, MS-275 and AZC, respectively. Both agents have been used as pharmacological approaches to alter gene expression especially in tumor cells (Piekarz and Bates, 2009). Treatment of the MCF-10A cells with either agent restored both a basal level of MT-3 mRNA expression and induction of expression by the heavy metal, Zn+2. It was also found that the MT-3 metal regulatory elements are potentially active binders of protein factors.
Genes vary considerably in their susceptibility to these agents and for the majority of genes, DNA methylation is considered dominant to that of histone acetylation (Brock et al., 2007). Exceptions do exist, however, where HDAC inhibitors are able to activate transcription of DNA-methylated loci (Dong et al., 2007) and MT-3 appears to be one of the exceptions to this general rule of dominance. The DNA demethylating agents and the inhibitors of histone deacetylation may potentially act through a common pathway to affect gene transcription, and HDAC inhibitors activate loci silenced by DNA methylation by attenuating the latter stages of chromatin compaction. The histone deacetylase inhibitors, such as MS-275, produce an increase in acetylation of lysine residues of histone tails, a posttranslational modification that is correlated to transcriptional activity of the region or gene. The mechanism of action is thought to be that the acetylation of lysine diminishes the positive charge on the ε-amino groups of lysine, allowing the DNA to unwind from the histone octamers (Shahbazian and Gunstein, 2007). The control of histone acetylation is managed by the activity of histone acetyltransferases (HATs) and histone deacetylases (HDAC) through the recruitment of chromatin remodeling complexes (Shahbazian and Grunstein, 2007). Histone tails are also acetylated during DNA replication and this might potentially be the pathway of histone acetylation in the region of highly compacted or highly silence genes by the exposure to HDAC inhibitors (Shahbazian and Grunstein, 2007; Ozdemir et al., 2006; Verreault, 2000). Demethylating agents like AZC block the methylation of cytosines of the newly synthesized DNA daughter strand at sites complementary to methylated CpG on the parent strand; leading to the activation of gene loci silenced by chromatin compaction into heterochromatin. Methylation of DNA confers this compact chromatin structure through methyl-DNA-binding proteins, such as MeCP2, which bind methyl-cytosines and recruit larger chromatin modifying complexes, some of which contain HDAC (Chiurazzi and Neri, 2003). Genes also vary in the ability of these two agents to synergize to activate transcription (Piekarz and Bates, 2009; Chiurazzi and Neri, 2003; Jain et al., 2009). The MT-3 gene in MCF-10A cells exhibited susceptibility to both agents, but did not exhibit synergy. While action of these agents is usually directly on the gene under examination, there is a possibility that the agents did not act directly on the MT-3 gene. It is possible to explain the current findings by proposing that these agents activated the transcription of MT-3 transacting factors. These genes might potentially code for, and act through, active MT-3 transcription factors. It is also possible, as is known to occur for some transcription factors that an alteration occurred in the acetylation status of nonhistone proteins (Drummond et al., 2005). The relatively specific HDAC inhibitor MS-275 is a member of the benzamide inhibitor class that was shown to primarily inhibit HDAC1, HDAC2, and HDAC3 (Bolden et al., 2006). Thus, the current results implicate these three ubiquitously expressed nuclear isoforms as potentially involved in the control of MT-3 gene expression.
