Abstract
Clothing use is an important modern behavior that contributed to the successful expansion of humans into higher latitudes and cold climates. Previous research suggests that clothing use originated anywhere between 40,000 and 3 Ma, though there is little direct archaeological, fossil, or genetic evidence to support more specific estimates. Since clothing lice evolved from head louse ancestors once humans adopted clothing, dating the emergence of clothing lice may provide more specific estimates of the origin of clothing use. Here, we use a Bayesian coalescent modeling approach to estimate that clothing lice diverged from head louse ancestors at least by 83,000 and possibly as early as 170,000 years ago. Our analysis suggests that the use of clothing likely originated with anatomically modern humans in Africa and reinforces a broad trend of modern human developments in Africa during the Middle to Late Pleistocene.
Keywords: lice, human evolution, isolation with migration
Hominins migrated out of Africa numerous times over the last two My (reviewed in Stringer 2002). Through the course of these migrations, archaic hominin populations occupied parts of Europe (e.g., Atapuerca, Spain; Carbonell et al. 2008) and Central Asia (e.g., the Altai Mountains, Siberia; Krause et al. 2010) that were cooler and increased their vulnerability to cold stress. Although evidence suggests that archaic hominins established long-lasting populations in these regions, anatomically modern humans (AMHs) likely outcompeted archaic hominins and were able to thrive despite the more seasonally variable climates (Gilligan 2010). A suite of complex behaviors and technologies associated with the transition of archaic to modern Homo sapiens, including improved clothing, are credited with facilitating the successful expansion of AMH out of Africa into higher latitudes. Critically, although clothing was likely a necessary technology for AMH, it is unknown whether clothing use originated early enough to play an important role in the expansion of archaic populations out of Africa.
Determining when clothing use began is challenging because early clothing (i.e., animal hides) would degrade rapidly, erasing any direct evidence of clothing use from the Late Pleistocene archeological record. The first evidence of tools used to scrape hides appears ∼780 Ka (Carbonell et al. 1999), but these very old dates do not necessarily signify clothing use. Animal hides had other uses besides clothing (e.g., providing shelter), although clothing is thought to be one of the earliest uses for skins. Eyed needles first appear in the archaeological record ∼40 Ka (Delson et al. 2000), but these signal the production of more complex clothing (e.g., tailored multilayered garment assemblages), which is undoubtedly a relatively recent innovation (Gilligan 2010). Importantly, the development of clothing likely occurred after humans lost their covering of body hair. Genetic data suggest that body hair was lost ∼1.2 Ma (Rogers et al. 2004), and an even older date (3 Ma) was hypothesized for the loss of body hair based on the origin of pubic lice in humans (Reed et al. 2007; Gilligan 2010). These studies suggest that clothing use may have evolved anywhere from 40 Ka to 3 Ma, and given the vastness of this time-span, alternative approaches for estimating the origin of clothing use are essential.
Parasites offer an ideal source of alternative data for determining when clothing use first began in hominins. Parasites can provide novel insights into the evolutionary history of their hosts, especially when the hosts exhibit low levels of genetic variation (Whiteman and Parker 2005). The parasitic sucking lice of primates (Phthiraptera: Anoplura) have cospeciated with their hosts and track both ancient (e.g., human–chimp split 5–7 Ma) and recent (e.g., expansion of AMHs ∼100 Ka) events in human evolution (Reed et al. 2004, 2007). The human louse (Pediculus humanus) is a single species that occurs as two ecological types (head and clothing lice) exhibiting morphological, behavioral, and ecological differences (Reed et al. 2004; Light et al. 2008). The loss of human body hair restricted P. humanus to the head, and the subsequent divergence of the two louse types is unlikely to have begun prior to the availability of the new clothing niche (Burgess 1995; Kittler et al. 2003). Thus, determining when head and clothing lice began to diverge provides a date by which clothing must have been in regular use by humans.
In this study, we analyzed a multilocus data set of clothing and head louse DNA sequences from three nuclear genes (18S ribosomal RNA [rRNA], nuclear elongation factor 1-α [EF-1α], and RNA polymerase II [RPII]) and the mitochondrial gene cytochrome c oxidase subunit I (COI). After estimating substitution rates for each locus based on the codivergence of human and chimpanzee lice with their primate hosts (Light and Reed 2009), we employed a multilocus Bayesian isolation-with-migration (IM) coalescent method (Nielsen and Wakeley 2001; Hey and Nielsen 2004; Hey 2005) to jointly estimate the divergence time (t), effective sizes (NeHEAD, NeCLOTH, NeANCESTRAL), and effective migration rates (mHEADtoCLOTH, mCLOTHtoHEAD) of head and clothing louse populations from our combined multilocus data set. This model is ideal for estimating the divergence of head and clothing lice because it assumes an ancestral population (i.e., head lice) diverged at some time t into two daughter populations, which then experience independent rates of exponential growth with migration between populations.
