Abstract
The human Ether-à-go-go related gene (hERG) potassium channel is responsible for the rapid delayed rectifier potassium current that plays a critical role in the repolarization of cardiomyocytes during the cardiac action potential. In humans, inhibition of hERG by drugs can prolong the electrocardiographic QT interval, which, in rare instance, leads to ventricular arrhythmia and sudden cardiac death. As such, several medications that block hERG channels in vitro have been withdrawn from the market due to QT prolongation and arrhythmias. The current FDA guidelines recommend that drug candidates destined for human use be evaluated for potential hERG activity (www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm074963.pdf). Here, we employed automated planar patch clamp (APPC), high-throughput fluorescent Tl+ flux, and moderate-throughput [3H]dofetilide competition binding assays to characterize a panel of 49 drugs for their activities at the hERG channel. Notably, we used the same HEK293-hERG cell line for all assays, facilitating comparisons of hERG potencies across screening platforms. In general, hERG inhibitors were most potent in APPC assays, intermediate potent in [3H]dofetilide binding assays, and least potent in Tl+ flux assays. Binding affinity constants (pKi values) and Tl+ flux potencies (pEC50 values) correlated well with APPC pEC50 values. Further, the inhibitory potencies of many known hERG inhibitors in APPC matched literature values from manual and/or automated patch clamp systems. We also developed a novel fluorescent Tl+ flux assays to measure the effects of drugs that modulate hERG trafficking and surface expression.
Introduction
The human Ether-à-go-go Related Gene (hERG, KCNH2) encodes the pore forming α subunit of the voltage-gated potassium channel Kv11.1 (hERG).1 The channel consists of four α subunits, each with six transmembrane domains. The hERG channel is responsible for the rapid component of the delayed rectifier current (IKr), which is critical for the repolarization phase of the cardiac action potential.
Improper myocardiocyte repolarization manifests on an electrocardiogram as a prolonged QT interval (i.e., the interval between the start of the QRS complex [ventricular depolarization] and the end of the T wave [ventricular repolarization]), known as long QT syndrome (LQTS). The syndrome, which can be either acquired (e.g., drug-induced) or inherited (genetic), can evolve into a lethal cardiac arrhythmia. Most, if not all, drugs that prolong the QT interval target hERG channels, either inhibiting hERG IKr or disrupting hERG trafficking to the plasma membrane.2–8 Genetic studies on patients with inherited LQTS support the role of hERG in modulating the QT interval: hERG loss-of-function mutations (e.g., dominant-negative mutations, mutations that interfere with protein folding, and/or channel trafficking and activity) cause inherited LQTS.6,9,10 In addition, hERG channel activators have been associated with drug-induced short QT syndrome, which can lead to potentially fatal arrhythmia.11,12 Regardless of the underlying molecular mechanisms for drug-induced long or short QT syndrome, an arrhythmogenic effect has been the single leading cause of drug withdrawal from the U.S. market in last 40 years,13 and 10 drugs have been withdrawn in the last 20 years due to drug-induced LQTS.14 The current FDA guidelines recommend that all drugs be evaluated for hERG modulation prior to human use (www.fda.gov/downloads/Drugs/GuidanceComplianceRegulatoryInformation/Guidances/ucm074963.pdf).
The measurement of action potential duration in tissue or isolated myocytes is time consuming, labor intensive, and expensive. The gold-standard technique for such measurements is manual patch clamp, a technically challenging approach. Like manual patch clamp, automated planar patch clamp (APPC) systems can measure current under voltage clamp, yet provide greater throughput due to automation of cell handling and dispensing, drug dispensing, and voltage adjustments. Nevertheless, APPC throughput is insufficient for rapid screening of large numbers of compounds. Some hindrances to the development of better techniques are the financial investment required to develop new systems, the cost of consumables, and the variable success rates of automated systems. In addition to APPC, several nonelectrophysiological assays have been developed and used on transfected cells for hERG screening at medium to high throughput,15,16 including radioligand competition binding assays with [3H]dofetilide17,18 or [3H]astemizole,19 rubidium ion efflux assay,20 and fluorescence assays of membrane potential.21,22 More recently, higher throughput assays have been developed; these include an antibody-based chemiluminescent assay (96-well format) for hERG trafficking inhibitors3 and a Tl+ flux assay (384-well format) for hERG blockers.23,24
The Psychoactive Drug Screening Program provides hERG screening assays using HEK293 cells stably expressing hERG channels. One of our goals is to develop and employ comprehensive and efficient screening assays to identify different types of hERG modulators, including hERG inhibitors, hERG activators, and hERG protein trafficking inhibitors. When screening large numbers of compounds against antitargets such as hERG, it is critical to maximize the sensitivity and minimize the rate of false-negatives. To optimize our hERG screening assays and compare nonelectrophysiological platforms with APPC, we assayed in parallel a panel of 49 drugs using APPC, Tl+ flux, and [3H]dofetilide binding assays. Most of the drugs we selected are known hERG inhibitors from different therapeutic categories and with a wide range of chemical structures, including all drugs withdrawn from the U.S. market in the last 20 years due to QT prolongation. In addition, we evaluated ouabain and PD-118057 as positive controls for inhibition of hERG trafficking3,25 and for hERG activation,26 respectively, to determine whether our screening assays can identify compounds with these activities. We also included less potent drugs such as amantadine (known to prolong the QT interval at high doses) and phenytoin and venlafaxin (known to be weak hERG inhibitors) to determine the cutoff threshold to reduce the false-negative rate. As a negative control, we selected clozapine-N-oxide (CNO), which we had characterized extensively and determined to be inactive at many G-protein-coupled receptors, transporters, and ion channels (including hERG) (Roth BL, unpublished observations).27 To our knowledge, this is the first side-by-side comparison of the three screening assays used to identify hERG modulators, and the first high-throughput screening assay (384-well format) for hERG trafficking inhibitors. The results we present in this study demonstrate that an academic lab in the public sector (e.g., the National Institute of Mental Health Psychoactive Drug Screening Program) can provide comprehensive hERG screening to identify potential inhibitors, activators, and trafficking inhibitors.
Materials and Methods
Reagents
The drugs used in this study were purchased from either Sigma (St. Louis, MO) or Tocris (Ellisville, MO). Drug stocks were made in dimethyl sulfoxide (DMSO) at 10 mM in 1.5 mL microcentrifuge tubes and stored at 4°C in small aliquots until use. Working solutions of drugs were prepared in assay-specific buffers (see below) immediately before use. The FluxOR potassium ion channel assay kit (#F10017) was obtained from Invitrogen (Madison, WI). [3H]dofetilide was purchased from PerkinElmer Life and Analytic Sciences (Waltham, MA). Most other chemicals used in this study were purchased from either Sigma or Fisher Scientific (Fair Lawn, NJ). Cell culture media, 100 × penicillin–streptomycin stock, G418, and trypsin–ethylenediaminetetraacetic acid solution were purchased from Mediatech, Inc. (Manassas, VA). Fetal bovine serum (FBS) was from Gibco/Invitrogen.
Cell Culture
A HEK 293 cell line stably expressing hERG was obtained from ChanTest (Cleveland, OH), maintained in T-75 flasks or 10-cm dishes in growth media (Dulbecco's modified Eagle's medium [DMEM] supplemented with 10% FBS, 1× Penicillin/Streptomycin), and kept under selection with 500 μg/mL G418. The cells were routinely passaged when they reached >80% confluency and were never overgrown. After 30 passages, the cells were discarded and a new stock was revived as recommended by ChanTest.
Membrane Preparation
To obtain membrane pellets for radioligand binding assays, hERG-expressing HEK 293 cells were subcultured into 15-cm dishes in growth media without G418. When they reached 90% confluency, cells were scraped off the plate and pelleted by centrifugation for 10 min at 3,200 g at 4°C. The pellet was then resuspended in hypotonic ice-cold 50 mM Tris-HCl buffer (pH 7.4), and the crude membrane suspension was aliquoted into 1.5 mL microcentrifuge tubes (1 mL per tube). The crude membrane suspension was centrifuged at 20,000 g for 20 min (4°C) to yield membrane pellets that were stored at −80°C until use. Usually, three pellets were prepared from one 15-cm dish.
Radioligand Binding Assays
Saturation assays were carried out in triplicate in 0.5-mL round-bottom polypropylene 96-well plates in a final volume of 250 μL. Total and nonspecific binding was determined for 11 concentrations of [3H]dofetilide (ranging from 0.01 to 16 nM) in the absence and presence of 10 μM cold dofetilide, respectively. The hERG binding buffer (hBB) contained 135 mM NaCl, 5 mM KCl, 0.8 mM MgCl2, 10 mM HEPES, 10 mM glucose, 1 mM EGTA, and 0.01% BSA, pH 7.4, as reported.18 In brief, 5 × [3H]dofetilide working stock was prepared in hBB, and the stock concentrations were adjusted as necessary after liquid scintillation counting of an aliquot of each stock. The working stocks (50 μL/well) were distributed to all wells, and then 50 μL of hBB (for total binding) or 50 μL of cold dofetilide (for nonspecific binding) were added to appropriate wells. Membrane pellets were resuspended in hBB and homogenized by repeated passage through a 26-gauge needle; after homogenization, membranes were diluted in hBB to a final volume of 16 mL. The membrane suspension was added to each well (150 μL, ∼50 μg of protein), and the plates were incubated for 90 min at room temperature in the dark. Reactions were terminated by filtration through 0.3% of polyethyleneimine-soaked glass fiber filters (Printed Filtermat A; PerkinElmer Life and Analytic Sciences) on a FilterMate Harvester (PerkinElmer Life and Analytic Sciences) followed by two washes with cold washing buffer (50 mM Tris-HCl, 10 mM MgCl2, and 0.1 mM ethylenediaminetetraacetic acid, pH 7.4). The filters were then microwaved briefly to dry them, and Meltilex scintillant (PerkinElmer Life and Analytic Sciences) was applied to the dried filters with a hot plate. Radioactivity retained on the filter was counted on a PerkinElmer 1450 MicroBeta Trilux plate counter.
In competition binding assays, 12-point concentration–response curves (maximum concentration 30 μM) were generated in triplicate. The assays were conducted in 0.5-mL round-bottom polypropylene 96-well plates in a final volume of 250 μL, and the final concentration of [3H]dofetilide was 1.5 nM. The test compounds were diluted in hBB from 2 mM DMSO stocks (1.5% DMSO does not affect [3H]dofetilide binding). Radioligand stocks were prepared and distributed in the plate as above. Dilutions of the test compounds, cold dofetilide (positive control), or hBB (negative control) were added to the appropriate wells (50 μL of 5 × solutions). Membrane pellets were prepared, homogenized, and distributed in the plate as above. Three membrane pellets were used for each plate. The reactions were incubated, harvested, and counted as above. The assay protocol table is included as Supplementary Table S1 (Supplementary Data are available online at www.liebertonline.com/adt).
Tl+ Flux Assays
Thallium flux assays were carried out using the FluxOR Potassium Ion Channel Assay kit (Invitrogen) after extensive modification and optimization. A large number of preliminary attempts were made to determine the optimal conditions for assaying Tl+ flux using HEK293 cells stably expressing hERG (results not shown). The conditions we modified included (1) assay buffer, such as the assay buffer from the kit, our own assay buffer (see below), and the external buffer used in the APPC assay (see below); (2) a variety of K+ concentrations (0–20 mM final) and Tl+ (0–8 mM final) in the stimulation buffer; (3) cell density (5,000–40,000 cells/well); (4) use of growing and cryopreserved cells; (5) varying drug incubation times (5–30 min) before stimulation cycle; (6) DMSO concentration; (7) and a variety of endpoint readouts such as initial slope, area under curve, and maximal fluorescence intensity. The final optimized protocol is detailed below.