The epithelial cells of the human kidney have basal expression of MT-3 in vivo and proximal tubule cell cultures derived from this organ retain the basal expression of MT-3 (Garrett et al., 1999, 2002). Functional studies using cultures of proximal tubule cells have implicated MT-3 in the regulation of vectorial active transport and in the regulation of expression of E- and N-cadherin (Kim et al., 2002; Bathula et al., 2008). In proximal tubule cells exposed to Cd+2, the expression of MT-3 was found to mediate the choice between an apoptotic or necrotic mechanism of cell death (Somji et al., 2004, 2006). In contrast, the functional significance of MT-3 expression in breast cancer has been difficult to define for lack of in vitro model systems that express MT-3 under basal conditions. The finding that MT-3 displayed no basal expression in MCF-10A cells, and that the cells could not be stably transfected to express MT-3 protein, is not a unique finding restricted to this cell line. This lab has performed parallel studies in human bladder cancer with similar results. In these studies, it was also shown that the normal urothelium displayed no expression of MT-3, but that a large % of archival specimens of human bladder cancers did immunostain for the MT-3 protein (Zhou et al., 2006; Sens et al., 2000). Similarly, there was no basal expression of MT-3 in a normal urothelial cell line model (UROtsa) and attempts to stably transfect the cell line with MT-3 resulted in expressed MT-3 mRNA, but no MT-3 protein (Garrett et al., 2005). In the transfection studies with the UROtsa cells, it was also demonstrated that deletion of the unique N-terminal sequence of the MT-3 gene, did allow the altered MT-3 protein to be expressed following stable transfection into UROtsa cells. This identical deletion was also shown to allow expression of the altered MT-3 protein in MCF-10A cells (unpublished study). An assessment of basal expression of MT-3 in cancer cell lines demonstrated a unique pattern of MT-3 expression between the in vitro and in vivo environment. Zhou et al. (2006) demonstrated that there was no basal expression of MT-3 under cell culture conditions in the breast cancer cell lines; MCF-7, T-47D, Hs578T and MDA-MB-231; the prostate cancer cell line, PC-3; and, in UROtsa cell lines malignantly transformed through exposure to Cd+2 or As+3. However, when any of these cell lines were subcutaneously heterotransplanted into immunocompromised mice, all the resulting tumors showed a majority of cells with moderate to intense immunostaining for the MT-3 protein (Zhou et al., 2006). These features of MT-3 expression in normal and malignant breast and bladder cell lines makes functional studies for each cancer difficult to perform, but the similarity of features may also indicate similar epigenetic mechanisms of regulation of MT-3 expression.
Acknowledgments
The project described was supported by grant numbers R01 CA098832 from the National Cancer Institute (NCI), NIH. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NCI, NIH.
Abbreviations
- MT-3
Metallothionein
- EMSA
Electrophoretic Mobility Shift Assay
- GIF
Growth Inhibitory Factor
- H&E
Hematoxylin and Eosin
- MTT
3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyltetrazolium bromide
- AZC
5-aza-2′-deoxycytidine
- IDC
Invasive Ductal Carcinomas
- ER
Estrogen Receptors
- PR
Progesterone Receptors
- g3pdh
lyceraldehyde 3-Phosphate Dehydrogenase
- MREs
Metal Regulatory Elements
- HATs
Histone Acetyltransferases
- HDAC
Histone Deacetylases
- UROtsa
Urothelial Cell Line Model
- MRE
Metal Response Element
References
- Andrews GK. Regulation of metallothionein gene expression by oxidative stress and metal ions. Biochem Pharmacol. 2000;59:95–104. doi: 10.1016/s0006-2952(99)00301-9. [DOI] [PubMed] [Google Scholar]
