Abstract
Background
The mechanisms underlying ozone (O3)-induced pulmonary inflammation remain unclear. Interleukin-10 (IL-10) is an anti-inflammatory cytokine that is known to inhibit inflammatory mediators.
Objectives
We investigated the molecular mechanisms underlying interleuken-10 (IL-10)–mediated attenuation of O3-induced pulmonary inflammation in mice.
Methods
Il10-deficient (Il10−/−) and wild-type (Il10+/+) mice were exposed to 0.3 ppm O3 or filtered air for 24, 48, or 72 hr. Immediately after exposure, differential cell counts and total protein (a marker of lung permeability) were assessed from bronchoalveolar lavage fluid (BALF). mRNA and protein levels of cellular mediators were determined from lung homogenates. We also used global mRNA expression analyses of lung tissue with Ingenuity Pathway Analysis to identify patterns of gene expression through which IL-10 modifies O3-induced inflammation.
Results
Mean numbers of BALF polymorphonuclear leukocytes (PMNs) were significantly greater in Il10−/− mice than in Il10+/+ mice after exposure to O3 at all time points tested. O3-enhanced nuclear NF-κB translocation was elevated in the lungs of Il10−/− compared with Il10+/+ mice. Gene expression analyses revealed several IL-10–dependent and O3-dependent mediators, including macrophage inflammatory protein 2, cathepsin E, and serum amyloid A3.
Conclusions
Results indicate that IL-10 protects against O3-induced pulmonary neutrophilic inflammation and cell proliferation. Moreover, gene expression analyses identified three response pathways and several genetic targets through which IL-10 may modulate the innate and adaptive immune response. These novel mechanisms of protection against the pathogenesis of O3-induced pulmonary inflammation may also provide potential therapeutic targets to protect susceptible individuals.
Keywords: air pollution, gene array, IL-10, inflammation, lung, ozone, pulmonary
Ozone (O3) is the most toxic oxidant gas in air pollution mixtures and has been associated with numerous health effects, including airway inflammation and hyperreactivity (Plopper et al. 1998; Toward and Broadley 2002), exacerbation of asthma (McConnell et al. 2002; Neuhaus-Steinmetz et al. 2000), and increased mortality and morbidity (Jerrett et al. 2009). O3-induced lung inflammation is characterized by polymorphonuclear leukocyte (PMN) infiltration (Mudway and Kelly 2000) and release of a number of proinflammatory cytokines, such as tumor necrosis factor-α (TNF-α) (Bhalla et al. 2002; Cho et al. 2001), interleukin-6 (IL-6) (Johnston et al. 2005), and the PMN chemoattractant macrophage inflammatory protein 2 (MIP-2) (Driscoll et al. 1993). O3 causes upregulation of inducible nitric oxide synthase (iNOS), which further contributes to lung injury (Fakhrzadeh et al. 2002; Inoue et al. 2000; Kleeberger et al. 2001). Lung injury after O3 exposure may directly or indirectly affect adaptive immune responses such as T-cell proliferation (Jakab et al. 1995), response to allergen (Depuydt et al. 2002), and upregulation of costimulatory molecules such as cellular differentiation factor 86 (CD86, B7.2) that contribute to T-cell activation (Koike et al. 2001).
Interleuken (IL)-10 is a pleiotropic cytokine that is produced by activated monocytes, macrophages, and helper T-cells, and B-cells. IL-10 reduces TNF-α and IL-6 production (Lang et al. 2002), and inhibits MIP-2 (Standiford et al. 1995). Previously, Reinhart et al. (1999) showed that intratracheal instillation of recombinant IL-10 in Sprague-Dawley rats before O3 exposure significantly reduced O3-induced PMN infiltration and pulmonary hyperpermeability responses [fibronectin, albumin, bronchoalveolar lavage fluid (BALF) protein]. However, the relationship between IL-10 and other inflammatory mediators and downstream molecular events has not been investigated. IL-10 inhibits inflammation by suppressing macrophage CD86 expression leading to anergic T-cells in a schistosomiasis model (Ding et al. 1993; Flores Villanueva et al. 1994). IL-10 also inhibits iNOS production in macrophages (Cunha et al. 1992) and blocks nuclear factor-κB (NF-κB) activation (Saadane et al. 2005). Tyrosine phosphorylation of the intracellular domains of the IL-10 receptor is known to activate the signaling transducer and activator of transcription-3 (STAT3) pathway (Donnelly et al. 1999). Tarzi et al. (2006) observed that peptide immunotherapy caused induction of IL-10 and suppressor of cytokine signaling 3 (SOCS3) in a bee venom model, and Gao and Ward (2007) has implicated SOCS3 in inflammatory diseases.
Polymorphisms in the human IL10 gene have been associated with the development of asthma (Chatterjee et al. 2005) and lipopolysaccharide (LPS) sensitivity (Schippers et al. 2005). The functional role of pulmonary IL-10 has been studied in experimental animals in response to LPS (Standiford et al. 1995), silica (Huaux et al. 1998), and infectious organisms (Higgins et al. 2003) and as a key mediator in allergy (Akdis et al. 2001) and cystic fibrosis (Saadane et al. 2005). However, the role of IL-10 in O3-induced lung inflammation and its underlying mechanism has not been sufficiently studied.
In this study, we tested the hypothesis that targeted deletion of Il10 in mice would enhance O3-induced pulmonary inflammation via modulation of expression of inflammatory mediators CD86 and MIP-2 and nuclear transcription factors NF-κB and STAT3. We compared O3-induced alterations in pulmonary injury phenotypes and putative downstream molecular events between Il10-sufficient (Il10+/+) and Il10-deficient (Il10−/−) mice. We also used global mRNA expression analyses of lung tissue to identify patterns of gene expression that are modulated by IL-10 after exposure to O3. Enhanced O3-induced inflammation phenotypes in Il10−/− mice compared with Il10+/+ mice were consistent with a protective role for IL-10. Microarray and pathway analyses identified significant differences in inflammatory mediator expression between Il10+/+ and Il10−/− mice in response to O3 and suggested novel genetic targets [e.g., cathepsin E (Ctse) and serum amyloid A3 (Saa3)] affiliated with Il10 expression and response to environmental oxidant exposure.