Our results indicate a small effective population size for ancestral P. humanus (NeANCESTRAL, median = 0.934 × 106; 95% highest probability density [HPD] = 0.011–3.161 × 106; table 1), which is consistent with either a bottleneck created by the loss of body hair or a bottleneck imposed in the ancestral human host population. The estimates of effective population size for both head and clothing lice were larger (NeHEAD = 7.008 × 106; 95% HPD = 1.991–16.998 × 106 and NeCLOTH = 4.038 × 106; 95% HPD = 0.461–10.676 × 106) and consistent with postbottleneck expansions. Estimates of s (∼0.24) indicate that a large fraction of the ancestral head louse population initially became clothing lice, perhaps rapidly exploiting new niche space.
Table 1.
Mean, Median, Mode, and 95% HPD for Parameters Estimated in IM.
| 95% HPD | Mean | Median | Mode | |
| NeHEADa | 1.991–16.998 × 106 | 8.033 × 106 | 7.008 × 106 | 4.893 × 106 |
| NeCLOTHb | 0.461–10.676 × 106 | 4.742 × 106 | 4.038 × 106 | 3.274 × 106 |
| NeANCESTRALc | 0.011–3.161 × 106 | 1.317 × 106 | 0.934 × 106 | 0.011 × 106 |
| tDivergenced | 29–691 Ka | 229 Ka | 170 Ka | 83 Ka |
| mHEADtoCLOTHe | 0.005–1.755 | 0.492 | 0.235 | 0.005 |
| mCLOTHtoHEADf | 0.495–3.785 | 1.836 | 1.615 | 1.335 |
| sg | 0.001–0.879 | 0.325 | 0.242 | 0.001 |
Effective population size of modern head lice (NeHEAD).
Effective population size of modern clothing lice (NeCLOTH).
Effective population size of ancestral head lice population (NeANCESTRAL).
Time of clothing and head lice divergence (tDivergence).
Migration from head to clothing lice populations (mHEADtoCLOTH).
Migration from clothing to head lice populations (mCLOTHtoHEAD).
Proportion of the ancestral head lice population that contributes to the modern head lice population (s).
Parameter estimates showed elevated rates of continuous migration in the direction of clothing to head lice (mCLOTHtoHEAD = 1.615, 95% HPD = 0.495–3.785) but notably less migration in the opposite direction (mHEADtoCLOTH = 0.235, 95% HPD = 0.005–1.755). These estimates contradict previous studies that found no migration between head and clothing lice based on microsatellite data (Leo et al. 2005). In addition, the direction of gene flow is unexpected given that head lice can readily colonize the clothing niche (Alpatov and Nastjukova 1955; Levene and Dobzhansky 1959; Li et al. 2010).
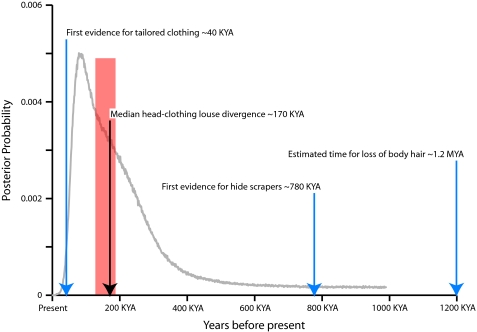
The posterior probability distribution for the head and clothing louse divergence time is characterized by a mode (i.e., the single estimate with the highest posterior probability) of 83 Ka and a median value of 170 Ka (95% HPD = 29–691 Ka; fig. 1). These dates are largely consistent with those estimated by Kittler et al. (2003, 2004), who analyzed a single mitochondrial gene using a distance-based method. However, the Bayesian multilocus IM method accounts for uncertainty in the model parameters as well as stochastic variation between loci, which provides a more robust and accurate parameter estimate (Edwards and Beerli 2000).
FIG. 1.
Divergence time of human head and clothing lice. The posterior distribution for the divergence of head and clothing lice (gray curve) places the median estimate for the origin of clothing lice at 170 Ka (black arrow). This estimate is substantially older than a previous estimate of 30–112 Ka from molecular data (Kittler et al. 2003) and is consistent with the relative antiquity of the first archaeological evidence for hide scrapers ∼780 Ka (Carbonell et al. 1999), the loss of human body hair by ∼1.2 Ma (Rogers et al. 2004), and the first evidence for tailored clothing ∼40 Ka (Delson et al. 2000), which are indicated by blue arrows. Furthermore, the median estimate lies within the ice age coincident with Marine Isotope Stage 6 ∼130–190 Ka (EPICA Community Members 2004), indicated by the red-shaded region.
During the latter part of the Middle Pleistocene (e.g., 83–170 Ka), archaic hominins lived in cold climates in Eurasia, whereas H. sapiens was still in Africa. Whether these archaic hominins had clothing is unknown because they left no clothing louse descendents that we can sample among living humans. All modern clothing lice are confined to a single mitochondrial clade that shows a contemporaneous population expansion with modern humans ∼100 Ka (Reed et al. 2004, 2007). Therefore, we are left to conclude that regular clothing use must have occurred in H. sapiens at least by 83 Ka and possibly as early as 170 Ka. Whether archaic hominins used clothing cannot be assessed from these lice and may require the collection of lice from archaic human remains, which is unlikely.