Before assay, 25,000 HEK 293-hERG cells were seeded into each well of a poly-L-Lys (50 μg/mL)-coated 384-well black clear bottom plate (Greiner Bio-one, Monroe, NC) in DMEM supplemented with 5% FBS without G418, and the plate was incubated overnight. The next day, Tl+ flux dye-loading solution was prepared by dilution of FluxOR reagent stock (1,000×, Component A) and PowerLoad (100×, Component C) in fresh assay buffer (1 × Hank's balanced salt solution, 20 mM HEPES, and 2.5 mM Probenecid, pH 7.40), and added (20 μL/well) in place of the growth media. The plate was then incubated for 90 min at room temperature in the dark. During the incubation, drug plates (8 or 16 concentration points in quadruplicate or triplicate, respectively) and stimulation solution were prepared in Tl+ flux assay buffer in polypropylene 384-well plates (Fisher Scientific). The stimulation solution was composed as follows: 2.5 mL deionized water, 1 mL FluxOR chloride-free buffer (5 × stock, Component E), 1 mL K2SO4 stock (125 mM, Component F), and 0.5 mL Tl2SO4 (50 mM, Component G). The K+ and Tl+ concentrations in the stimulation working solution are 50 and 10 mM, respectively. At the end of dye loading period, the dye was removed, and a FLIPRTETRA (MDS Analytical Technologies, Sunnyvale, CA) was used to transfer drugs from drug plates into cell plates (25 μL/well of 1.25× of final concentrations). The cells were incubated with the drugs for 15 min at room temperature in the dark. Stimulation solution (6.3 μL/well) was added with the FLIPRTETRA. The fluorescence intensity (excitation at 490 nm and emission at 525 nm) in each well was measured every second for 10 s before addition of stimulation solution, and for 90 s thereafter by the FLIPRTETRA using ScreenWorks 2.0 software. When cryopreserved hERG HEK 293 cells (stored in DMEM + 10% FBS and 10% DMSO) were plated for acute Tl+ flux assays, cells were first washed once to remove DMSO, then plated at 30,000 cells/well in DMEM + 5% FBS, and finally assayed 5 h after plating. Results using cryopreserved cells were not different from those obtained using fresh cells. The above protocol was designed to measure acute effect of drugs on hERG activity (such as hERG inhibitors and activators). For those drugs without acute inhibitory effect in the Tl+ flux assay, we modified the standard protocol to conduct a longer chronic study to identify compounds that might be acting through indirect mechanisms (e.g., hERG trafficking inhibition, hERG internalization, and Na+-K+ ATPase inhibition). Briefly, cells were first treated with drugs for desired time period (up to 16 h) and were washed once with assay buffer before dye loading. The dye solution was replaced with assay buffer and the fluorescence intensity was measured upon addition of stimulation solution per the standard protocol. The assay protocol table is included as Supplementary Table S4.
APPC: PatchXpress Electrophysiology
External and internal solutions
On the day of patch clamp assays, fresh external buffer (137 mM NaCl, 4 mM KCl, 1.8 mM CaCl2, 1 mM MgCl2, 10 mM HEPES, and 10 mM glucose, pH 7.4 by NaOH) and fresh internal buffer (15 mM NaCl, 70 mM KF, 60 mM KCl, 1 mM MgCl2, 5 mM HEPES, 5 mM EGTA, 4 mM ATP, and 0.4 mM GTP, pH 7.2 by KOH) were prepared at room temperature. The KF-based internal buffer was adopted from a report by Zeng et al.,28 in which they reported a greater success rate with a KF-based internal buffer than with a traditional KCl internal solution. We added ATP, GTP, and Mg2+ to the KF-based internal solution to prevent potential current run-down. Osmolarity of the buffers was determined with a VAPRO 5520 Vapor Pressure Osmometer (Wescor, Inc., Logan, UT). The osmolarity was usually 285 ± 10 mmol/kg for the external buffer and 295 ± 10 mmol/kg for the internal buffer. Then, the buffers were vacuum filtered to remove any air bubbles or small particles.
Cell preparation
For patch clamp assays, the hERG HEK 293 cells were maintained as described above and subcultured into 10-cm dishes 2 days before scheduled assays at 1–2 million cells per dish in growth media without G418. Cells were not used if they reach >90% confluency. To prepare the cells for patch clamping, we followed ChanTest's recommendations with minor modifications. The goal was to prepare a clean, fresh cell suspension immediately before loading cells onto the PatchXpress. Cells were briefly washed with PBS, treated with Accutase (2.5 mL per 10-cm dish; Sigma) for 4 min at room temperature to detach them, gently transferred and suspended in 20 mL growth media without G418 in a 50-mL centrifuge tube, and allowed to recover from detachment for 30 min at 37°C in CO2 incubator. At the end of the incubation period, 1 million cells were transferred into another 50 mL centrifuge tube and pelleted by centrifugation at 250 g for 2.5 min at room temperature. The cell pellet was then gently resuspended into 170 μL of the external buffer, transferred into a 1.5 mL mcirocentrifuge tube, and loaded onto the PatchXpress 7000A (MDS Analytical Technologies) in waiting mode for cells. To minimize delay and cell clumping, the APPC system must be started well in advance of use and primed with fresh external and internal solutions before preparing cells. The patch clamping procedure should be started with a new SealChip right at the end of the 30 min incubation and recovery period. By the time the cell suspension is ready to load into the APPC system, the system should then be in waiting mode for receiving the cells.
Drug plate preparation
While cells were in the incubation and recovery periods, drug solutions were prepared in 0.5 mL polypropylene round-bottom 96-well drug plate (Fisher Scientific). Drugs in 10 mM DMSO stocks were diluted in the external buffer at a final volume of 360 μL each well, in which is sufficient to test two cells (triple addition at 50 μL each for one cell per concentration). The final concentration of DMSO was 0.3% (v/v) for all dilutions (except for several drugs that required 3% DMSO as indicated in the Results). For initial assays, 8-point (ranging from 30 nM to 30 μM with 0.3% DMSO in external buffer as a negative control) concentration–response curves were generated. For subsequent assays, the concentration range was adjusted as necessary to give full concentration–response curves (0%–100% inhibition). The drug plate setup information was manually entered into the APPC system before starting the procedure. The assay was initiated within 15 min, and the highest concentration was tested within 90 min. A new drug plate was prepared for each new SealChip.
APPC procedures
The APPC procedure started with manual loading of a SealChip16™ (AVIVA Biosciences, San Diego, CA), and was executed automatically at room temperature. The system requires ∼7 min to (1) dry and load the SealChip onto the recording station; (2) add external and internal solutions; (3) check the quality of each of the 16 chambers; and (4) enter waiting mode and prompt for cell loading, at which point the cell suspension prepared above was immediately loaded. After loading, the single-cell suspension was triturated briefly and aliquoted (35 μL, ∼20,000 cells) by the on-board robotic Cavro pipette to each chamber on the SealChip. First, positive pressure (6 mmHg) and then brief suction with negative pressure (−45 mmHg) were applied to help the cells descend quickly, and a single cell was drawn onto the top of the electrode hole of each chamber. A negative pressure ramp to 75 mmHg (at 5 mmHg/s) was then applied repeatedly until the seal resistance reached 1GΩ (Gigaohm) or greater for Giga-seals, and over 200 MΩ (Megaohm) for 2nd seals. After obtaining a seal, another negative pressure ramp from −40 to −250 mmHg was applied repeatedly to rupture the patched membrane and achieve whole-cell configuration. Chambers with low seal resistances (i.e., no cell detection) were terminated within 90 s; chambers that did not form seals within 5 min were terminated by either a built-in script or by user intervention. After achieving whole cell configuration, the voltage protocol (Fig. 1C) was activated, beginning with a 2-min washing-out and 5-min stabilization period. After stabilization, cells with <0.2 nA tail current amplitude were terminated. The voltage protocol consisted following steps: (1) depolarization from a holding potential of −80 to −50 mV for 50 ms to measure leak current without activation of the hERG channel; (2) further depolarization to + 20 mV for 5 s to activate the hERG channel (hERG channels were activated and quickly inactivated); (3) repolarization to −50 mV for 1.7 s to remove inactivation and elicit outward hERG tail current; and (4) repolarization to a holding potential of −80 mV to keep hERG channels closed. The pulse pattern was applied repeatedly every 10 s (0.1 Hz). The instantaneous current at −50 mV before stepping to +20 mV was designated as leak current and was subtracted from corresponding peak tail current for data processing. Each concentration–response experiment started with a buffer control to determine the maximal hERG tail current (0% inhibition) in the absence of drugs. Each dose was applied three times (50 μL each at 25 μL/s) with 11 s between additions, and each addition was preceded by aspiration of buffer or previous drug solution from the chamber down to a 5 μL dead volume. This triple addition (Fig. 1D, three green lines) and aspiration protocol fully exchanges the solution in the chamber with negligible dilution within 1 min. The built-in DataStable script was activated between each dose. When either drug effect reached steady state (<0.1% difference from last measurement) or after a maximum of 5 min, the next dose was queued for delivery. At the end of each concentration–response trial, a 7-min wash-out was applied to monitor recovery of the hERG channels from inhibition. The assay protocol table is included as Supplementary Table S5.
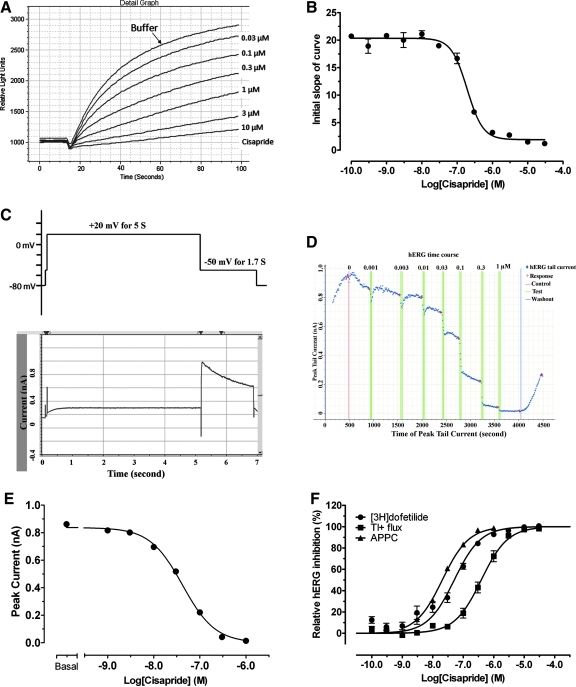
Fig. 1.
(A) Raw fluorescence signal traces recorded on fluorometric imaging plate reader (FLIPR) with HEK 293 cells stably expressing human Ether-à-go-go related gene (hERG) channels. Recording started 10 s before addition of stimulation solution and continued for another 90 s. Cisapride was shown to reduce signals in a dose-dependent manner (error bars were not shown). Initial slope of curves was plotted against concentrations of cisapride (triplicate set) and fitted to a four-parameter logistic function (B) in the Prism 5.0. (C) Voltage protocol (upper) and corresponding hERG tail current recording (lower) for the automated planar patch clamp (APPC) assay. (D) The captured screen in the DataXpress program of a representative whole-cell patch clamp recordings of hERG tail currents in the absence and presence of cisapride. Red vertical line indicated addition of buffer control and green vertical triple lines indicated triple additions of drugs. Cisapride concentrations (μM) are listed above the green lines. Red cross indicates the time point when hERG tail current was measured by the system. After washout (blue vertical line), hERG channel was shown to partially recover from complete inhibition. The hERG tail currents were extracted from the DataXpress, plotted against cisapride concentrations, and fitted to a four-parameter logistic function (E) in the Prism 5.0. (F) Side-by-side comparison of dose-dependent relationships of cisapride in (•) [3H]dofetilide competition binding assay, (▪) Tl+ flux assay, and (▴) APPC assay. Results from multiple assays (n ≥ 2) were normalized to percentage inhibition and pooled for curve-fitting in Prism. The potency or affinity value and corresponding Hill slope from the best fitting model were reported in Table 2.