Materials and Methods
Animals
We purchased male Il10+/+ (C57BL/6) and Il10−/− (B6.129P2-Il10tm1Cgn/J) mice (6–8 weeks) from Jackson Laboratories (Bar Harbor, ME). We provided mice with water and pelleted open-formula rodent diet NIH-31 (Zeigler Brothers, Gardners, PA) ad libitum. All experimental procedures were conducted in accordance with approved guidelines from the National Institutes of Health (Institute of Laboratory Animal Resources 1996) and the American Physiological Society (2002). Animals were treated humanely and with regard for alleviation of suffering.
O3 exposure
We placed mice in individual stainless steel wire cages within a Hazelton 1000 chamber (Lab Products, Maywood, NJ) equipped with a charcoal and high-efficiency particulate air–filtered air supply. We exposed mice to 0.3 ppm O3 or filtered air for 24, 48, or 72 hr (23.5 hr/day) as described previously (Cho et al. 2007).
Necropsy and BALF analyses
We euthanized mice (intraperitoneal sodium pentobarbital, 104 mg/kg) immediately after exposure. We lavaged the right lung with Hanks’ balanced salt solution and processed the lung for cell and total protein (a marker of lung permeability) analyses following Cho et al. (2001). The left lung was snap-frozen in liquid nitrogen for molecular analyses.
Histological analysis
We inflated lavaged right lungs with 10% formalin, removed en bloc, and immersed the lungs in 10% formalin. Details of histological analyses are in the Supplemental Material (doi:10.1289/ehp.1002182). Immunohistological staining was done using a specific antibody against Ki-67 (ab15580; Abcam, Cambridge, MA); we followed the procedures outlined by Cho et al. (2007).
Real-time quantitative reverse-transcriptase polymerase chain reaction
We isolated total RNA from left lung homogenates using the RNeasy Midi Kit (Qiagen Inc., Valencia, CA) following the manufacturer’s instructions and as described by Cho et al. (2007). Details of quantitative reverse-transcriptase polymerase chain reaction (RT-PCR) procedures are outlined in the Supplemental Material (doi:10.1289/ehp.1002182).
Western blot analysis
We prepared total lung protein in RIPA buffer containing protease and phosphatase inhibitors. Details of Western blot procedures and analyses are presented in the Supplemental Material (doi:10.1289/ehp.1002182).
Enzyme-linked immunosorbant assay for MIP-2 and NF-κB subunit, p65
We used 50 μL BALF from each mouse for enzyme-linked immunosorbant assay (ELISA). The procedures were performed following manufacturer’s instructions (R&D Systems, Minneapolis, MN). Processing and analysis procedures are detailed in the Supplemental Material (doi:10.1289/ehp.1002182).
Nuclear protein isolation and electrophoretic mobility shift assay for NF-κB
We prepared nuclear extracts from right lung lobes using a Nuclear Extraction Kit (Active Motif, Carlsbad, CA). Details of the electrophoretic mobility shift assay (EMSA) procedures are in the Supplemental Material (doi:10.1289/ehp.1002182).
Gene array analysis
RNA isolation and Affymetrix GeneChip hybridization
We isolated total RNA from left lung lobes, and the RNA was used after passing quality testing using an Agilent Bioanalyzer 2100 (Agilent Technologies, Inc., Santa Clara, CA, USA). For each treatment group, we performed GeneChip analysis (Affymetrix, Santa Clara, CA) analyses in duplicate (air controls) or triplicate (O3 exposed). Further details of RNA processing and hybridization are in the Supplemental Material (doi:10.1289/ehp.1002182).
Data analysis
We normalized and summarized the resulting files (in CEL format) with the Robust Multichip Average method using RMAExpress (http://rmaexpress.bmbolstad.com/). We exported log2 values for analysis using the Spotfire DecisionSite (TIBCO Spotfire, Somerville, MA). Hierarchical clustering identified one outlier, a sample from an Il10−/− mouse exposed to O3 for 24 hr, which was not included in subsequent analysis. We also applied Ingenuity Pathway Analysis (IPA) software (Ingenuity Systems, Inc., Redwood City, CA), a structured network knowledge-based approach, to evaluate functions and to elect putative interaction and signaling mechanisms through which IL-10 plays a role in pulmonary pathogenesis in response to O3. We further analyzed statistically the CEL files by two-way analysis of variance (ANOVA; p < 0.01) using genotype (Il10+/+, Il10−/−) and exposure (air, 24-hr O3, 48-hr O3, 72-hr O3) as the variables in GeneSpring GX 11.0 Expression Analysis software program (Agilent Technologies). We deposited the raw data discussed in this publication into the National Center for Biotechnology Information’s Gene Expression Omnibus data repository (http://www.ncbi.nlm.nih.gov/geo/, series GSE25095) and the National Institute of Environmental Health Sciences Chemical Effects in Biological Systems database (http://cebs.niehs.nih.gov/, accession no. 005-00003-0070-000-5). Details of the processes used to identify genes with altered transcript levels as a function of exposure, strain, and/or time are in the Supplemental Material (doi:10.1289/ehp.1002182). After consolidating probe sets, we loaded the resulting list of 165 genes and annotation into IPA (version 7.4, April 2009) to identify unique gene expression pathways that reflected an interaction between strain and treatment effect.
Statistics
We expressed all data as group means ± SE. We log-transformed PMN data to ensure normal data distribution and equal variance. We assessed differences in effects of O3 and IL-10 on response phenotypes by two-factor ANOVA. The factors were exposure (air or O3) and genotype (Il10+/+ or Il10−/−). Dependent variables were BALF protein concentration, BALF cells, mRNA expression, and protein levels. We used the Student Newman–Keuls post hoc test to compare group means. We performed all statistical analyses using the SigmaStat statistics package (version 3; Systat Software, Inc., Point Richmond, CA). We accepted statistical significance at p < 0.05. Sample sizes are included in the figure captions.