- Andrews GK. Cellular zinc sensors: MTF-1 regulation of gene expression. Biometals. 2001;14:223–237. doi: 10.1023/a:1012932712483. [DOI] [PubMed] [Google Scholar]
- Antila E, Mussalo-Rauhamaa H, Kantola M, Atroshi F, Westermarch T. Association of cadmium with human breast cancer. Sci Total Environ. 1996;186:251–256. doi: 10.1016/0048-9697(96)05119-4. [DOI] [PubMed] [Google Scholar]
- Bathula CS, Garrett SH, Zhou XD, Sens MA, Sens DA, Somji S. Cadmium, vectorial active transport, and MT-3-dependent regulation of cadherin expression in human proximal tubular cells. Toxicol Sci. 2008;102:210–318. doi: 10.1093/toxsci/kfn004. [DOI] [PubMed] [Google Scholar]
- Bier B, Douglas-Jones A, Totsch M, Dockhorn-Dworniczak B, Bocker W, Jasani B, Schmid KW. Immunohistochemical demonstration of metallothionein in normal human breast tissue and benign and malignant lesions. Breast Cancer Res Treat. 1994;30:213–221. doi: 10.1007/BF00665963. [DOI] [PubMed] [Google Scholar]
- Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- Brama M, Gnessi L, Basciani S, Cerulli N, Politi L, Spera G, Mariani S, Cherubini S, dAbusco AS, Scandurra R, Migliaccio S. Cadmium induces mitogenic signaling in breast cancer cells by an ER alpha-dependent mechanism. Mol Cell Endocrinol. 2007;264:102–108. doi: 10.1016/j.mce.2006.10.013. [DOI] [PubMed] [Google Scholar]
- Brock MV, Herman JG, Baylin SB. Cancer as a manifestation of aberrant chromatin structure. Cancer J. 2007;13:3–8. doi: 10.1097/PPO.0b013e31803c5415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiurazzi P, Neri G. Reactivation of silenced genes and transcriptional therapy. Cytogenet Genome Res. 2003;100:56–64. doi: 10.1159/000072838. [DOI] [PubMed] [Google Scholar]
- Clemens S. Toxic metal accumulation, responses to exposure and mechanisms of tolerance in plants. Biochimie. 2006;88:1707–1719. doi: 10.1016/j.biochi.2006.07.003. [DOI] [PubMed] [Google Scholar]
- Cousins RJ. Metallothionein-aspects related to copper and zinc metabolism. J Inherited Metab Dis. 1983;6:15–21. doi: 10.1007/BF01811318. [DOI] [PubMed] [Google Scholar]
- Darbre PD. Metalloestrogens: an emerging class of inorganic xenoestrogens with potential to add to the estrogenic burden of the human breast. J Appl Toxicol. 2006;26:191–197. doi: 10.1002/jat.1135. [DOI] [PubMed] [Google Scholar]
- Detre S, Jotti GS, Dowsett M. A “quickscore” method for immunohistochemical semiquantitation: validation for oestrogen receptor in breast carcinomas. J Clin Pathol. 1995;48:876–878. doi: 10.1136/jcp.48.9.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong E, Guidotti A, Grayson DR, Costa E. Histone hyperacetylation induces demethylation of reelin and 67-kDa glutamic acid decarboxylase promoters. Proc Natl Acad Sci U S A. 2007;104:4676–4681. doi: 10.1073/pnas.0700529104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DC, Noble CO, Kirpotin DB, Guo Z, Scott GK, Benz CC. Clinical development of histone deacetylase inhibitors as anticancer agents. Annu Rev Pharmacol Toxicol. 2005;45:495–528. doi: 10.1146/annurev.pharmtox.45.120403.095825. [DOI] [PubMed] [Google Scholar]
- Dutta R, Sens DA, Somji S, Sens MA, Garrett SH. Metallothionein isoform 3 expression inhibits cell growth and increases drug resistance of PC-3 prostate cancer cells. Prostate. 2002;52:89–97. doi: 10.1002/pros.10097. [DOI] [PubMed] [Google Scholar]
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. doi: 10.1111/j.1365-2559.1991.tb00229.x. [DOI] [PubMed] [Google Scholar]
- Franz E, Romkens P, van Raamsdonk L, van der Fels-Flerz I. A chain modeling approach to estimate the impact of soil cadmium pollution on human dietary exposure. J Food Prot. 2008;71:2504–2513. doi: 10.4315/0362-028x-71.12.2504. [DOI] [PubMed] [Google Scholar]