Data Analysis
GraphPad Prism V5.0 for Windows (San Diego, CA) was used for normalization, curve-fitting, and correlation analysis. For saturation binding assays, the data (cpm per well) were analyzed by nonlinear regression with Prism's built-in one-site saturation binding function to obtain Kd values for [3H]dofetilide. The average Kd value of 4.7 nM (n = 5, each measured in triplicate) was then used in the competition binding assays to obtain Ki values according to the Cheng-Prusoff equation.29 For competition binding assays, the data (cpm per well) from each plate were transformed and normalized to percentage inhibition (i.e., total binding = 0% inhibition, nonspecific binding = 100% inhibition).
For Tl+ flux assays, the ScreenWorks program generated fluorescence intensity time–course curves and calculated the slope of the curve for the first 15 s after addition of the stimulation buffer (Fig. 1A). Then, Prism was used to transform and normalize the slope values to percent inhibition (buffer = 0% inhibition, 10 μM cisapride = 100% inhibition) (Fig. 1B, F).
For APPC assays, data were collected and deposited into the database program DataXpress V2.0 (MDS Analytic Technologies). If a cell had a leak current that was as much as 1/3 of the total tail current, the data set for that cell was excluded. If a cell showed current run-down of >25% of the initial total tail current at the first (lowest) drug concentration, the data set for that cell was excluded. The drug concentration range was usually adjusted after an initial assay so that in subsequent assays the lowest drug concentration would inhibit hERG by <10%. The hERG tail currents were transformed and normalized to percent inhibition with a built-in script in the DataXpress program (total initial tail current = 0% inhibition, no current = 100% inhibition) (Fig. 1D–F).
For all three assays, normalized results from multiple assays were pooled and analyzed with Prism's built-in four-parameter logistic functions (Fig. 1F). A built-in statistical comparison function in Prism was then used to determine if a model with a variable Hill slope fit the data better than a model with a standard Hill slope (slope of 1); a P value < 0.05 was considered significant. Finally, the potency value and corresponding Hill slope from the best-fit model were reported.
Results
We selected 49 drugs for study, most of which are known hERG blockers used routinely in electrophysiology and/or are formerly FDA-approved drugs that were withdrawn from the market because of their induction of LQTS. We measured in parallel the potencies of these drugs using [3H]dofetilide competition binding, Tl+ flux assay, and APPC. As indicated in Figure 1, cisapride inhibited hERG activity in a dose-dependent manner in the Tl+ flux assay (Fig. 1A, B) and in the APPC assay (Fig. 1C–E). The assay parameters are summarized in Table 1. The results are summarized in Table 2 (corresponding potency values, in μM, are listed in Supplementary Table S1).
Table 1.
The Assay Parameters for [3H]Dofetilide Binding Assays, Tl+ Flux Assays, and Automated Planar Patch Clamp Assays
| [3H]Dofetilide Binding Assay | Tl+ Flux Assay | APPC Assay |
|---|---|---|
| Average total binding: 798 ± 26 cpm/well |
Average initial slope: 33.8 ± 1.6 |
Seals with initial Rm over 200 MΩ (n = 1,245 cells) |
| Average nonspecific binding: 31 ± 1 cpm/well |
Slope with 10 μM Cisapride: 2.4 ± 0.1 |
• Overall seal rate: 81% • Rseal = 1,121 ± 27 MΩ • Rm = 729 ± 14 MΩ • Ra = 6.78 ± 0.14 MΩ |
| Z′ score: 0.83 ± 0.01 |
Z′ score: 0.69 ± 0.02 |
Average tail current: 723 ± 18 pA (n = 958 cells) |
| Total of 113 × 96-well plates | Total of 91 × 384-well plates | Overall successful rate: 59%, for these successful cells (n = 893 cells, based on 94 SealChips) • Rseal = 1,033 ± 30 MΩ • Rm = 692 ± 17 MΩ • Ra = 6.81 ± 0.16 MΩ • Time = 73.2 ± 0.4 min |
Values represent mean ± SEM. Rm is for membrane resistance; Rseal for seal resistance; Ra for access resistance. Seal rate was based on total number of seals with initial Rm over 200 MΩ. A successful cell was defined as being capable of finishing at least 5 points of 7-point dose-responses.
APPC, automated planar patch clamp.
Table 2.
Summary of Pharmacological Parameters from Automated Planar Patch Clamp, Tl+ Flux, and [3H]Dofetilide Binding Assays
| |
|
Tl+ Flux Assays |
APPC Assays |
APPC Over (Fold) |
||||
|---|---|---|---|---|---|---|---|---|
| Drugs | [3H]Dofetilide (3H) pKi | pEC50 | Hill Slope | pEC50 | Emax (%) | Hill Slope | 3H | Tl+ |
| Amantadine |
4.98 ± 0.10 |
NI |
NA |
4.38 ± 0.20 |
55.4 ± 8.0 |
1 |
0.3 |
NA |
| Amiodarone |
6.23 ± 0.08 |
5.56 ± 0.03 |
1 |
5.97 ± 0.08 |
∼100.0 |
0.80 ± 0.07 |
0.6 |
2.6 |
| Amitriptyline |
5.05 ± 0.08 |
4.55 ± 0.04 |
1.39 ± 0.17 |
5.87 ± 0.04 |
∼100.0 |
1.11 ± 0.08 |
6.6 |
20.9 |
| Aripiprazole |
5.99 ± 0.06 |
5.09 ± 0.03 |
1.72 ± 0.18 |
6.62 ± 0.22 |
84.0 ± 15.3 |
1 |
4.3 |
33.9 |
|
Asetmiozole |
8.50 ± 0.06 |
5.68 ± 0.01 |
2.21 ± 0.13 |
7.67 ± 0.12 |
95.3 ± 5.7 |
1 |
0.2 |
97.7 |
| Bepridil |
6.29 ± 0.07 |
5.36 ± 0.03 |
2.61 ± 0.41 |
6.41 ± 0.10 |
∼100.0 |
1 |
1.3 |
11.2 |
| Chlorpheniramine |
5.26 ± 0.08 |
4.83 ± 0.04 |
1.42 ± 0.17 |
5.67 ± 0.08 |
98.4 ± 5.0 |
1 |
2.6 |
6.9 |
| Chlorpromazine |
5.68 ± 0.08 |
5.18 ± 0.03 |
1.70 ± 0.14 |
5.67 ± 0.09 |
∼100.0 |
1 |
1.0 |
3.1 |
| Ciclopirox |
4.77 ± 0.07 |
4.03 ± 0.16a |
1 |
3.58 ± 0.20 |
∼100.0 |
1 |
0.1 |
0.4 |
|
Cisapride |
7.41 ± 0.05 |
6.47 ± 0.03 |
1 |
7.69 ± 0.03 |
∼100.0 |
1 |
1.9 |
16.6 |
| Clomipramine |
5.52 ± 0.07 |
4.85 ± 0.02 |
2.14 ± 0.16 |
6.00 ± 0.00 |
∼100.0 |
1 |
3.0 |
14.1 |
| Clozapine |
5.46 ± 0.06 |
4.93 ± 0.03 |
1.19 ± 0.10 |
6.09 ± 0.09 |
∼100.0 |
1 |
4.3 |
14.5 |
| Clozapine-N-oxide |
5.03 ± 0.14 |
NI |
NA |
NI |
NA |
NA |
NA |
NA |
|
Dofetilide |
8.39 ± 0.03 |
6.64 ± 0.03 |
1 |
7.75 ± 0.06 |
∼100.0 |
1.00 |
0.2 |
12.9 |
| Doxepin |
5.08 ± 0.11 |
4.67 ± 0.03 |
1.22 ± 0.12 |
5.67 ± 0.10 |
96.1 ± 5.8 |
1.00 |
3.9 |
10.0 |
| Droperidol |
6.54 ± 0.05 |
6.33 ± 0.04 |
1 |
7.34 ± 0.12 |
95.4 ± 5.9 |
1 |
6.3 |
10.2 |
| E-4031 |
7.66 ± 0.04 |
6.72 ± 0.03 |
1.42 ± 0.13 |
7.57 ± 0.06 |
∼100.0 |
1.80 ± 0.41 |
0.8 |
7.1 |
| Fluoxetine |
5.37 ± 0.06 |
5.14 ± 0.03 |
1.55 ± 0.17 |
6.44 ± 0.04 |
∼100.0 |
1 |
11.8 |
20.0 |
| Fluvoxamine |
5.12 ± 0.07 |
4.72 ± 0.04 |
1.52 ± 0.18 |
5.50 ± 0.08 |
∼100.0 |
1 |
2.4 |
6.0 |
|
Grepafloxacin |
4.77 ± 0.10 |
NI |
NA |
NI |
NA |
NA |
NA |
NA |
| Haloperidol |
6.98 ± 0.06 |
6.57 ± 0.02 |
0.86 ± 0.03 |
7.50 ± 0.07 |
98.9 ± 3.9 |
1 |
3.3 |
8.5 |
| Imipramine |
5.46 ± 0.09 |
4.50 ± 0.03 |
1.53 ± 0.16 |
6.29 ± 0.21 |
∼100.0 |
0.64 ± 0.12 |
6.8 |
61.7 |
|
LAAM |
5.49 ± 0.07 |
5.24 ± 0.03 |
1.45 ± 0.11 |
6.14 ± 0.13 |
98.4 ± 7.7 |
1 |
4.5 |
7.9 |
| Lidoflazine |
7.22 ± 0.04 |
5.52 ± 0.03 |
2.11 ± 0.30 |
7.01 ± 0.11 |
∼100.0 |
1 |
0.6 |
30.9 |
| Loratadine |
5.26 ± 0.07 |
4.66 ± 0.04 |
2.21 ± 0.31 |
5.82 ± 0.23 |
∼100.0 |
0.71 ± 0.17 |
3.6 |
14.5 |
| Maprotilene |
5.36 ± 0.06 |
4.56 ± 0.03 |
1.86 ± 0.23 |
5.29 ± 0.19 |
∼100.0 |
1 |
0.9 |