Results
Lung inflammatory responses
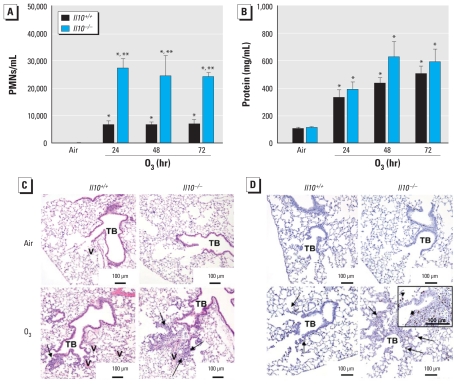
O3-induced increases in total numbers of BALF cells were significantly greater in Il10−/− compared with Il10+/+ mice [see Supplemental Material, Table 1 (doi:10.1289/ehp.1002182)] and largely attributable to differences in total numbers of PMNs. Compared with Il10+/+ mice, the numbers of PMNs were significantly greater (~3-fold) in Il10−/− mice at each O3 exposure time point (Figure 1A). Total BALF protein concentrations also increased significantly in both genotypes after O3, but we found no significant differences between the two genotypes (p = 0.055; Figure 1B).
Figure 1.
Changes in number of PMNs (A) and total protein concentration (B) recovered in BALF from Il10+/+ and Il10−/− mice in response to air or 0.3 ppm O3 (means ± SE; n = 8/group). *p < 0.05, air versus 0.3 ppm O3; **p < 0.05, Il10+/+ versus Il10−/−. (C) H&E staining of 5-μm lung sections from Il10+/+ and Il10−/− mice after air or 0.3 ppm O3 (72 hr). Arrows illustrate areas of greatest neutrophilic inflammation, particularly around terminal bronchioles (TB) and blood vessels (V). Sections are representative of mice from each treatment group. (D) Ki67 immunostaining of 5-μm lung sections from Il10+/+ and Il10−/− mice after air or 0.3 ppm O3 (72 hr). Arrows indicate foci of cellular proliferation (arrows indicate the arrowheads shown in the inset). Sections are representative of mice from each treatment group.
Histopathological analysis of lung injury
Consistent with BALF phenotypes, we found greater O3-induced inflammation in perivascular, peribronchiolar, and terminal bronchial regions in hemotoxylin and eosin (H&E)-stained lung tissue sections from Il10−/− mice compared with Il10+/+ mice (Figure 1C). Further, the density of Ki67-positive cells at distal perivascular-peribronchiolar areas and centriacinar regions was more prominent in Il10−/− than in Il10+/+ mice exposed to O3, indicating enhanced cellular proliferation (Figure 1D).
Inflammatory mediator production
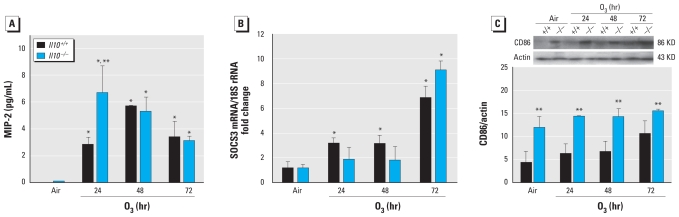
We did not detect Il10 mRNA expression in Il10−/− mouse lung homogenates after air or O3 exposure (data not shown). However, Il10 mRNA was significantly elevated relative to air-exposed controls after 24-hr O3 (4.4-fold; p < 0.05) but was not significantly different between air- and O3-exposed Il10+/+ mice after 48- and 72-hr exposures (data not shown). Relative to respective air controls, O3 significantly increased mean BALF concentration of the PMN chemoattractant MIP-2 in both genotypes and was significantly higher in Il10−/− compared with Il10+/+ mice at 24 hr (Figure 2A). However, these genotype-dependent differences were not evident at 48- and 72-hr O3. TNF-α and iNOS mRNA expression levels were significantly increased in both strains during O3, but we found no significant genotypic differences (data not shown).
Figure 2.
(A) MIP-2 protein levels in BALF recovered from Il10+/+ and Il10−/− mice exposed to air and 0.3 ppm O3 (means ± SE; n = 4 per group). *p < 0.05, air versus O3; **p < 0.05, Il10+/+ versus Il10−/−. (B) Socs3 mRNA expression in Il10+/+ and Il10−/− mice in response to air or 0.3 ppm O3. We determined the ratio of Socs3 to 18S RNA from whole-lung homogenates from PCR (means ± SE; n = 3 per group). *p < 0.05, air versus 0.3 ppm O3. (C) Lung CD86 protein expression in Il10+/+ and Il10−/− mice exposed to air and 0.3 ppm O3 (means ± SE; n = 3–5 per group). We detected CD86 expression from whole-lung homogenates by Western blot analysis and reprobed blots with actin for normalization. +p < 0.05.
SOCS3 expression
Because SOCS3 has been proposed as a mechanism of IL-10–mediated protection against inflammatory processes in other models of lung disease (Gao and Ward 2007), we asked whether SOCS3 was differentially expressed in Il10−/− and Il10+/+ mice. Relative to air controls, Socs3 mRNA expression increased significantly in Il10+/+ mice after 24-, 48-, and 72-hr O3 (Figure 2B); Socs3 expression in Il10−/− mice was increased significantly only after 72-hr O3.
CD86 production
Compared with Il10+/+ mice, we found significantly higher levels of lung CD86 proteins in Il10−/− mice after air and O3 (Figure 2C). However, O3 did not increase CD86 in either genotype, and we found no interaction between genotype and exposure.