- Garcia-Morales P, Saceda M, Kennedy N, Kim N, Salomon DS, Gottardis MM, Solomon HB, Sholler PF, Jordan VC, Martin MB. Effect of cadmium on estrogen receptor levels and estrogen-induced responses in human breast cancer cells. J Biol Chem. 1994;269:16896–16901. [PubMed] [Google Scholar]
- Garrett SH, Sens MA, Todd JH, Somji S, Sens DA. Expression of MT-3 protein in the human kidney. Toxicol Lett. 1999;105:207–214. doi: 10.1016/s0378-4274(99)00003-x. [DOI] [PubMed] [Google Scholar]
- Garrett SH, Phillips V, Somji S, Sens MA, Dutta R, Park S, Kim D, Sens DA. Transient induction of metallothionein isoform 3 (MT-3), c-fos, c-jun and c-myc in human proximal tubule cells exposed to cadmium. Toxicol Lett. 2002;126:69–80. doi: 10.1016/s0378-4274(01)00448-9. [DOI] [PubMed] [Google Scholar]
- Garrett SH, Park S, Sens MA, Somji S, Singh RK, Namburi VBRK, Sens DA. Expression of metallothoinein isoform 3 is restricted at the post-transcriptional level in human bladder epithelial cells. Toxicol Sci. 2005;87:66–74. doi: 10.1093/toxsci/kfi231. [DOI] [PubMed] [Google Scholar]
- Goering PL, Klaassen CD. Altered subcellular distribution of cadmium following cadmium pretreatment: possible mechanism of tolerance to cadmium-induced lethality. Toxicol Appl Pharmacol. 1983;70:195–203. doi: 10.1016/0041-008x(83)90095-9. [DOI] [PubMed] [Google Scholar]
- Gurel V, Sens DA, Somji S, Garrett SH, Nath J, Sens MA. Stable transfection and overexpression of metallothionein isoform 3 inhibits the growth of MCF-7 and Hs578T cells but not that of T-47D or MDA-MB-231 cells. Breast Cancer Res Treat. 2003;80:181–191. doi: 10.1023/A:1024520801262. [DOI] [PubMed] [Google Scholar]
- Gurel V, Sens DA, Somji S, Garrett SH, Weiland T, Sens MA. Post-transcriptional regulation of metallothionein isoform 1 and 2 expression in the human breast and the MCF-10A cell line. Toxicol Sci. 2005;85:906–915. doi: 10.1093/toxsci/kfi155. [DOI] [PubMed] [Google Scholar]
- Hamer DH. Metallothionein. Ann Rev Biochem. 1986;55:913–951. doi: 10.1146/annurev.bi.55.070186.004405. [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer. IARC Monographs on the Evaluation of the Carcinogenic Risk of Chemicals to Man. Vol. 23. Lyon, France: IARC; 1980. Some Metals and Metallic Compounds. [Google Scholar]
- International Agency for Research on Cancer. International Agency for Research on Cancer Monographs on the Evaluation of the Carcinogenic Risks to Humans. Vol. 58. Lyon, France: IARC; 1993. Beryllium, cadmium, mercury, and exposure in the glass manufacturing industry. [PMC free article] [PubMed] [Google Scholar]
- Jahroudi N, Foster R, Price-Haughey J, Beitel G, Gedamu L. Cell-type specific and differential regulation of the human metallothionein genes. J Biol Chem. 1990;265:6506–6511. [PubMed] [Google Scholar]
- Jain N, Rossi A, Garcia-Manero G. Epigenetic therapy of leukemia: An update. Int J Biochem Cell Biol. 2009;41:72–80. doi: 10.1016/j.biocel.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasani B, Schmid KW. Significance of metallothionein overexpression in human tumours. Histopathology. 1997;31:211–214. doi: 10.1046/j.1365-2559.1997.2140848.x. [DOI] [PubMed] [Google Scholar]
- Jin R, Bay BH, Chow VTK, Tan PH, Dheen T. Significance of metallothionein expression in breast myoepithelial cells. Cell Tissue Res. 2001;303:221–226. doi: 10.1007/s004410000310. [DOI] [PubMed] [Google Scholar]
- Johnson MD, Kenney N, Stoica A, Hilakivi-Clarke L, Singh B, Chepko G, Clarke R, Sholler PF, Lirio AA, Foss C, Reiter R, Truck B, Paik S, Martin MB. Cadmium mimics the in vivo effects of estrogen in the uterus and mammary gland. Nature Med. 2003;9:1081–1084. doi: 10.1038/nm902. [DOI] [PubMed] [Google Scholar]