5.4 |
| Mefloquine |
5.32 ± 0.08 |
4.60 ± 0.02 |
2.17 ± 0.26 |
5.66 ± 0.12 |
∼100.0 |
1 |
2.2 |
11.5 |
|
Mibefradil |
6.02 ± 0.07 |
5.17 ± 0.03 |
2.48 ± 0.29 |
6.03 ± 0.12 |
99.4 ± 7.5 |
1 |
1.0 |
7.2 |
| Nortriptyline |
5.32 ± 0.08 |
4.53 ± 0.02 |
1.57 ± 0.11 |
5.82 ± 0.18 |
85.9 ± 11.3 |
1 |
3.2 |
19.5 |
| Olanzapine |
5.20 ± 0.07 |
4.28 ± 0.05 |
1 |
5.63 ± 0.26 |
∼100.0 |
0.57 ± 0.09 |
2.7 |
22.4 |
| Ouabain |
4.75 ± 0.08 |
NI |
NA |
NI |
NA |
NA |
NA |
NA |
| PD-118057 |
4.91 ± 0.07 |
4.28 ± 0.07a |
1 |
Activator |
NA |
1 |
NA |
NA |
| Perphenazine |
5.98 ± 0.06 |
5.21 ± 0.04 |
1.41 ± 0.17 |
5.94 ± 0.14 |
∼100.0 |
1 |
0.9 |
5.4 |
| Phenytoin |
4.74 ± 0.12 |
NI |
NA |
6.67 ± 0.51 |
59.8 ± 13.0 |
0.47 ± 0.16 |
85.1 |
NA |
| Pimozide |
7.63 ± 0.04 |
5.79 ± 0.05 |
1.40 ± 0.20 |
7.16 ± 0.07 |
∼100.0 |
1 |
0.3 |
23.4 |
| Propafenone |
5.74 ± 0.06 |
5.42 ± 0.03 |
1.92 ± 0.25 |
7.07 ± 0.06 |
∼100.0 |
1 |
21.4 |
44.7 |
| Pyrilamine |
5.23 ± 0.08 |
5.05 ± 0.04 |
1 |
5.71 ± 0.06 |
∼100.0 |
1 |
3.0 |
4.6 |
| Quetiapine |
5.35 ± 0.06 |
4.94 ± 0.05 |
1 |
5.60 ± 0.11 |
∼100.0 |
1 |
1.8 |
4.6 |
| Quinidine |
5.56 ± 0.07 |
5.20 ± 0.04 |
1 |
6.58 ± 0.62 |
∼100.0 |
0.51 ± 0.16 |
10.5 |
24.0 |
| Risperidone |
6.00 ± 0.07 |
5.78 ± 0.05 |
1 |
6.55 ± 0.07 |
∼100.0 |
1.33 ± 0.23 |
3.6 |
5.9 |
|
Sertindole |
6.89 ± 0.04 |
5.52 ± 0.03 |
1.74 ± 0.17 |
6.39 ± 0.09 |
∼100.0 |
1.30 ± 0.25 |
0.3 |
7.4 |
| Tamoxifen |
6.02 ± 0.07 |
4.65 ± 0.08 |
1 |
5.17 ± 0.16 |
∼100.0 |
1 |
0.1 |
3.3 |
|
Terfenadine |
6.87 ± 0.05 |
5.30 ± 0.04 |
1 |
6.87 ± 0.49 |
∼100.0 |
0.59 ± 0.18 |
1.0 |
37.2 |
|
Terodiline |
5.67 ± 0.06 |
5.37 ± 0.02 |
1.60 ± 0.11 |
6.49 ± 0.35 |
∼100.0 |
0.60 ± 0.17 |
6.6 |
13.2 |
|
Thioridazin |
5.92 ± 0.08 |
4.95 ± 0.03 |
2.92 ± 0.70 |
6.29 ± 0.08 |
∼100.0 |
1 |
2.3 |
21.9 |
| Trifluoperazine |
6.01 ± 0.07 |
4.76 ± 0.02 |
2.45 ± 0.17 |
5.81 ± 0.11 |
99.2 ± 6.9 |
1 |
0.6 |
11.2 |
| Venlafaxine |
4.78 ± 0.07 |
NI |
NA |
4.67 ± 0.07 |
95.9 ± 4.2 |
|
0.8 |
NA |
| Verapamil |
5.81 ± 0.07 |
5.55 ± 0.02 |
1.78 ± 0.17 |
6.24 ± 0.08 |
∼100.0 |
1 |
2.7 |
4.9 |
| Way 161503 | 5.09 ± 0.06 | 4.35 ± 0.03 | 1 | 5.45 ± 0.10 | 97.2 ± 7.3 | 1 | 2.3 | 12.6 |
Affinity was expressed in pKi (−log of Ki from binding assays) and potency was expressed in pEC50 (−log of EC50 from Tl+ flux or APPC assays). Results represented best-fit values ± SE taken directly from curve-fittings in Prism. Multiple assays (n ≥ 2) were normalized to percentage inhibition and pooled for analysis. All [3H]dofetilide competition binding curves favored (P < 0.05) four-parameter logistic model with Hill slope of 1. For Tl+ flux and APPC assays, if a curve favored (P < 0.05) variable slope model, variable slope and corresponding potency values were reported; otherwise, Hill slope of 1 and corresponding potency value were reported. The maximal inhibition in competition binding and Tl+ flux assays were 100% and not listed in the table (see Results for PD-118057 and ciclopirox). The last two columns list potency difference (in fold) between APPC over [3H]-dofetilide binding assay (3H) or Tl+ flux assay. In average, the APPC assay is 17-fold more potent than Tl+ assay and 5-fold more potent than the [3H]-dofetilide binding assay. Those drug names in bold are the 10 withdrawn drugs from the market since 1990s.
Indicated potencies for human Ether-à-go-go related gene activator activity in the Tl+ flux or APPC assay.
NI, no inhibition at up to 100 μM; NA, not applicable.
The pEC50 values from APPC ranged from 3.58 (ciclopirox) to 7.75 (dofetilide), with an average pEC50 of 6.17 (0.67 μM). The pEC50 from Tl+ flux assays ranged from 4.03 (ciclopirox) to 6.72 (E-4031) with an average pEC50 of 5.17 (6.69 μM). The pKi determined by [3H]dofetilide binding assays ranged from 4.74 (phenytoin) to 8.50 (astemizole) with an average pKi of 5.58 (1.41 μM). Notably, for many of the 49 drugs, the hERG inhibitory potencies in APPC were comparable to those summarized in a recent review.30 Among the 49 tested drugs, 28 were more potent in the APPC assay than in the [3H]dofetilide binding assay; 13 drugs were more potent in the [3H]dofetilide binding assay than in the APPC assay; and three drugs were similarly potent in the [3H]dofetilide binding assays and APPC assays. On the basis of the average potency values, the APPC assay is 5-fold more sensitive than the [3H]dofetilide binding assay and about 17-fold more sensitive than the Tl+ flux assay for detecting hERG inhibitory potency. These results indicated that, of the three assays, APPC is the most sensitive and the Tl+ flux assay is the least sensitive. Notably, sensitivity decreases with throughput.
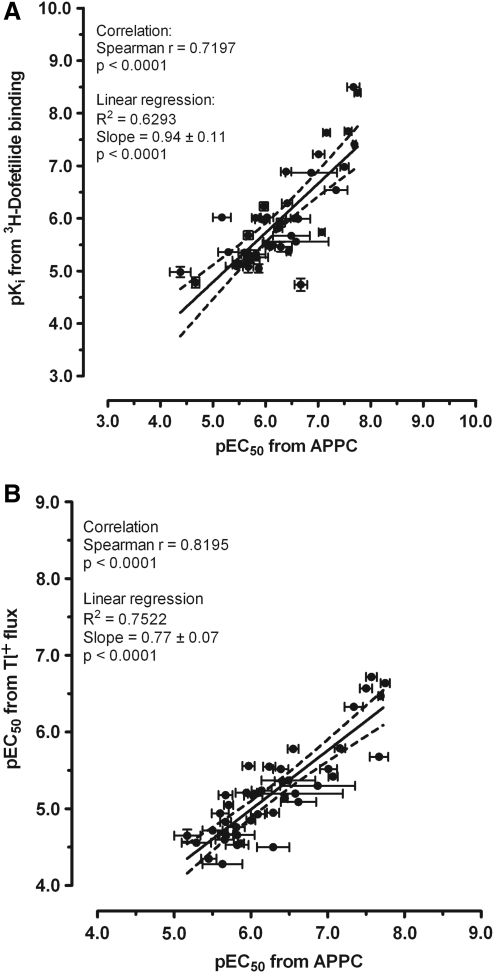
The pEC50 values from APPC assays were then plotted against the pKi values from [3H]dofetilide binding assays or the pEC50 values from Tl+ flux assays (Fig. 2). Linear regression showed a statistically significant, though modest, correlation in both cases. Among the 49 drugs tested, six drugs did not inhibit hERG activity in the Tl+ flux assay (Table 2): amantadine, CNO, phenytoin, venlafaxine, ouabain, and grepafloxacin. Additionally, CNO, ouabain, and grepafloxacin showed no inhibition in the APPC assays. The drug PD-118057 was a hERG activator in both Tl+ flux and APPC assays. Ciclopirox was found to activate hERG in Tl+ flux but not in APPC. CNO, an inert metabolite of clozapine, was chosen as a negative control. As expected, CNO did not inhibit hERG channels either in Tl+ flux at up to 100 μM (−6.5% ± 6.5% inhibition, n = 2, vs. − 8.2% ± 2.4% by 1% DMSO, n = 3) or in APPC assays at up to 30 μM (36.2% ± 7.4% inhibition, n = 4 cells, vs. 32.1% ± 7.4% inhibition with 0.3% DMSO, n = 5 cells). This small reduction in tail current measured in APPC was not greater than the current run-down observed with external buffer alone (24.2% ± 4.9%, n = 9 cells). However, CNO inhibited [3H]dofetilide binding with an affinity of 9.33 μM; therefore, we conclude that CNO is a false-negative.
Fig. 2.
Correlation between the inhibition potencies from APPC assay versus the binding affinities from [3H]dofetilide binding assays (A) or inhibition potencies from Tl+ flux assays (B). Means ± SE were taken from Table 2, except those without inhibitory activity in either Tl+ flux or APPC assays. The solid line represents linear regression; dashed lines represent 95% confidence intervals.
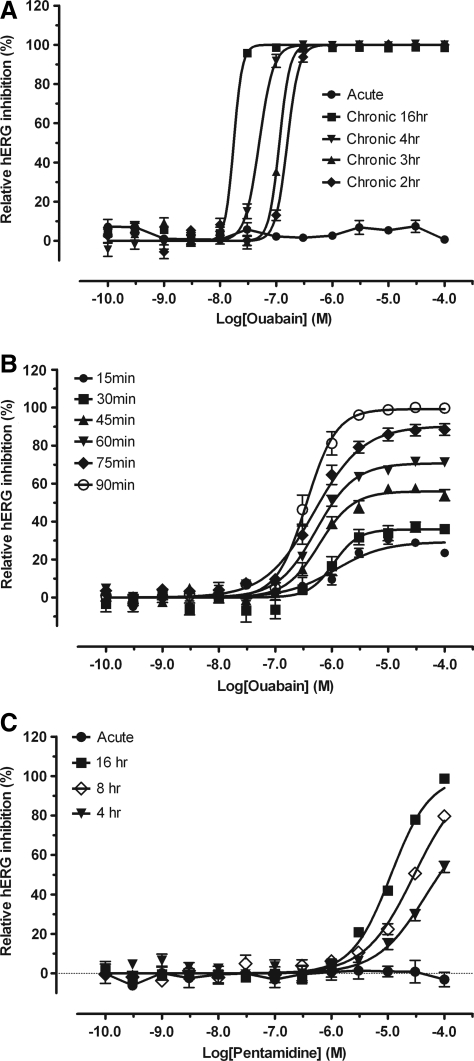
Ouabain is an Na+-K+ ATPase inhibitor and a cardiac glycoside that potently inhibits hERG trafficking without acute effects on hERG activity.25 We included ouabain in this study as a positive control for inhibition of hERG trafficking to determine whether our screening can detect such compounds. As expected, ouabain did not display acute inhibition of hERG channel activity in either Tl+ flux assay (Fig. 3) at up to 100 μM (0.7% ± 1.8%, n = 4) or APPC assays at up to 300 μM (44.6% ± 6.0% inhibition, n = 6 cells, vs. 40.2% ± 1.9% inhibition with 3% DMSO, n = 5 cells).
Fig. 3.