Nuclear NF-κB activity and STAT3 activation
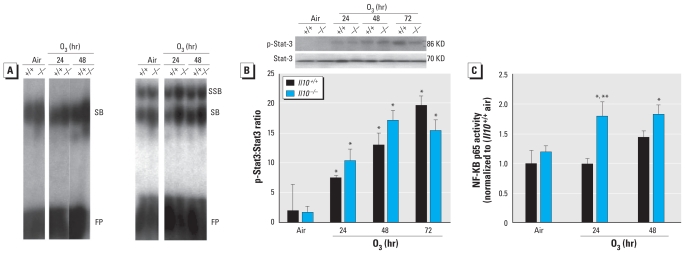
To determine whether the protective effect of IL-10 may be mediated in part by differential NF-κB activity, we evaluated nuclear DNA binding activity of total NF-κB and specific p50, a subunit of NF-κB and p65 κB activity in Il10+/+ and Il10−/− mice. Interestingly, baseline binding activity of total NF-κB and specific p50 κB was slightly greater in Il10−/− mice than in Il10+/+ mice (Figure 3A). O3 effects on activation of total NF-κB and specific p50 κB binding activity were more marked in Il10−/− mice than in Il10+/+ mice (Figure 3A). Specific p65 subunit NF-κB activity was significantly increased only in Il10−/− mice after O3 (Figure 3A). The ratio of phosphorylated STAT3 (p-STAT3) to STAT3 protein in the lung increased significantly after O3 compared with air controls in both genotypes, signifying an increase in activated STAT3 protein in response to O3 (Figure 3B). However, we found no differences in O3-induced increases in the p-STAT3:STAT3 ratio between Il10−/− and Il10+/+ mice.
Figure 3.
(A) EMSA to determine DNA binding activity of total NF-κB (top left) and specific p50 κB (top right) after 0.3 ppm O3 in Il10+/+ and Il10−/− mice. Each lane represents nuclear protein pooled from three representative animals of each treatment group and was repeated three times. SB, shifted band; SSB, supershifted band; FP, free probe. Quantified p65 κB determined by ELISA is presented below (means ± SE; n = 3 per group). *p < 0.05, air versus 0.3 ppm O3; **p < 0.05, Il10+/+ versus Il10−/−. (B) Phosphorylated STAT3 (p-STAT3) in Il10+/+ and Il10−/− mice in response to air or 0.3 ppm O3. We determined the ratio of p-STAT3 to total STAT3 from whole-lung homogenates by Western blot analysis (means ± SE; n = 3 per group). *p < 0.05, air versus 0.3 ppm O3.
O3-induced gene expression profiles are altered in Il10+/+ compared with Il10−/− mice
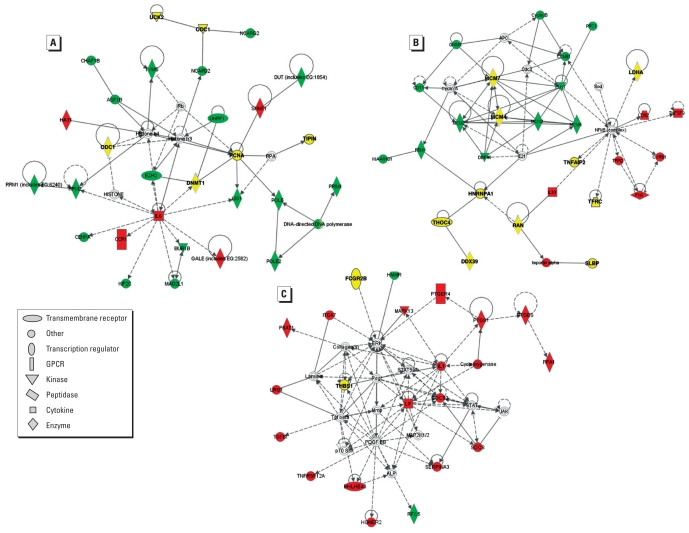
We first used unsupervised analyses of gene expression profiles in Il10−/− and Il10+/+ mice to provide additional insight into the means through which IL-10 protects the lung against O3-induced inflammation. Using k-means clustering analysis, we identified differential gene expression profile patterns between Il10+/+ and Il10−/− mice that suggested genotype-independent and genotype-specific O3 effects [for gene expression clusters and gene lists, see Supplemental Material (doi:10.1289/ehp.1002182)]. IPA revealed distinct gene expression pathways for specific O3-induced changes that displayed similar kinetics between Il10+/+ and Il10−/− mice [Figure 4, red shapes; see also Supplemental Material, Table 2, Figure 5A (doi:10.1289/ehp.1002182)]. For example, Gale (galatose-4-epimerase, UDP), Hat1 (histone acetyl transferase 1), and Ccr1 [chemokine (C-C motif) receptor-1] expression was altered after O3 (Figure 4A, red shapes). In addition, we observed O3-induced gene expression profile differences in Il33 in Il10−/− and Il10+/+ mice (Figure 4B, red shapes), Socs3, and Il1 (Figure 4C, red shapes). This is consistent with Figure 2B that shows parallel increases in Socs3 transcript level in both strains, although statistical testing revealed a significant increase in Il10+/+ mice earlier than in the Il10−/− mice. These genes are associated with stress and inflammatory pathways [see Supplemental Material, Table 3 (doi:10.1289/ehp.1002182)]. Moreover, we found O3-induced gene expression patterns that were altered temporally in Il10−/− compared with Il10+/+ mice [Figure 4, green shapes; see also Supplemental Material, Figure 5D, Table 2 (doi:10.1289/ehp.1002182)].
Figure 4.
IPA illustrates the three most highly associated networks (A–C) of IL-10–dependent genes in response to O3. Red symbols, genes whose profiles were derived from Supplemental Material Figure 5A [see Supplemental Material, Figure 5, Table 2 (doi:10.1289/ehp.1002182)]; yellow symbols, genes with profiles that followed the pattern illustrated in Supplemental Material Figure 5B (see Supplemental Material, Figure 5, Table 2); green symbols, genes with profiles that differ between Il10+/+ and Il10−/− mice [see Supplemental Material, Figure 5D, Table 2 (doi:10.1289/ehp.1002182)]. Gene abbreviations are identified in Supplemental Material, Table 2. GPCR, G-protein–coupled receptor.