- Kägi JHR. Evolution, structure and chemical activity of class I metallothioneins: An overview. In: Suzuki KT, Imura M, Kimura M, editors. Metallothionein III. Biological Roles and Medical Implications. Berlin: Birkhauser Verlag; 1993. pp. 29–56. [Google Scholar]
- Kägi JHR, Hunziker P. Mammalian metallothionein. Biol Trace Element Res. 1989;21:111–118. doi: 10.1007/BF02917243. [DOI] [PubMed] [Google Scholar]
- Kim D, Garrett SH, Sens MA, Somji S, Sens DA. Metallothionein isoform 3 and proximal tubule vectorial active transport. Kidney Int. 2002;61:464–472. doi: 10.1046/j.1523-1755.2002.00153.x. [DOI] [PubMed] [Google Scholar]
- Koizumi S, Suzuki K, Ogra Y, Yamada H, Otsuka F. Transcriptional activity and regulatory protein binding of metal-responsive elements of the human metallothionein-IIA gene. Eur J Biochem. 1999;259:635–642. doi: 10.1046/j.1432-1327.1999.00069.x. [DOI] [PubMed] [Google Scholar]
- McElroy JA, Shafer MM, Trentham-Dietz A, Hampton JM, Newcomb PA. Cadmium exposure and breast cancer risk. J Natl Cancer Inst. 2006;98:869–873. doi: 10.1093/jnci/djj233. [DOI] [PubMed] [Google Scholar]
- Mididoddi S, McGuirt JP, Sens MA, Todd JH, Sens DA. Isoform-specific expression of metallothionein mRNA in the developing and adult human kidney. Toxicol Lett. 1996;85:17–27. doi: 10.1016/0378-4274(96)03632-6. [DOI] [PubMed] [Google Scholar]
- Oskarsson A, Widell A, Olsson IM, Grawe KP. Cadmium in food chain and health effects in sensitive population groups. Biometals. 2004;17:531–534. doi: 10.1023/b:biom.0000045733.38583.8e. [DOI] [PubMed] [Google Scholar]
- Ozdemir A, Masumoto H, Fitzjohn P, Verreault A, Logie C. Histone H3 lysine 56 acetylation: a new twist in the chromosome cycle. Cell Cycle. 2006;5:2602–2608. doi: 10.4161/cc.5.22.3473. [DOI] [PubMed] [Google Scholar]
- Palmiter RD, Findley SD, Whitmore TE, Durnam DM. MT-III, a brain- specific member of the metallothionein gene family. Proc Natl Acad Sci USA. 1992;89:6333–6337. doi: 10.1073/pnas.89.14.6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels M, Van Weyenbergh J, Soumillion A, Proost P, DeLey M. Induction by zinc of specific metallothionein isoforms in human monocytes. Eur J Biochem. 1994;220:105–110. doi: 10.1111/j.1432-1033.1994.tb18603.x. [DOI] [PubMed] [Google Scholar]
- Piekarz RL, Bates SE. Epigenetic modifiers: basic understanding and clinical development. Clin Cancer Res. 2009;15:3918–3926. doi: 10.1158/1078-0432.CCR-08-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quaife CJ, Findley SD, Erickson JC, Froelick GJ, Kelly EJ, Zambrowicz BP, Palmiter RD. Induction of a new metallothionein isoform (MT-IV) occurs during differentiation of stratified squamous epithelia. Biochemistry. 1994;33:7250–7259. doi: 10.1021/bi00189a029. [DOI] [PubMed] [Google Scholar]
- Richards RI, Heguy A, Karin M. Structural and functional analysis of the human metallothionein-IA gene: differential induction by metal ions and glucocorticoids. Cell. 1984;37:263–272. doi: 10.1016/0092-8674(84)90322-2. [DOI] [PubMed] [Google Scholar]
- Rossi MR, Somji S, Garrett SH, Sens MA, Nath J, Sens DA. Expression of hsp 27, hsp 60, hsc 70 and hsp 70 stress response genes in cultured human urothelial cells (UROtsa) exposed to lethal and sublethal concentrations of sodium arsenite. Environ Health Persp. 2002;110:1225–1232. doi: 10.1289/ehp.021101225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhu C, Gedamu L. Regulation of human metallothionein (MT) genes. J Biol Chem. 1988;263:2679–2684. [PubMed] [Google Scholar]