Acute and chronic effect of hERG trafficking inhibitor ouabain (A and B) and pentadimine (C) on hERG channel activity. Activity of ouabain and pentamidine was determined with acute Tl+ flux assay (•), 2–16 h chronic Tl+ assay (A and C: ▪, 16 h; ◊, 8 h; ▾, 4 h; ▴, 3 h; ♦, 2 h, n ≥ 2 assays and each in quadruplicate set), and 15–90 min chronic Tl+ assay (B: ○, 90 min; ♦, 75 min; ▾, 60 min; ▴, 45 min; ▪, 30 min; •, 15 min, n ≥ 2 assay and each in quadruplicate set). Cells were first incubated with ouabain or pentamidine for desired time period at 37°C in the incubator; the remaining hERG activity was determined with Tl+ flux assays. Multiple sets of results were normalized and pooled for curve-fitting. Potency, maximum inhibition, and Hill slope values are reported in the Table 3.
Inhibitors of hERG trafficking can have effects similar to hERG blockers in vivo. Therefore, we developed a novel paradigm to measure inhibition of hERG trafficking using the Tl+ flux assay. Cells were first incubated with ouabain for up to 16 h, and then the total remaining hERG channel activity was measured. As indicated in Figure 3 and Table 3 (also Supplementary Table S2), incubation of cells with ouabain for up to 16 h reduced hERG channel activity in a concentration-dependent manner with a potency of 158 nM (n = 2, each in quadruplicate) after a 2-hr treatment, 114 nM (n = 2) after a 3-hr treatment, 50 nM (n = 4) after a 4-hr treatment, and 18 nM (n = 4) after a 16-hr treatment. Comparison of the four curves indicated that the increased inhibitory potency is significant (P < 0.0001). Our results with ouabain are consistent with previous studies examining surface expression of hERG in stably transfected HEK 293 cells after overnight treatment with ouabain.25 However, the finding that sub-μM concentrations of ouabain that are inactive in acute paradigms completely inhibit hERG activity within 2 h is novel. We therefore examined the time-dependent inhibition of hERG by ouabain. As indicated in Figure 3 and Table 3, ouabain displayed significant hERG inhibition (29%) after 15 min (EC50 = 1.16 μM) and reached its maximal inhibition (99%) after 90 min treatment (EC50 = 0.36 μM). Overnight incubation of the cells with ouabain, as with DMSO and CNO, had no effect on cell viability as assessed by baseline fluorescence after dye loading (data not shown). In addition, CNO had no inhibitory effect on hERG activity after overnight incubation (3.4% ± 4.2% inhibition at 30 μM and − 11.4% ± 5.5% inhibition at 100 μM, n = 4). Similarly, DMSO showed a small stimulatory effect at 0.3% (−8.7% ± 1.4% inhibition, n = 6) and 1% (corresponding to the concentration of 100 μM drugs) (−28.6% ± 2.3% inhibition, n = 6). To examine whether pentamidine, another well-known hERG trafficking inhibitor,4,31 behaves similarly to ouabain, we tested the drug in both acute and chronic Tl+ flux assays. As indicated in Figure 3C and Table 3, pentamidine did not have an inhibitory effect on hERG channel activity in acute Tl+ flux assays at up to 100 μM, but displayed dose- and time-dependent inhibition in chronic Tl+ flux assays with a shorter t1/2 and lower potency compared to ouabain. The potency of pentamidine for hERG inhibition after overnight incubation is 11.3 μM (Table 3 and Fig. 3C), consistent with the reported potency of 5.1 μM for inhibition of hERG surface expression and of 7.8 μM in inhibition of hERG tail currents.4 These results indicate that ouabain and pentamidine are not acute hERG inhibitors, but they are hERG trafficking inhibitors, probably with differing mechanisms of reducing hERG surface expression.
Table 3.
Chronic Effect of Ouabain and Pentamidine on Human Ether-à-go-go Related Gene Activity as Determined in Chronic Tl+ Flux Assays
| Time (min) | Maximum Inhibition (%) | pEC50 | Hill Slope |
|---|---|---|---|
| Ouabain | |||
| 15 |
29.3 ± 2.1 |
5.94 ± 0.16 |
1 |
| 30 |
35.8 ± 2.2 |
5.97 ± 0.09 |
1.87 ± 0.81 |
| 45 |
55.9 ± 1.6 |
6.22 ± 0.05 |
1.46 ± 0.20 |
| 60 |
70.7 ± 1.3 |
6.28 ± 0.04 |
1.28 ± 0.11 |
| 75 |
88.4 ± 1.9 |
6.34 ± 0.04 |
1.19 ± 0.12 |
| 90 |
99.2 ± 1.9 |
6.45 ± 0.04 |
1.64 ± 0.20 |
| 120 |
∼100.0 |
6.80 ± 0.02 |
4.16 ± 0.43 |
| 180 |
∼100.0 |
6.94 ± 0.03 |
4.66 ± 2.10 |
| 240 |
∼100.0 |
7.30 ± 0.02 |
3.43 ± 0.27 |
| 960 (16 h) |
∼100.0 |
7.76 ± 0.02 |
5.72 ± 0.48 |
| Pentamidine | |||
| 240 |
78.7 ± 10.3 |
4.33 ± 0.13 |
1 |
| 480 |
∼100.0 |
4.53 ± 0.08 |
1 |
| 960 | ∼100.0 | 4.95 ± 0.04 | 1.26 ± 0.06 |
Results represent best-fit values ± SE. Cells were first incubated with ouabain or pentamidine for desired time (minutes to hours) at 37°C in cell incubator, washed once with assay buffer, and loaded with Tl+ flux dye solution for 90 min at room temperature in the dark, remaining total human Ether-à-go-go related gene activity was then determined. Initial slope of the curves was normalized to percentages with buffer control as 0% inhibition and complete inhibition (initial slope of around 2 with 10 μM cisapride) as 100% and pooled for analysis in the Prism with built-in four-parameter logistic function. Figures were presented in Figure 3. Potency, Hill slope, and maximum inhibition were reported directly from Prism. While curve-fitting, bottom was constrained to 0%, top was constrained to < 100%, and Hill slope was determine by the built-in comparison function in the Prism. Intracellular (if they entered cells) and residual drugs might be still working after removal of drug solution and during 90 min incubation with Tl+ flux dye.
Potent hERG inhibitors (such as those with EC50 values < 10 μM or pEC50 > 5.0 from the Tl+ assay in the Table 2), once detected and confirmed, would probably be eliminated for further consideration as drug candidates. To reduce false-negative rate in antitarget hERG screening, it is perhaps more important to identify less potent hERG inhibitors than to determine potency values for potent hERG inhibitors with accuracy and precision. With this in mind, we focused on several less potent drugs, such as those detailed below, to ensure that our assays have the requisite sensitivity to detect low-potency hERG modulators.
Amantadine, an antiparkinsonism as well as an antiviral drug that is known to cause significant QT prolongation at high doses,32 was therefore included in our study. Amantadine inhibited [3H]dofetilide binding with Ki of 10.5 μM, but failed to show inhibitory activity in Tl+ flux assays at up to 100 μM (9.4% ± 4.8% inhibition, n = 3) despite its clinically relevant action at hERG. In the APPC, amantadine inhibited hERG channel activity by 52.1% ± 1.1% (n = 5 cells) at 300 μM, only slightly higher than 3% DMSO vehicle control (40.2% ± 1.9% inhibition, n = 5 cells, P = 0.0315, unpaired t-test). The potency value is 41.3 μM (Hill slope of 1) with a maximum inhibition of 55% (Table 2). To determine whether amantadine inhibits hERG trafficking, we studied time-dependent hERG inhibition using the chronic Tl+ flux assay. Overnight incubation with amantadine (30 μM) had no inhibitory effect on hERG activity (−7.8% ± 2.8%, n = 4).
In HEK293-hERG cells, conventional patch clamp measurements revealed that venlafaxine, a serotonin and norepinephrine reuptake inhibitor antidepressant, inhibits hERG channels with a potency of 28 μM.33 In our study, venlafaxine inhibited [3H]dofetilide binding with Ki of 16.6 μM, but failed to inhibit hERG in acute Tl+ flux assays at concentrations up to 100 μM (10.2% ± 2.6%, n = 4). In APPC, venlafaxine inhibited hERG activity with a potency of 21.4 μM, consistent with the aforementioned report. We also tested whether venlafaxine inhibits hERG trafficking using the paradigm described above and found the drug to be inactive (−8.6% ± 4.2% inhibition at 100 μM, n = 4).
Phenytoin, an antiepileptic drug and a nonselective cation channel blocker, is reported to inhibit hERG channels stably expressed in HEK293 cells with a potency of 242 μM as measured by manual patch clamp.34 In our assays, phenytoin inhibited [3H]dofetilide binding with Ki of 18.2 μM and inhibited hERG channels in APPC with a potency of 0.21 μM and a maximum inhibition of 60% (n = 11 cells). However, phenytoin barely inhibited hERG channels in Tl+ flux assays (7.9% ± 2.3% inhibition at 100 μM, n = 5). We also tested whether phenytoin perturbs hERG trafficking and found the drug to be inactive (−32.5% ± 4.2% inhibition, n = 4), compared to vehicle (−28.6% ± 2.3% inhibition, n = 6).
We speculate that phenytoin, a known inducer of LQTS, did not show inhibition in Tl+ flux assays because the drug prefers the closed state of the hERG channel in a use-dependent manner.35 Thus, we tested the known closed-state hERG inhibitor BeKm-1.36–38 We reasoned that if phenytoin was inactive in Tl+ flux assays due to preferential inhibition of the closed state of hERG channels, then BeKm-1 would also be inactive. However, BeKm-1 was the most potent hERG inhibitor we characterized with a potency of 9.5 nM (Hill slope of 0.51 ± 0.04, n = 2). Therefore, use-dependent but not closed-state preference might be the reason that phenytoin did not inhibit hERG activity in the Tl+ flux assay.
Grepafloxacin is a fluoroquinolone antibiotic that induces LQTS. The drug was removed from the market in 1999. In manual patch clamp recordings of transfected CHO cells, grepafloxacin inhibited hERG channel activity with a potency of 50–100 μM.39,40 In another manual patch clamp study using transfected HEK293 cells, grepafloxacin was shown not to inhibit hERG at 100 μM, and to cause 25% inhibition of hERG at 300 μM (with 14% inhibition by vehicle alone41). In this study, we tested grepafloxacin as well as nine additional withdrawn drugs. As indicated in Table 2, of the 10 withdrawn drugs, grepafloxacin was exceptional in that it showed low binding affinity (17 μM) in [3H]dofetilide binding assays. Also, grepafloxacin failed to inhibit hERG activity in Tl+ flux assays at concentrations up to 100 μM (19.5% ± 4.7% inhibition, n = 4, vs. − 8.2% ± 2.4% inhibition by 1% DMSO, n = 3). Grepafloxacin was also inactive at hERG as measured by APPC (27.1% ± 3.9% inhibition at 100 μM, n = 11 cells; 37.2% ± 3.5% inhibition at 300 μM, n = 11 cells, vs. 40.2% ± 1.9% inhibition by 3% DMSO, n = 5 cells). Our APPC results were consistent with those from manual patch clamp recordings.41 We also examined whether grepafloxacin interfered with hERG trafficking and found the drug to be inactive in our paradigm (−14.3% ± 3.7% inhibition at 100 μM, n = 4).