We also applied supervised analysis using GeneSpring to identify genes that varied statistically significantly between Il10+/+ and Il10−/− mice during 24- to 72-hr O3 [see Supplemental Material, Figure 6, Table 4 (doi:10.1289/ehp.1002182)]. Genes > 2-fold higher in Il10−/− mice than in Il10+/+ mice included those encoding CTSE, WDFY1 (WD repeat and FYVE domain containing 1), IL-1β, SAA3, nicotinamide nucleotide transhydrogenase (NNT), and peptidylglycine α-amidating monooxygenase (PAM). Many of these genes were also elucidated by unsupervised analysis [see Supplemental Material, Table 4 (doi:10.1289/ehp.1002182)]. We validated a subset of the genes (n = 4) listed in the Supplemental Material, Table 4 (doi:10.1289/ehp.1002182), by measuring mRNA expression in lung homogenates from air- and O3-exposed Il10−/− and Il10+/+ mice. We found that mRNA expression of Ctse (24 hr), Wdfy1 (24, 48 hr), Saa3 (48 hr), and S100a14 (S100 calcium binding protein A14; 24, 48, and 72 hr) was significantly (p < 0.05) greater in Il10−/− than in Il10+/+ lung homogenates [see Supplemental Material, Figure 7 (doi:10.1289/ehp.1002182)].
Discussion
We investigated the mechanisms of IL-10–mediated reductions in O3-induced airway neutrophilia in mice. Targeted deletion of Il10 significantly enhanced O3-induced pulmonary cellular inflammation, as indicated by significant differences in numbers of infiltrating neutrophils and enhanced cellular proliferation in centriacinar regions compared with Il10+/+ controls. Nuclear activity of NF-κB and expression of CD86 proteins were also greater in lungs of Il10−/− mice. Furthermore, MIP-2 protein was significantly elevated in response to 24-hr O3 and was exacerbated in Il10−/− compared with Il10+/+ mice. Results support a role for IL-10 in protection against O3-induced inflammation associated with diminished NF-κB activity and MIP-2 production. Nonbiased (visual, unsupervised) and supervised (ANOVA) gene array analysis identified novel cellular targets that may contribute to the protective mechanism of IL-10 in O3-induced inflammation.
O3-induced increases in BALF neutrophils and lung cell proliferation were significantly enhanced in Il10−/− mice relative to Il10+/+ wild-type controls, but the lung permeability response to O3 was not significantly different between the two genotypes. Interestingly, Reinhart et al. (1999) found that intratracheal instillation of recombinant IL-10 into Sprague-Dawley rats significantly attenuated by 25% protein permeability responses induced by acute O3 exposure (0.8 ppm, 3 hr) compared with controls. An explanation for the disparate observations is not entirely clear, but the role of IL-10 in the hyperpermeability response may depend on the concentration and duration of exposure to O3 (0.8 ppm for 3 hr compared with 0.3 ppm for 24, 48, or 72 hr) or may be species specific. Moreover, these results are consistent with previous studies suggesting that inflammatory and permeability responses in this model are regulated through different mechanisms (e.g., Cho et al. 2007).
Upregulation of the PMN chemoattractant MIP-2 has been suggested to regulate initial neutrophilic infiltration in response to O3. O3-induced increases in MIP-2 were potentiated in the absence of Il10 at 24 hr before the peak PMN influx and therefore may be a mechanism through which IL-10 protects against O3-induced inflammation and cell proliferation. Similarly, neutralization of IL-10 prior to LPS administration can attenuate production of MIP-2 and associated neutrophilia (Standiford et al. 1995). We observed no significantly different O3-induced changes in TNF-α and iNOS between Il10−/− and Il10+/+ mice, despite genotype-specific differences in NF-κB after O3. These results suggest that O3-induced increases in TNF-α and iNOS, which are thought to be NF-κB dependent (e.g., Cho et al. 2007), are not modulated by IL-10. These results are inconsistent with those of Lang et al. (2002) and Standiford et al. (1995), who found that TNF-α production is IL-10 dependent in responses to endotoxin. Collectively, these studies may suggest important differences in pulmonary cellular responses to endotoxin and O3 and warrant further investigation.
We also found that lung CD86 protein levels were enhanced in Il10−/− compared with Il10+/+ mice, independent of O3 exposure. Soltys et al. (2002) showed that CD86 in alveolar macrophages was significantly enhanced in Il10−/− mice at baseline and suggested that low levels of alveolar macrophage costimulatory molecule expression are maintained at least in part by endogenous IL-10 activity. Koike et al. (2001) found that approximately 8% of alveolar macrophages obtained from rats exposed to 1 ppm O3 for 3 days were CD86 positive and suggested that small increases in costimulatory activity may be sufficient to regulate O3-induced immune responses. Our data similarly indicate that Il10 deficiency permits endogenous CD86 expression, which could explain why we did not detect subtle increases in CD86 in Il10−/− mice after O3 exposure.
To investigate the potential signaling pathway through which IL-10 protects against O3-induced inflammation, we evaluated NF-κB activation in Il10+/+ and Il10−/− mice. NF-κB is a nuclear protein that regulates transcription of many gene products that activate major pulmonary inflammatory pathways, including responses to O3 (Cho et al. 2007). IL-10 inhibits NF-κB (Saadane et al. 2005; Spight et al. 2005). We found that O3-induced p50 levels and p65 κB binding activity were significantly greater in Il10−/− than in Il10+/+ mice. The mechanism through which IL-10 inhibits NF-κB may occur indirectly, such as through inhibition of inhibitory kappa kinase, as demonstrated in response to LPS (Saadane et al. 2005; Spight et al. 2005).
STAT3 was associated with IL-10–mediated inhibition of alveolar macrophage activation by LPS (Berlato et al. 2002). Stat3 has also been shown to mediate O3-induced inflammation (Laskin et al. 2002) and pulmonary inflammation and hyperresponsiveness associated with asthmatic responses (Corry 2002). Interestingly, we found no significant genotype-specific differences in the p-STAT3:STAT3 ratio (Figure 3B). Similarly, gene array analysis did not detect an interaction among O3 exposure, Il10 deficiency, and Stat3 gene expression at these time points. These results suggest that O3-induced STAT3 activation occurs in an IL-10–independent manner.
To further elucidate the potential mechanism through which IL-10 protects the lung against O3-induced inflammation, we used k-means clustering and Ingenuity analyses of microarray expression data. We identified three distinct clusters of molecules, representing three pathways and numerous gene targets. Several genes identified in this analysis (e.g., Il6, Il1ra, Il1) contribute to O3-induced pulmonary inflammation in mice (Figure 4).