- Schmidt CJ, Hamer DH. Cell specificity and an effect of ras on human metallothionein gene expression. Proc Natl Acad Sci USA. 1986;83:3346–3350. doi: 10.1073/pnas.83.10.3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sens MA, Somji S, Lamm DL, Garrett SG, Slovinsky F, Todd JH, Sens DA. Metallothionein Isoform 3 as a potential biomarker for human bladder cancer. Environ Health Perspect. 2000;108:413–418. doi: 10.1289/ehp.00108413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sens MA, Somji S, Garrett SH, Sens DA. Metallothionein isoform 3 (MT-3) overexpression is associated with breast cancers having a poor prognosis. Am J Pathol. 2001;159:21–26. doi: 10.1016/S0002-9440(10)61668-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahbazian MD, Grunstein M. Functions of site-specific histone acetylation and deacetylation. Annu Rev Biochem. 2007;76:75–100. doi: 10.1146/annurev.biochem.76.052705.162114. [DOI] [PubMed] [Google Scholar]
- Somji S, Garrett SH, Sens MA, Gurel V, Sens DA. Expression of metallothionein isoform 3 (MT-3) determines the choice between apoptotic or necrotic cell death in Cd+2-exposed human proximal tubule cells. Toxicol Sci. 2004;80:358–366. doi: 10.1093/toxsci/kfh158. [DOI] [PubMed] [Google Scholar]
- Somji S, Garrett SH, Sens MA, Sens DA. The unique N-terminal sequence of metallothionein-3 is required to regulate the choice between apoptotic or necrotic cell death of human proximal tubule cells exposed to Cd+2. Toxicol Sci. 2006;90:369–376. doi: 10.1093/toxsci/kfj089. [DOI] [PubMed] [Google Scholar]
- Soumillion A, Van Damme J, DeLey M. Cloning and specific polymerized-chain- reaction amplification of a third charge-separable human metallothionein isoform. Eur J Biochem. 1992;209:999–1004. doi: 10.1111/j.1432-1033.1992.tb17374.x. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Holloway AF, Hamilton J, West AK. Characterization of six additional human metallothionein genes. Biochim Biophys Acta. 1994;1218:357–365. doi: 10.1016/0167-4781(94)90189-9. [DOI] [PubMed] [Google Scholar]
- Stoica A, Katzenellenbogen BS, Martin MB. Activation of estrogen receptor-alpha by the heavy metal cadmium. Mol Endocrinol. 2000;14:545–553. doi: 10.1210/mend.14.4.0441. [DOI] [PubMed] [Google Scholar]
- Transcriptional Element Search System, University of Pennsylvania. http://www.cbil.upenn.edu/cgi-bin/tess/tess.
- Tsuji S, Kobayashi H, Uchida Y, Ihara Y, Miyatake T. Molecular cloning of human growth inhibitory factor cDNA and its down-regulation in Alzheimer’s disease. EMBO J. 1992;11:4843–4850. doi: 10.1002/j.1460-2075.1992.tb05590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida Y, Takio K, Titani K, Ihara Y, Tomonaga M. The growth inhibitory factor that is deficient in Alzheimer’s disease is a 68 amino acid metallothionein-like protein. Neuron. 1991;7:337–347. doi: 10.1016/0896-6273(91)90272-2. [DOI] [PubMed] [Google Scholar]
- Vasconcelos MH, Tam SC, Beattie JH, Hesketh J. Evidence for differences in the post-transcriptional regulation of rat metallothionein isoforms. Biochem J. 1996;315:665–671. doi: 10.1042/bj3150665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasconcelos MH, Tam SC, Hesketh J, Reid M, Beattie JH. Metal- and tissue- dependent relationship between metallothionein mRNA and protein. Toxicol Appl Pharmacol. 2002;182:91–97. doi: 10.1006/taap.2002.9428. [DOI] [PubMed] [Google Scholar]
- Verreault A. De novo nucleosome assembly: new pieces in an old puzzle. Genes Dev. 2000;14:1430–1438. [PubMed] [Google Scholar]
- West AK, Stallings R, Hildebrand CE, Chiu R, Karin M, Richards R. Human metallothionein genes: structure of the functional locus at 16q13. Genomics. 1990;8:513–518. doi: 10.1016/0888-7543(90)90038-v. [DOI] [PubMed] [Google Scholar]
- Zhou XD, Sens MA, Garrett SH, Somji S, Park S, Gurel V, Sens DA. Enhanced expression of metallothionein isoform 3 (MT-3) protein in tumor heterotransplants derived from As+3 and Cd+2 transformed human urothelial cells. Toxicol Sci. 2006;93:322–330. doi: 10.1093/toxsci/kfl065. [DOI] [PubMed] [Google Scholar]