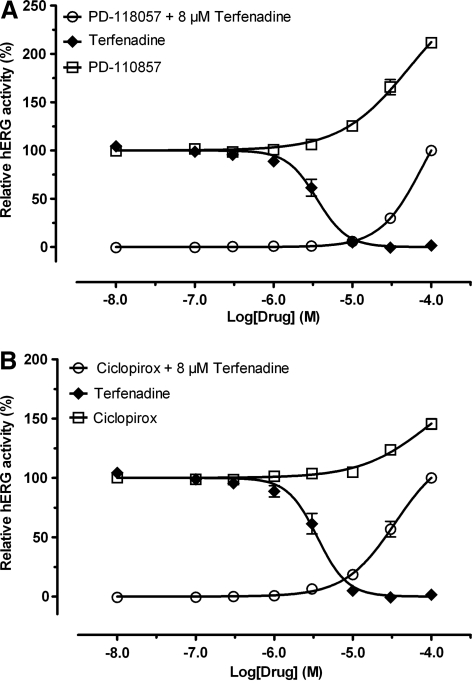
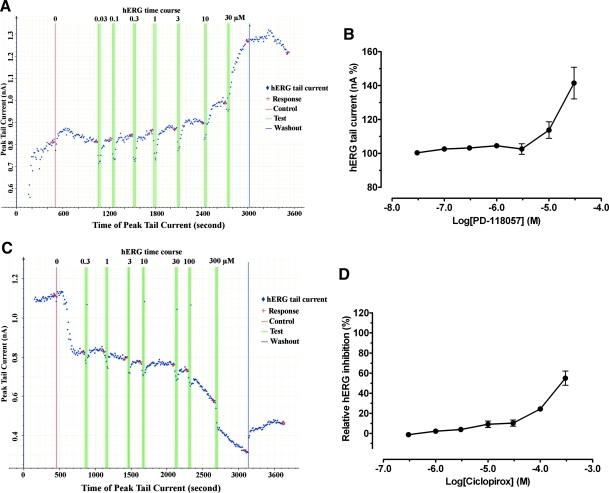
The compound PD-118057 is a known hERG activator that stimulates channel activity by reducing the rate of inactivation.26 We included the drug in this study as a control to assess whether the screening assays we employed can detect hERG activators. The drug PD-118057 inhibited [3H]dofetilide binding with an affinity of 12.3 μM. As expected, it activated the hERG channel in Tl+ flux assay with a potency of 52.5 μM (maximal activation = 270.8% ± 14.1% above the basal level of 100%, n = 4 assays and each in quadruplicate) (Fig. 4). Further, PD-118057 completely reversed hERG inhibition by 8 μM terfenadine in Tl+ flux assays with a potency of 76.1 μM (Hill slope = 1.63 ± 0.18, maximal activation = 164.1% ± 28.1% above basal of 0%, n = 4 assays and each measured in quadruplicate). In APPC, PD-118057 (Fig. 5) also displayed hERG channel activation in a concentration-dependent manner.
Fig. 4.
Activation of hERG channels in the Tl+ flux assays. Terfenadine (♦) completely inhibited hERG channel activity in a dose-dependent manner, whereas PD-118057 (A) and ciclopirox (B) were able to activate the hERG channel in the absence (○) and presence (•) of 8 μM terfenadine. For assays done in the presence of terfenadine, terfenadine was added 5 min before PD-118057 or ciclopirox. For hERG activator activities, they were normalized to their corresponding basal activities in percentage: basal activity as 100% in the absence of terfenadine and basal activity as 0% in the presence of terfenadine. The dose-dependent hERG activation by PD-118057 or ciclopirox was fitted to a four-parameter logistic function without constraining the maximal activation (top). PD-118057 showed maximal activation of 270.8% and potency of 52.5 μM in the absence of terfenadine and maximal activation of 164.1% and potency of 76.1 μM in the presence of terfenadine. Ciclopirox showed maximal activation of 188.5% and potency of 93.2 μM in the absence of terfenadine and maximal activation of 123.6% and potency of 34.4 in the presence of terfenadine.
Fig. 5.
Activation of hERG channels by PD-118057 and inhibition of hERG channels by ciclopirox in APPC assays. The capture screen from DataXpress of a representative whole-cell patch clamp recording showed activation of hERG channels by PD-118057 (A) and inhibition by ciclopirox (C). Red vertical line indicated addition of buffer control and green vertical triple lines indicated triple additions of drugs. Drug concentrations (μM) were listed at the top of the green lines. Red cross indicated the time point when hERG tail current was measured by the system. Upon washout (blue vertical line), hERG tail current reduced from activation by PD-118057 or recovered from inhibition by ciclopirox, indicating that activation PD-118057 or inhibition by ciclopirox was not due to current run-up or run-down, respectively. (B) The corrected tail currents (nA) in the presence of PD-1158057 were extracted from DataXpress program, and normalized to basal currents (with buffer = 100%), and plotted again PD-118057 concentrations (n = 3). (D) The tail currents in the presence of ciclopirox were extracted from DataXpress, normalized to percentage inhibition, and plotted for against ciclopirox concentrations (n = 4).
Ciclopirox is an antiviral and anticancer drug, the antitumor activity of which is reportedly due to its ability to chelate intracellular iron.42,43 We included ciclopirox in this study for further characterization. In [3H]dofetilide binding assays, ciclopirox displayed an affinity of 17.0 μM, similar to the known hERG activator PD-118057. Further, ciclopirox activated the hERG channel in Tl+ flux assays with a potency of 93.2 μM (Fig. 4) (maximal activation = 188.5% ± 17.7% above basal of 100%, n = 4 assays and each measured in quadruplicate). Like PD-118057, ciclopirox completely reversed hERG inhibition by 8 μM terfenadine with a potency of 34.4 μM (Hill slope = 1.36 ± 0.18, maximal activation = 123.6% ± 11.2% above basal of 0%, n = 3 assays and each measured in quadruplicate). However, in APPC, ciclopirox inhibited hERG channels by 55.0% ± 7.1% (n = 4 cells) at 300 μM (Fig. 5). The reason for the disparate Tl+ flux and APPC results is not clear; however, our results indicate that ciclopirox clearly modulates hERG channel activity. Since hERG channels are overexpressed in many tumor cells and modulation of hERG channel activity has resulted in inhibition of tumor growth,44,45 it is tempting to speculate that the antitumor activity of ciclopirox is at least partially due to modulation of hERG channel activity.
Discussion
The major findings of this study are as follows: (1) hERG inhibitor potencies measured via [3H]dofetilide binding and Tl+ flux correlated significantly, though modestly, with potencies from APPC assays (Fig. 2); (2) hERG inhibitors in general were most potent in APPC assays, of intermediate potency in [3H]dofetilide binding assays, and least potent in Tl+ flux assays. The sensitivity of the assays to measure hERG modulation varies inversely with throughput. We also discovered that because some drugs behave as false-negatives in Tl+ flux assays (e.g., phenytoin, venlafaxine, and amantadine), this higher-throughput approach cannot be reliably used to predict hERG inhibitory potency, especially for the less potent hERG inhibitors. However, in contrast to the [3H]dofetilide binding assay, there were no false-positives from the Tl+ flux assay, and all hits were confirmed by APPC. Therefore, the Tl+ flux assay can be used as a primary high-throughput platform to eliminate relatively potent hERG inhibitors from further consideration at an early stage of drug development. Certainly for public sector drug discovery initiatives, this represents a cost-effective triaging approach. Further, we make the novel observation that the Tl+ flux assay can be used to screen for hERG trafficking inhibitors and hERG activators, two additional types of arrhythmogenic hERG modulators that have received increased attention by the U.S. FDA in recent years.
A recent comprehensive review of hERG modulators by Polak et al.30 listed 240 drug potencies as measured by automated planar or manual patch clamp. Notably, for most of the known hERG inhibitors and/or drugs that induce LQTS in Table 2, the potency values we obtained by APPC recapitulated those published by Polak et al., validating the APPC platform. Although sensitive and reproducible, APPC does not afford the throughput necessary for rapidly screening large numbers of compounds for hERG modulators.
To ascertain whether alternative, higher-throughput methods can be used to reliably identify hERG modulators, we measured the potencies of the 49 drugs by [3H]dofetilide competition binding. As summarized in Table 2, potencies in radioligand binding assays, which were on average about fivefold less sensitive than APPC, ranged from 30 nM (astemizole) to 18 μM (phenytoin). Some drugs known to inhibit hERG and/or induce LQTS, such as grepafloxacin and venlafaxine, displayed potencies >10 μM in [3H]dofetilide binding assays. Twenty-five of the 49 drugs had potencies between 1 and 10 μM. Thus, to reduce the false-negative rate when screening compounds for hERG modulators via [3H]dofetilide binding, a low threshold should be applied. On the basis of the data in Table 2, a potency cut-off of 30 μM incorporates all of the known hERG inhibitors we tested; this corresponds to 30% inhibition of [3H]dofetilide binding by a drug at 10 μM.
We also measured the potencies of the 49 drugs using Tl+ flux assays. The Tl+ flux assay afforded greater throughput but was about 17-fold less sensitive on average than APPC (Table 2). Drug potencies in Tl+ flux assays ranged from 200 nM (E-4031) to 52 μM (olanzapine). Eighteen of the drugs known to inhibit hERG and/or cause LQTS displayed potencies >10 μM, and 19 of them exhibited potencies between 1 and 10 μM. Olanzapine was the least potent of the known hERG inhibitors and/or LQTS inducers in Tl+ flux assays (52 μM). Thus, a low-stringency cut-off (such as 30% inhibition at 10 μM) is likely necessary to minimize false-negatives.
Despite the lower sensitivity of Tl+ flux assays for detecting hERG modulators (see above), the assay is not without advantages for screening large compound collections. First, Tl+ flux assays—unlike APPC and radioligand binding—are compatible with the 384- and 1,586-well formats, greatly increasing assay throughput. Second, because Tl+ flux assays indirectly reflect channel activity, drug libraries can be screened for inhibitors and activators simultaneously and with high throughput; this contrasts with radioligand binding data, which only reflect direct or indirect inhibition of [3H]dofetilide binding. Third, as we show for ouabain, Tl+ flux assays—unlike radioligand binding assays—can be used to identify compounds that interfere with hERG plasma membrane expression. The facility of detecting hERG trafficking inhibitors is of particular importance, as an accumulating body of evidence suggests that many drugs affect channel activity in vivo by decreasing cell surface expression of mature hERG.3
The best way to screen large compound sets to identify hERG modulators remains unclear. Our data show that, while drug potencies in radioligand binding and Tl+ flux assays correlate with potencies measured by APPC, the higher-throughput approaches are less sensitive, with known hERG modulators escaping detection (i.e., false-negatives). Of the 49 drugs we tested, most of which are known hERG modulators and/or drugs that induce LQTS, all had measurable (though often low) potency in [3H]dofetilide binding assays. In Tl+ flux assays, the drugs amantadine and phenytoin were inactive, despite having measurable activity in APPC.
Among the three methods we evaluated, [3H]dofetilide binding is the easiest and most cost-effective. However, radioligand binding-based hERG screening is not without drawbacks. The assay cannot illuminate hERG function (current flow) or channel trafficking (plasma membrane expression). Thus, the activities of drugs like PD-118057 (a hERG activator) and ouabain (a hERG trafficking inhibitor) would go unnoticed in [3H]dofetilide binding assays. Radioligand binding-based screens for hERG ligands are also confounded by the channel's notorious pharmacological promiscuity, which likely stems from the channel's large hydrophobic intracellular binding pocket46 and its multiple activity states (open, inactive, and closed). With this in mind, we endeavored to test chemically and functionally diverse drugs and found that all of them (including negative controls such as CNO and ouabain) measurably inhibit [3H]dofetilide binding with potencies similar to several of the positive controls (e.g., amitriptyline and doxepin). Therefore, our data suggest that large-scale screening using [3H]dofetilide binding will likely identify several false-positives. Thus, a mechanism to resolve the false-positives from a [3H]dofetilide binding screen results is desirable.