Other identified genes represent novel targets for O3-induced inflammation. For example, both gene expression analyses identified Ctse as differentially expressed between genotypes after exposure to O3. Ctse is a an intracellular aspartic protease that is found in immune system cells such as dendritic cells and macrophages and has been implicated in major histocompatibility complex (MHC) class II pathway antigen processing (e.g., Zaidi and Kalbacher 2008). Although Ctse has not been associated with O3-induced inflammation, MHC class II molecules have been implicated recently in the PMN response to O3 exposure in the mouse (Bauer et al. 2009), and it is plausible that CTSE could also be involved in this pathway. SAA and S100A14 have been implicated in inflammatory processes (e.g., Han et al. 2007) or inflammation-related diseases (Chen et al. 2009) and thus provide a rationale for a role in O3-induced inflammation.
Our gene array analysis identified Socs3 as being induced in response to O3, compared with air controls. RT-PCR analyses confirmed O3-induced upregulation of Socs3 and identified a small but statistically significant difference in expression kinetics between Il10+/+ and Il10−/− mice. Members of the SOCS signaling family, including SOCS3, regulate activation and function of STAT3 (Gao and Ward 2007). Emerging data on activation and function of STAT3 and SOCS3 in the lung during acute inflammation suggest that these molecules may regulate pulmonary inflammation (Gao and Ward 2007), and they have specifically been implicated in allergic airway inflammation (Paul et al. 2009). Given that SOCS3 is a proximal mediator associated with IL-10 ligand binding, the role of SOCS3 in O3-induced inflammation necessitates further investigation to determine its specific contribution to the adaptive/innate immune response.
The array analysis implicated a number of novel genes for response to O3 in Il10+/+ and Il10−/− mice, including Il33 and Hat1. IL-33 is a member of the IL-1 superfamily and is expressed on epithelial cells. This cytokine has been identified in joints of arthritis patients (Palmer et al. 2009) and is implicated in development and regulation of type 2 T-helper cell–dependent immune responses in sites of mucosal immunity, which may suggest a link between innate and adaptive immune responses after O3 exposure (Saenz et al. 2008). HAT1, an acetylation enzyme, controls access to DNA transcription by controlling histone protein activation during chromatin assembly (Parthun 2007). Although HAT1 expression has been linked to inflammatory mediators such as interferon-γ and TNF-α (Keslacy et al. 2007), the specific role of HAT1 in response to O3 remains unclear.
Conclusion
This study supports the hypothesis that IL-10 protects against O3-induced lung inflammation and identified several potential mechanisms involved in this response. IL-10 deficiency enhanced O3-induced neutrophilic inflammation and injury at the centriacinar region of the lung. Results also suggest that in response to O3 the effect of IL-10 may be mediated, in part, via modulation of NF-κB, MIP-2, and CD86. Gene array analysis identified three IL-10–mediated expression pathways with genes known previously to be affected by O3 and novel genes that may contribute significantly to the pathogenesis of O3-induced pulmonary inflammation. The present investigation identified novel molecular mechanisms of pulmonary O3 toxicity, which may provide possible therapeutic targets for attenuating the effects of O3 in susceptible individuals.
Footnotes
Supplemental Material is available online (doi:10.1289/ehp.1002182 via http://dx.doi.org/).
The authors acknowledge A. Jedlicka for her assistance with the microarray analyses.
This work was supported by the Division of Intramural Research, National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health, Department of Health and Human Services. D.B.P. was supported by a grant from NIEHS (R01ES012706).
References
- Akdis CA, Joss A, Akdis M, Blaser K. Mechanism of IL-10-induced T cell inactivation in allergic inflammation and normal response to allergens. Int Arch Allergy Immunol. 2001;124((1–3)):180–182. doi: 10.1159/000053704. [DOI] [PubMed] [Google Scholar]
- American Physiological Society. Appendix C. Guiding Principles for Research Involving Animals and Human Beings. Recommendations From The Revised Declaration of Helsinki by the World Medical Association Regarding Human Subjects. 2002. [(accessed 21 September 2010)]. Available: http://www.the-aps.org/about/opguide/appendix.htm#guiding.
- Bauer AK, Travis EL, Malhotra SS, Rondini EA, Walker C, Cho HY, et al. Identification of novel susceptibility genes in ozone-induced inflammation in mice. Eur Respir J. 2009;36((2)):428–437. doi: 10.1183/09031936.00145309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlato C, Cassatella MA, Kinjyo I, Gatto L, Yoshimura A, Bazzoni F. Involvement of suppressor of cytokine signaling-3 as a mediator of the inhibitory effects of IL-10 on lipopolysaccharide-induced macrophage activation. J Immunol. 2002;168((12)):6404–6411. doi: 10.4049/jimmunol.168.12.6404. [DOI] [PubMed] [Google Scholar]
- Bhalla DK, Reinhart PG, Bai C, Gupta SK. Amelioration of ozone-induced lung injury by antitumor necrosis factor-alpha. Toxicol Sci. 2002;69((2)):400–408. doi: 10.1093/toxsci/69.2.400. [DOI] [PubMed] [Google Scholar]