The Tl+-based fluorescence assay47 was recently employed for high-throughput hERG screening using hERG-expressing U2OS cells.23,24 Using our HEK293-hERG cells, we routinely obtained specific hERG signals (defined using 10 μM cisapride) in excess of 20-fold over baseline—about threefold greater than those reported for hERG-expressing U2OS cells. As mentioned above, we were able to identify hERG inhibitors as well as hERG activators using the Tl+ flux assay. The identification of hERG activators is important because such drugs can induce short QT syndrome and result in sudden death.11,12 We identified ciclopirox as a hERG activator with activity similar to that of the known hERG activator PD-118057.26,48 In addition, ciclopirox completely reversed the inhibition of hERG by terfenadine. However, ciclopirox was a hERG inhibitor in APPC assays. One possible reason for this discrepancy is that the activity of ciclopirox may be state dependent. The voltage protocol in APPC allows measurement of tail current under conditions where hERG channels are open and not inactivated (repolarized cells), whereas the Tl+ flux assay does not permit the strict control of membrane potential. Also, the cycling of the voltage in APPC might provide multiple opportunities for drugs to bind to different activity states. In addition, the potency of drugs in patch clamp recordings in the whole cell configuration is sensitive to many factors, including cell type, temperature, and the patch clamp system and voltage pulse protocol used.38,49,50
The Na+-K+ ATPase inhibitor ouabain (as well as several other glycosides) has been shown to reduce cell surface expression of fully glycosylated hERG channels with a potency of 9.8 nM and to inhibit hERG tail current with a potency of 15.2 nM after an overnight treatment.25 Here, we showed that acute ouabain treatment does not inhibit hERG tail current (Table 2). To examine inhibition of hERG trafficking, we adapted the original Tl+ flux assay protocol to study the chronic effects of ouabain on hERG activity. We show that 90 min of ouabain exposure eliminates specific Tl+ flux in HEK293-hERG cells, whereas short ouabain exposure is without effect (Fig. 3, Table 3). Given the hERG protein half-life of 11 h in HEK293-hERG cells,51,52 the maximal inhibition of hERG activity after 90 min suggests that ouabain promotes hERG channel internalization, degradation, and/or inactivation, in addition to its established effect on hERG trafficking. The notion that ouabain might cause hERG channel internalization is consistent with the finding that a potassium-free extracellular solution inhibits Na+-K+ APTase activity53 and dramatically increases hERG channel internalization and degradation in HEK293 cells.54,55 Other mechanisms also regulate hERG protein levels/stability: Walker et al. found that Hsp40 chaperones promote hERG degradation in HEK293 cells.56 Together, our data and others' emphasize the importance of considering drug-sensitive alterations in hERG expression and/or trafficking when designing screens for hERG modulators.
Our APPC results for phenytoin and grepafloxacin, two known inducers of LQTS, are noteworthy. In an earlier assay with neuroblastoma cells (expressing endogenous hERG channels), phenytoin was reported to inhibit steady state potassium currents with a maximal inhibition of 71% and potency of 31 μM.35 The inhibitory effect was determined at + 50 mV from a holding potential of − 60 mV. Similarly, in another manual patch clamp study using HEK293-hERG cells,34 phenytoin exhibited a maximal inhibition of around 52% and a potency of 47 μM. In our APPC assays, phenytoin showed a maximal inhibition of 60% (similar to the aforementioned studies) and a potency of 0.21 μM (two orders of magnitude more potent than the studies referenced above). Possible reasons for the increased phenytoin potency we observed are differences in intracellular solution composition (we used a KF-containing internal buffer) and voltage protocol.
Grepafloxacin was withdrawn from the market due to induction of LQTS, presumably via hERG channel blockade. However, published conventional patch clamp studies examining grepafloxacin activity at hERG channels are discrepant. Using CHO-FLAG-hERG cells, Bischoff et al. found that grepafloxacin inhibited hERG with a potency of 100 μM.39 However, using HEK293-hERG cells and a different voltage protocol, Lu et al. saw no hERG inhibition by grepafloxacin at concentrations up to 100 μM and only minor inhibition (25% for grepafloxacin vs. 14% for control) at 300 μM.41 Similarly, grepafloxacin did not increase action potential duration in isolated rabbit heart.41 Our APPC results for grepafloxacin are odds with those of Bischoff and similar to those of Lu et al.: we observed no inhibition of hERG by the drug at concentrations up to 300 μM. Thus, whether grepafloxacin induces QT prolongation via hERG inhibition remains unclear.
In summary, our results with 49 drugs suggest that the combination of radioligand binding assays and the novel, chronic Tl+ flux assay we developed enables the efficient, high-throughput initial screening of compounds for various types of hERG modulators (acute inhibitors, activators, and trafficking inhibitors). Importantly, APPC remains the gold standard, and whether the higher-throughput assays can provide data as valuable as those obtained via APPC requires further investigation. Notably, as we show, APPC is not immune to false-negatives: acute ouabain and grepafloxacin (two known inducers of LQTS) were each inactive in patch clamp as well as in Tl+ flux assays. Thus, strategies to identify hERG modulators via HTS should allow for the complex activities of drugs in addition to simple blockade or stimulation of the channel. In the vein, we would recommend using both [3H]dofetilide binding and Tl+ flux assays as primary screening assays, with 30% inhibition and 10% activation at 10 μM as thresholds for hERG blockers and activators, respectively. All potential hits should be confirmed and characterized using concentration–response analysis in [3H]dofetilide binding and Tl+ flux assays. The hits with potency and/or affinity of 30 μM or less should be tested using APPC.
Supplementary Material
Abbreviations:
- APPC
automated planar patch clamp
- CNO
clozapine-N-oxide
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
dimethyl sulfoxide
- FBS
fetal bovine serum
- hBB
hERG binding buffer
- hERG
human Ether-à-go-go related gene
- LQTS
long QT syndrome
- QRS
Complex, the name for the consecutive Q, R, and S waves on an electrocardiograph
- QT
interval, the interval between the start of the QRS complex and the end of T wave on an electrocardiograph
Acknowledgments
The authors would like to thank Sarah Rogan and Wes Kroeze for careful reading of the article and making suggestions. Authors would also like to thank Drs. Richard Kondo, Robert Rosenberg, and Thomas Kash for technical support in setting up and optimization of the PatchXpress system. Supported by the National Institute of Mental Health Psychoactive Drug Screening Program and the Michael Hooker Distinguished Professorship in Protein Therapeutics and Translational Proteomics.
Author Disclosure Statement
B.L.R. has served as a consultant to Invitrogen.
References
- 1.Warmke JW. Ganetzky B. A family of potassium channel genes related to eag in Drosophila and mammals. Proc Natl Acad Sci USA. 1994;91:3438–3442. doi: 10.1073/pnas.91.8.3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Recanatini M. Poluzzi E. Masetti M. Cavalli A. De Ponti F. QT prolongation through hERG K(+) channel blockade: current knowledge and strategies for the early prediction during drug development. Med Res Rev. 2005;25:133–166. doi: 10.1002/med.20019. [DOI] [PubMed] [Google Scholar]
- 3.Wible BA. Hawryluk P. Ficker E. Kuryshev YA. Kirsch G. Brown AM. HERG-Lite: a novel comprehensive high-throughput screen for drug-induced hERG risk. J Pharmacol Toxicol Methods. 2005;52:136–145. doi: 10.1016/j.vascn.2005.03.008. [DOI] [PubMed] [Google Scholar]
- 4.Kuryshev YA. Ficker E. Wang L. Hawryluk P. Dennis AT. Wible BA. Brown AM. Kang J. Chen XL. Sawamura K. Reynolds W. Rampe D. Pentamidine-induced long QT syndrome and block of hERG trafficking. J Pharmacol Exp Ther. 2005;312:316–323. doi: 10.1124/jpet.104.073692. [DOI] [PubMed] [Google Scholar]
- 5.Rajamani S. Eckhardt LL. Valdivia CR. Klemens CA. Gillman BM. Anderson CL. Holzem KM. Delisle BP. Anson BD. Makielski JC. January CT. Drug-induced long QT syndrome: hERG K + channel block and disruption of protein trafficking by fluoxetine and norfluoxetine. Br J Pharmacol. 2006;149:481–489. doi: 10.1038/sj.bjp.0706892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perrin MJ. Subbiah RN. Vandenberg JI. Hill AP. Human ether-a-go-go related gene (hERG) K + channels: function and dysfunction. Prog Biophys Mol Biol. 2008;98:137–148. doi: 10.1016/j.pbiomolbio.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 7.Roden DM. Cellular basis of drug-induced torsades de points. Br J Pharmacol. 2008;154:1502–1507. doi: 10.1038/bjp.2008.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dennis A. Wang L. Wan X. Ficker E. hERG channel trafficking: novel targets in drug-induced long QT syndrome. Biochem Soc Trans. 2007;35:1060–1063. doi: 10.1042/BST0351060. [DOI] [PubMed] [Google Scholar]
- 9.Curran ME. Splawski I. Timothy KW. Vincent GM. Green ED. Keating MT. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 10.Sanguinetti MC. Tristani-Firouzi M. hERG potassium channels and cardiac arrhythmia. Nature. 2006;440:463–469. doi: 10.1038/nature04710. [DOI] [PubMed] [Google Scholar]
- 11.Brugada R. Hong K. Cordeiro JM. Dumaine R. Short QT syndrome. CMAJ. 2005;173:1349–1354. doi: 10.1503/cmaj.050596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah RR. Drug-induced QT interval shortening: potential harbinger of proarrhythmia and regulatory perspectives. Br J Pharmacol. 2010;159:58–69. doi: 10.1111/j.1476-5381.2009.00191.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piccini JP. Whellan DJ. Berridge BR. Finkle JK. Pettit SD. Stockbridge N. Valentin JP. Vargas HM. Krucoff MW. Current challenges in the evaluation of cardiac safety during drug development: translational medicine meets the Critical Path Initiative. Am Heart J. 2009;158:317–326. doi: 10.1016/j.ahj.2009.06.007. [DOI] [PubMed] [Google Scholar]
- 14.Shah RR. Can pharmacogenetics help rescue drugs withdrawn from the market? Pharmacogenomics. 2006;7:889–908. doi: 10.2217/14622416.7.6.889. [DOI] [PubMed] [Google Scholar]
- 15.Netzer R. Bischoff U. Ebneth A. HTS techniques to investigate the potential effects of compounds on cardiac ion channels at early-stages of drug discovery. Curr Opin Drug Discov Dev. 2003;6:462–469. [PubMed] [Google Scholar]
- 16.Fermini B. Fossa AA. The impact of drug-induced QT interval prolongation on drug discovery and development. Nat Rev Drug Discov. 2003;2:439–447. doi: 10.1038/nrd1108. [DOI] [PubMed] [Google Scholar]