- Chatterjee R, Batra J, Kumar A, Mabalirajan U, Nahid S, Niphadkar PV, et al. Interleukin-10 promoter polymorphisms and atopic asthma in North Indians. Clin Exp Allergy. 2005;35((7)):914–919. doi: 10.1111/j.1365-2222.2005.02273.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Yu D, Luo A, Tan W, Zhang C, Zhao, et al. Functional role of S100A14 genetic variants and their association with esophageal squamous cell carcinoma. Cancer Res. 2009;69((8)):3451–3457. doi: 10.1158/0008-5472.CAN-08-4231. [DOI] [PubMed] [Google Scholar]
- Cho HY, Morgan DL, Bauer AK, Kleeberger SR. Signal transduction pathways of tumor necrosis factor--mediated lung injury induced by ozone in mice. Am J Respir Crit Care Med. 2007;175((8)):829–839. doi: 10.1164/rccm.200509-1527OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho HY, Zhang LY, Kleeberger SR. Ozone-induced lung inflammation and hyperreactivity are mediated via tumor necrosis factor-alpha receptors. Am J Physiol Lung Cell Mol Physiol. 2001;280((3)):L537–L546. doi: 10.1152/ajplung.2001.280.3.L537. [DOI] [PubMed] [Google Scholar]
- Corry DB. Emerging immune targets for the therapy of allergic asthma. Nat Rev Drug Discov. 2002;1((1)):55–64. doi: 10.1038/nrd702. [DOI] [PubMed] [Google Scholar]
- Cunha FQ, Moncada S, Liew FY. Interleukin-10 (IL-10) inhibits the induction of nitric oxide synthase by interferon-gamma in murine macrophages. Biochem Biophys Res Commun. 1992;182((3)):1155–1159. doi: 10.1016/0006-291x(92)91852-h. [DOI] [PubMed] [Google Scholar]
- Depuydt PO, Lambrecht BN, Joos GF, Pauwels RA. Effect of ozone exposure on allergic sensitization and airway inflammation induced by dendritic cells. Clin Exp Allergy. 2002;32((3)):391–396. doi: 10.1046/j.1365-2222.2002.01364.x. [DOI] [PubMed] [Google Scholar]
- Ding L, Linsley PS, Huang LY, Germain RN, Shevach EM. IL-10 inhibits macrophage costimulatory activity by selectively inhibiting the up-regulation of B7 expression. J Immunol. 1993;151((3)):1224–1234. [PubMed] [Google Scholar]
- Donnelly RP, Dickensheets H, Finbloom DS. The interleukin-10 signal transduction pathway and regulation of gene expression in mononuclear phagocytes. J Interferon Cytokine Res. 1999;19((6)):563–573. doi: 10.1089/107999099313695. [DOI] [PubMed] [Google Scholar]
- Driscoll KE, Simpson L, Carter J, Hassenbein D, Leikauf GD. Ozone inhalation stimulates expression of a neutrophil chemotactic protein, macrophage inflammatory protein 2. Toxicol Appl Pharmacol. 1993;119((2)):306–309. doi: 10.1006/taap.1993.1074. [DOI] [PubMed] [Google Scholar]
- Fakhrzadeh L, Laskin JD, Laskin DL. Deficiency in inducible nitric oxide synthase protects mice from ozone-induced lung inflammation and tissue injury. Am J Respir Cell Mol Biol. 2002;26((4)):413–419. doi: 10.1165/ajrcmb.26.4.4516. [DOI] [PubMed] [Google Scholar]
- Flores Villanueva PO, Reiser H, Stadecker MJ. Regulation of T helper cell responses in experimental murine schistosomiasis by IL-10. Effect on expression of B7 and B7–2 costimulatory molecules by macrophages. J Immunol. 1994;153((11)):5190–5199. [PubMed] [Google Scholar]
- Gao H, Ward PA. STAT3 and suppressor of cytokine signaling 3: potential targets in lung inflammatory responses. Expert Opin Ther Targets. 2007;11((7)):869–880. doi: 10.1517/14728222.11.7.869. [DOI] [PubMed] [Google Scholar]
- Han CY, Subramanian S, Chan CK, Omer M, Chiba T, Wight TN, et al. Adipocyte-derived serum amyloid A3 and hyaluronan play a role in monocyte recruitment and adhesion. Diabetes. 2007;56((9)):2260–2273. doi: 10.2337/db07-0218. [DOI] [PubMed] [Google Scholar]
- Higgins SC, Lavelle EC, McCann C, Keogh B, McNeela E, Byrne P, et al. Toll-like receptor 4-mediated innate IL-10 activates antigen-specific regulatory T cells and confers resistance to Bordetella pertussis by inhibiting inflammatory pathology. J Immunol. 2003;171((6)):3119–3127. doi: 10.4049/jimmunol.171.6.3119. [DOI] [PubMed] [Google Scholar]
- Huaux F, Louahed J, Hudspith B, Meredith C, Delos M, Renauld JC, et al. Role of interleukin-10 in the lung response to silica in mice. Am J Respir Cell Mol Biol. 1998;18((1)):51–59. doi: 10.1165/ajrcmb.18.1.2911. [DOI] [PubMed] [Google Scholar]
- Inoue H, Aizawa H, Nakano H, Matsumoto K, Kuwano K, Nadel JA, et al. Nitric oxide synthase inhibitors attenuate ozone-induced airway inflammation in guinea pigs. Possible role of interleukin-8. Am J Respir Crit Care Med. 2000;161((1)):249–256. doi: 10.1164/ajrccm.161.1.9804096. [DOI] [PubMed] [Google Scholar]
- Institute of Laboratory Animal Resources. Guide for the Care and Use of Laboratory Animals. 7th ed. Washington, DC: National Academy Press; 1996. [Google Scholar]
- Jakab GJ, Spannhake EW, Canning BJ, Kleeberger SR, Gilmour MI. The effects of ozone on immune function. Environ Health Perspect. 1995;103((suppl 2)):77–89. doi: 10.1289/ehp.95103s277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerrett M, Burnett RT, Pope CA, III, Ito K, Thurston G, Krewski D, et al. Long-term exposure to ozone and mortality. N Engl J Med. 2009;360((11)):1085–1095. doi: 10.1056/NEJMoa0803894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston RA, Schwartzman IN, Flynt L, Shore SA. Role of interleukin-6 in murine airway responses to ozone. Am J Physiol Lung Cell Mol Physiol. 2005;288((2)):L390–L397. doi: 10.1152/ajplung.00007.2004. [DOI] [PubMed] [Google Scholar]
- Keslacy S, Tliba O, Baidouri H, Amrani Y. Inhibition of tumor necrosis factor-alpha-inducible inflammatory genes by interferon-gamma is associated with altered nuclear factor-kappaB transactivation and enhanced histone deacetylase activity. Mol Pharmacol. 2007;71((2)):609–618. doi: 10.1124/mol.106.030171. [DOI] [PubMed] [Google Scholar]
- Kleeberger SR, Reddy SP, Zhang LY, Cho HY, Jedlicka AE. Toll-like receptor 4 mediates ozone-induced murine lung hyperpermeability via inducible nitric oxide synthase. Am J Physiol Lung Cell Mol Physiol. 2001;280((2)):L326–L333. doi: 10.1152/ajplung.2001.280.2.L326. [DOI] [PubMed] [Google Scholar]
- Koike E, Kobayashi T, Shimojo N. Ozone exposure enhances expression of cell-surface molecules associated with antigen-presenting activity on bronchoalveolar lavage cells in rats. Toxicol Sci. 2001;63((1)):115–124. doi: 10.1093/toxsci/63.1.115. [DOI] [PubMed] [Google Scholar]
- Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol. 2002;169((5)):2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- Laskin DL, Fakhrzadeh L, Heck DE, Gerecke D, Laskin JD. Upregulation of phosphoinositide 3-kinase and protein kinase B in alveolar macrophages following ozone inhalation. Role of NF-kappaB and STAT-1 in ozone-induced nitric oxide production and toxicity. Mol Cell Biochem. 2002;234–235(1–2):91–98. [PubMed] [Google Scholar]
- McConnell R, Berhane K, Gilliland F, London SJ, Islam T, Gauderman WJ, et al. Asthma in exercising children exposed to ozone: a cohort study. Lancet. 2002;359((9304)):386–391. doi: 10.1016/S0140-6736(02)07597-9. [DOI] [PubMed] [Google Scholar]
- Mudway IS, Kelly FJ. Ozone and the lung: a sensitive issue. Mol Aspects Med. 2000;21((1–2)):1–48. doi: 10.1016/s0098-2997(00)00003-0. [DOI] [PubMed] [Google Scholar]
- Neuhaus-Steinmetz U, Uffhausen F, Herz U, Renz H. Priming of allergic immune responses by repeated ozone exposure in mice. Am J Respir Cell Mol Biol. 2000;23((2)):228–233. doi: 10.1165/ajrcmb.23.2.3898. [DOI] [PubMed] [Google Scholar]
- Palmer G, Talabot-Ayer D, Lamacchia C, Toy D, Seemayer CA, Viatte S, et al. Inhibition of interleukin-33 signaling attenuates the severity of experimental arthritis. Arthritis Rheum. 2009;60((3)):738–749. doi: 10.1002/art.24305. [DOI] [PubMed] [Google Scholar]
- Parthun MR. Hat1: the emerging cellular roles of a type B histone acetyltransferase. Oncogene. 2007;26((37)):5319–5328. doi: 10.1038/sj.onc.1210602. [DOI] [PubMed] [Google Scholar]
- Paul B, Mishra V, Chaudhury B, Awasthi A, Das AB, Saxena U, et al. Status of Stat3 in an ovalbumin-induced mouse model of asthma: analysis of the role of Socs3 and IL-6. Int Arch Allergy Immunol. 2009;148((2)):99–108. doi: 10.1159/000155740. [DOI] [PubMed] [Google Scholar]
- Plopper CG, Hatch GE, Wong V, Duan X, Weir AJ, Tarkington BK, et al. Relationship of inhaled ozone concentration to acute tracheobronchial epithelial injury, site-specific ozone dose, and glutathione depletion in rhesus monkeys. Am J Respir Cell Mol Biol. 1998;19((3)):387–399. doi: 10.1165/ajrcmb.19.3.3183. [DOI] [PubMed] [Google Scholar]
- Reinhart PG, Gupta SK, Bhalla DK. Attenuation of ozone-induced lung injury by interleukin-10. Toxicol Lett. 1999;110((1–2)):35–42. doi: 10.1016/s0378-4274(99)00136-8. [DOI] [PubMed] [Google Scholar]
- Saadane A, Soltys J, Berger M. Role of IL-10 deficiency in excessive nuclear factor-kappaB activation and lung inflammation in cystic fibrosis transmembrane conductance regulator knockout mice. J Allergy Clin Immunol. 2005;115((2)):405–411. doi: 10.1016/j.jaci.2004.10.044. [DOI] [PubMed] [Google Scholar]
- Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunol Rev. 2008;226:172–190. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schippers EF, van ‘t Veer C, van Voorden S, Martina CA, Huizinga TW, le Cessie S, et al. IL-10 and toll-like receptor-4 polymorphisms and the in vivo and ex vivo response to endotoxin. Cytokine. 2005;29((5)):215–228. doi: 10.1016/j.cyto.2004.12.005. [DOI] [PubMed] [Google Scholar]
- Soltys J, Bonfield T, Chmiel J, Berger M. Functional IL-10 deficiency in the lung of cystic fibrosis (cftr(−/−)) and IL-10 knockout mice causes increased expression and function of B7 costimulatory molecules on alveolar macrophages. J Immunol. 2002;168((4)):1903–1910. doi: 10.4049/jimmunol.168.4.1903. [DOI] [PubMed] [Google Scholar]
- Spight D, Zhao B, Haas M, Wert S, Denenberg A, Shanley TP. Immunoregulatory effects of regulated, lung-targeted expression of IL-10 in vivo. Am J Physiol Lung Cell Mol Physiol. 2005;288((2)):L251–L265. doi: 10.1152/ajplung.00122.2004. [DOI] [PubMed] [Google Scholar]
- Standiford TJ, Strieter RM, Lukacs NW, Kunkel SL. Neutralization of IL-10 increases lethality in endotoxemia. Cooperative effects of macrophage inflammatory protein-2 and tumor necrosis factor. J Immunol. 1995;155((4)):2222–2229. [PubMed] [Google Scholar]
- Tarzi M, Klunker S, Texier C, Verhoef A, Stapel SO, Akdis CA, et al. Induction of interleukin-10 and suppressor of cytokine signalling-3 gene expression following peptide immunotherapy. Clin Exp Allergy. 2006;36((4)):465–474. doi: 10.1111/j.1365-2222.2006.02469.x. [DOI] [PubMed] [Google Scholar]
- Toward TJ, Broadley KJ. Airway function, oedema, cell infiltration and nitric oxide generation in conscious ozone-exposed guinea-pigs: effects of dexamethasone and rolipram. Br J Pharmacol. 2002;136((5)):735–745. doi: 10.1038/sj.bjp.0704764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaidi N, Kalbacher H. Cathepsin E: a mini review. Biochem Biophys Res Commun. 2008;367((3)):517–522. doi: 10.1016/j.bbrc.2007.12.163. [DOI] [PubMed] [Google Scholar]