- 17.Finlayson K. Pennington AJ. Kelly JS. [3H]dofetilide binding in SHSY5Y and HEK293 cells expressing a HERG-like K + channel? Eur J Pharmacol. 2001;412:203–212. doi: 10.1016/s0014-2999(01)00731-2. [DOI] [PubMed] [Google Scholar]
- 18.Diaz GJ. Daniell K. Leitza ST. Martin RL. Su Z. McDermott JS. Cox BF. Gintant GA. The [3H]dofetilide binding assay is a predictive screening tool for hERG blockade and proarrhythmia: comparison of intact cell and membrane preparations and effects of altering K + o. J Pharmacol Toxicol Methods. 2004;50:187–199. doi: 10.1016/j.vascn.2004.04.001. [DOI] [PubMed] [Google Scholar]
- 19.Chiu PJ. Marcoe KF. Bounds SE. Lin CH. Feng JJ. Lin A. Cheng FC. Crumb WJ. Mitchell R. Validation of a [3H]-astemizole binding assay in HEK293 cells expressing HERG K + channels. J Pharmacol Sci. 2004;95:311–319. doi: 10.1254/jphs.fpe0040101. [DOI] [PubMed] [Google Scholar]
- 20.Cheng CS. Alderman D. Kwash J. Dessaint J. Patel R. Lescoe MK. Kinrade MB. Yu W. A high-throughput HERG potassium channel function assay: an old assay with a new look. Drug Dev Ind Pharm. 2002;28:177–191. doi: 10.1081/ddc-120002451. [DOI] [PubMed] [Google Scholar]
- 21.Baxter DF. Kirk M. Garcia AF. Raimondi A. Holmqvist MH. Flint KK. Bojanic D. Distefano PS. Curtis R. Xie Y. A novel membrane potential-sensitive fluorescent dye improves cell-based assays for ion channels. J Biomol Screen. 2002;7:79–85. doi: 10.1177/108705710200700110. [DOI] [PubMed] [Google Scholar]
- 22.Falconer M. Smith F. Surah-Narwal S. Congrave G. Liu Z. Hayter P. Ciaramella G. Keighley W. Haddock P. Waldron G. Sewing A. High-throughput screening for ion channel modulators. J Biomed Screen. 2002;7:460–465. doi: 10.1177/108705702237678. [DOI] [PubMed] [Google Scholar]
- 23.Titus SA. Beacham D. Shahane SA. Southall N. Xia M. Huang R. Hooten E. Zhao Y. Shou L. Austin CP. Zheng W. A new homogeneous high-throughput screening assay for profiling compound activity on the human ether-a-go-go-related gene channel. Anal Biochem. 2009;394:30–38. doi: 10.1016/j.ab.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beacham DW. Blackmer T. O'Grady M. Hanson GT. Cell-based potassium ion channel screening using the FluxOR assay. J Biomol Screen. 2010;15:441–446. doi: 10.1177/1087057109359807. [DOI] [PubMed] [Google Scholar]
- 25.Wang L. Wible BA. Wan X. Ficker E. Cardiac glycosides as novel inhibitors of human ether-a-go-go-related gene channel trafficking. J Pharmacol Exp Ther. 2007;320:525–534. doi: 10.1124/jpet.106.113043. [DOI] [PubMed] [Google Scholar]
- 26.Zhou J. Augelli-Szafran CE. Bradley JA. Chen X. Koci BJ. Volberg WA. Sun Z. Cordes JS. Novel potent human ether-a-go-go-related gene (hERG) potassium channel enhancers and their in vitro antiarrhythmic activity. Mol Pharmacol. 2005;68:876–884. doi: 10.1124/mol.105.014035. [DOI] [PubMed] [Google Scholar]
- 27.Armbruster BN. Li X. Pausch MH. Herlitze S. Roth RL. Evolving the lock to fit the key to create a family of G protein-coupled receptors potently activated by an inert ligand. Proc Natl Acad Sci USA. 2007;104:5163–5168. doi: 10.1073/pnas.0700293104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeng H. Penniman JR. Kinose F. Kim D. Trepakova ES. Malik MG. Dech SJ. Balasubramanian B. Salata JJ. Improved throughput of PatchXpress hERG assay using intracellular potassium fluoride. Assay Drug Dev Technol. 2008;6:235–241. doi: 10.1089/adt.2007.116. [DOI] [PubMed] [Google Scholar]
- 29.Cheng Y. Prusoff WH. Relationship between the inhibition constant (K1) and the concentration of inhibitor which causes 50 per cent inhibition (I50) of an enzymatic reaction. Biochem Pharmacol. 1973;22:3099–3108. doi: 10.1016/0006-2952(73)90196-2. [DOI] [PubMed] [Google Scholar]
- 30.Polak S. Wisniowska B. Brandys J. Collation, assessment and analysis of literature in vitro data on hERG receptor blocking potency for subsequent modeling of drugs' cardiotoxic properties. J Appl Toxicol. 2009;29:183–206. doi: 10.1002/jat.1395. [DOI] [PubMed] [Google Scholar]
- 31.Cordes JS. Sun Z. Lloyd DB. Bradley JA. Opsahl AC. Tengowski MW. Chen X. Zhou J. Pentamidine reduces hERG expression to prolong the QT interval. Br J Pharmacol. 2005;145:15–23. doi: 10.1038/sj.bjp.0706140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz M. Patel M. Kazzi Z. Morgan B. Cardiotoxicity after massive amantadine overdose. J Med Toxicol. 2008;4:173–179. doi: 10.1007/BF03161197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fossa AA. Gorczyca W. Wisialowski T. Yasgar A. Wang E. Crimin K. Volberg W. Zhou J. Electrical alternans and hemodynamics in the anesthetized guinea pig can discriminate the cardiac safety of antidepressants. J Pharmacol Toxicol Methods. 2007;55:78–85. doi: 10.1016/j.vascn.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 34.Danielsson BR. Lansdell K. Patmore L. Tomson T. Phenytoin and phenobarbital inhibit human HERG potassium channels. Epilepsy Res. 2003;55:147–157. doi: 10.1016/s0920-1211(03)00119-0. [DOI] [PubMed] [Google Scholar]
- 35.Nobile M. Vercellino P. Inhibition of delayed rectifier K + channels by phenytoin in rat neuroblastoma cells. Br J Pharmacol. 1997;120:647–652. doi: 10.1038/sj.bjp.0700969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Milnes JT. Dempsey CE. Ridley JM. Crociani O. Arcangeli A. Hancox JC. Witchel HJ. Preferential closed channel blockade of HERG potassium currents by chemically synthesised BeKm-1 scorpion toxin. FEBS Lett. 2003;547:20–26. doi: 10.1016/s0014-5793(03)00662-8. [DOI] [PubMed] [Google Scholar]
- 37.Zhang M. Korolkova YV. Liu J. Jiang M. Grishin EV. Tseng GN. BeKm-1 is a HERG-specific toxin that shares the structure with ChTx but the mechanism of action with ErgTx1. Biophys J. 2003;84:3022–3036. doi: 10.1016/S0006-3495(03)70028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yao JA. Du X. Lu D. Baker RL. Daharsh E. Atterson P. Estimation of potency of HERG channel blockers: impact of voltage protocol and temperature. J Pharmacol Toxicol Methods. 2005;52:146–153. doi: 10.1016/j.vascn.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 39.Bischoff U. Schmidt C. Netzer R. Pongs O. Effects of fluoroquinolones on HERG currents. Eur J Pharmacol. 2000;406:341–343. doi: 10.1016/s0014-2999(00)00693-2. [DOI] [PubMed] [Google Scholar]
- 40.Kang J. Chen XL. Wang L. Rampe D. Interactions of the antimalarial drug mefloquine with the human cardiac potassium channels KvLQT1/minK and HERG. J Pharmacol Exp Ther. 2001;299:290–296. [PubMed] [Google Scholar]
- 41.Lu HR. Vlaminckx E. Van de Water A. Rohrbacher J. Hermans A. Gallacher DJ. In-vitro experimental models for the risk assessment of antibiotic-induced QT prolongation. Eur J Pharmacol. 2007;577:222–232. doi: 10.1016/j.ejphar.2007.07.070. [DOI] [PubMed] [Google Scholar]
- 42.Eberhard Y. McDermott SP. Wang X. Gronda M. Venugopal A. Wood TW. Hurren R. Datti A. Batey RA. Wrana J. Antholine WE. Dick JE. Schimmer AD. Chelation of intracellular iron with the antifungal agent ciclopirox olamine induces cell death in leukemia and myeloma cells. Blood. 2009;114:3064–3073. doi: 10.1182/blood-2009-03-209965. [DOI] [PubMed] [Google Scholar]
- 43.Hoque M. Hanauske-Abel HM. Palumbo P. Saxena D. D'Alliessi Gandolfi D. Park MH. Peery T. Mathews MB. Inhibition of HIV-1 gene expression by Ciclopirox and Deferiprone, drugs that prevent hypusination of eukaryotic initiation factor 5A. Retrovirology. 2009;6:90. doi: 10.1186/1742-4690-6-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dolderer JH. Schuldes H. Bockhorn H. Altmannsberger M. Lambers C. von Zabern D. Jonas D. Schwegler H. Linke R. Schroder UH. HERG1 gene expression as a specific tumor marker in colorectal tissues. Eur J Surg Oncol. 2009;36:72–77. doi: 10.1016/j.ejso.2009.05.009. [DOI] [PubMed] [Google Scholar]
- 45.Ganapathi SB. Kester M. Elmslie KS. State-dependent block of HERG potassium channels by R-roscovitine: implications for cancer therapy. Am J Physiol Cell Physiol. 2009;296:C701–C710. doi: 10.1152/ajpcell.00633.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitcheson JS. Chen J. Lin M. Culberson C. Sanguinetti MC. A structural basis for drug-induced long QT syndrome. Proc Natl Acad Sci USA. 2000;97:12329–12333. doi: 10.1073/pnas.210244497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weaver CD. Harden D. Dworetzky SI. Robertson B. Knox RJ. A thallium-sensitive, fluorescence-based assay for detecting and characterizing potassium channel modulators in mammalian cells. J Biomol Screen. 2004;9:671–677. doi: 10.1177/1087057104268749. [DOI] [PubMed] [Google Scholar]
- 48.Perry M. Sachse FB. Abbruzzese J. Sanguinetti MC. PD-118057 contacts the pore helix of hERG1 channels to attenuate inactivation and enhance K + conductance. Proc Natl Acad Sci USA. 2009;106:20075–20080. doi: 10.1073/pnas.0906597106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kirsch GE. Trepakova ES. Brimecombe JC. Sidach SS. Erickson HD. Kochan MC. Shyjka LM. Lacerda AE. Brown AM. Variability in the measurement of hERG potassium channel inhibition: effects of temperature and stimulus pattern. J Pharmacol Toxicol Methods. 2004;50:93–101. doi: 10.1016/j.vascn.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 50.Milnes JT. Witchel HJ. Leaney JL. Leishman DJ. Hancox JC. Investigating dynamic protocol-dependence of hERG potassium channel inhibition at 37 degrees C: cisapride versus dofetilide. J Pharmacol Toxicol Methods. 2010;61:178–191. doi: 10.1016/j.vascn.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 51.Ficker E. Dennis AT. Wang L. Brown AM. Role of the cytosolic chaperones Hsp70 and Hsp90 in maturation of the cardiac potassium channel HERG. Circ Res. 2003;92:e87–e100. doi: 10.1161/01.RES.0000079028.31393.15. [DOI] [PubMed] [Google Scholar]
- 52.Chen J. Sroubek J. Krishnan Y. Li Y. Bian J. McDonald TV. PKA phosphorylation of HERG protein regulates the rate of channel synthesis. Am J Physiol Heart Circ Physiol. 2009;296:H1244–H1254. doi: 10.1152/ajpheart.01252.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orlov SN. Thorin-Trescases N. Kotelevtsev SV. Tremblay J. Hamet P. Inversion of the intracellular Na + /K + ratio blocks apoptosis in vascular smooth muscle at a site upstream of caspase-3. J Biol Chem. 1999;274:16545–16552. doi: 10.1074/jbc.274.23.16545. [DOI] [PubMed] [Google Scholar]
- 54.Guo J. Massaeli H. Xu J. Jia Z. Wigle JT. Mesaeli N. Zhang S. Extracellular K + concentration controls cell surface density of IKr in rabbit hearts and of the HERG channel in human cell lines. J Clin Invest. 2009;119:2745–2757. doi: 10.1172/JCI39027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Massaeli H. Guo J. Xu J. Zhang S. Extracellular K + is a prerequisite for the function and plasma membrane stability of HERG channels. Circ Res. 2010;106:1072–1082. doi: 10.1161/CIRCRESAHA.109.215970. [DOI] [PubMed] [Google Scholar]
- 56.Walker VE. Wong MJ. Atanasiu R. Hantouche C. Young JC. Shrier A. Hsp40 chaperones promote degradation of the HERG potassium channel. J Biol Chem. 2010;285:3319–3329. doi: 10.1074/jbc.M109.024000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.