Even though archaic hominins dispersed into cold climates hundreds of thousands of years before AMH, modern humans are often credited with outcompeting contemporary archaic species due to increased fitness stemming from a suite of “modern” behaviors and technologies that include the use of clothing (Gilligan 2010). Interestingly, we estimated that clothing may have been in use as early as 170 Ka, which corresponds to the rapid onset of an ice age, Marine Isotope Stage 6 (∼190–130 Ka; EPICA Community Members 2004), that would have caused cold stress for populations living outside the tropics and could have led to the initial use of clothing by modern humans. Our estimate for the origin of clothing use suggests that one of the technologies necessary for successful dispersal into colder climates was already available to AMH prior to their emergence out of Africa.
Methods
All available DNA sequences for COI (108 head and 58 clothing lice), 18S rRNA (10 head and 12 clothing lice), EF-1α (25 head and 9 clothing lice), and RPII (25 head and 10 clothing lice) for P. humanus and the outgroup P. schaeffi (chimpanzee louse) were downloaded from GenBank (available as supplementary table S1, Supplementary Material online). All sequences were aligned by hand using Se-Al v2.01a11 (http://tree.bio.ed.ac.uk/software/seal/), with the 18S rDNA aligned to secondary structure (Gillespie 2004; Gillespie et al. 2005).
Substitution rates (table 2) for the four genes were estimated in BEAST v.1.5.3 (Drummond and Rambaut 2007). Rates were calibrated by placing an exponential prior distribution (lower bound = 5 Ma, mean = 5.5 Ma) on the divergence of P. humanus (human) and P. schaeffi (chimpanzee) lice that reflects conservatively recent estimates for the divergence of their hosts (Kumar and Hedges 1998). Each gene was analyzed using a range of substitution and clock models, as well as tree priors, with posterior estimates made from the model that best fit the data as determined by marginal likelihoods estimated in the program Tracer v1.5 (http://tree.bio.ed.ac.uk/software/tracer). Markov chains were run for at least 100 million generations, sampled every 10,000 generations, and the first 10% of samples were discarded as burn-in. All runs were duplicated to ensure convergence.
Table 2.
Models of Substitution, Likelihood Scores, and Mean Substitution Rates Per Gene Calculated in BEAST.
| Gene | Substitution Model | Clock Model | Tree Prior | Marginal Likelihood | Mean Substitution Rate (95% HPD) |
| COI | GTR + CPa | UCEDb | BSPc | −1318.728 | 6.28 × 10−8 (3.36–9.64 × 10−8) |
| 18s rDNA | HKY + Gd + Ie | UCED | BSP | −3087.519 | 7.19 × 10−9 (3.64–11.01 × 10−9) |
| EF-1α | GTR + CP + G | UCED | BSP | −853.401 | 7.89 × 10−9 (3.90–12.622 × 10−9) |
| RPII | GTR + CP + G | UCED | Constantf | −1144.325 | 1.26 × 10−8 (0.46–2.34 × 10−8) |
Between site rate variation partitioned by codon position (CP).
Uncorrelated exponentially distributed relaxed clock (UCED, Drummond et al. 2006).
Bayesian skyline plot tree prior (BSP, Drummond et al. 2005).
Gamma distribution of between site rate variation (G).
Invariate proportion of sites (I).
Constant population size tree prior.
Multilocus Bayesian IM coalescent analysis was performed on the P. humanus sequences using the program IM (Hey 2005). All analyses used the HKY substitution model, whereas priors on model parameters were broad uniform distributions conservatively estimated from preliminary runs. Markov chains were run for >200 million generations and replicated 8 times to ensure convergence. A louse generation time of 21 days (18 generations per year) and the substitution rates in table 2 were used to convert parameter estimates from mutational to demographic units.
Supplementary Material
Supplementary table S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for valuable comments. We acknowledge the University of Florida High-Performance Computing Center for providing computational support. This work was supported by grants to D.L.R. from the University of Florida Research Opportunity SEED Fund and the National Science Foundation (DEB 0555024, DEB 0717165, and DEB 0845392).
References
- Alpatov WW, Nastjukova OK. Transformation of the head form of Pediculus humanus L. into the body form under the influence of changed living conditions. Bull Soc Nat Moscow. 1955;60:79–92. [Google Scholar]
- Burgess IF. Human lice and their management. Advances in Parasitology. 1995;36:271–342. doi: 10.1016/s0065-308x(08)60493-5. [DOI] [PubMed] [Google Scholar]
- Carbonell E, de Castro JMB, Pares JM, et al. (30 co-authors) The first hominin of Europe. Nature. 2008;452:U465–U467. doi: 10.1038/nature06815. [DOI] [PubMed] [Google Scholar]
- Carbonell E, Garcia-Anton MD, Mallol C, Mosquera M, Olle A, Rodriguez XP, Sahnouni M, Sala R, Verges JM. The TD6 level lithic industry from Gran Dolina, Atapuerca (Burgos, Spain): production and use. J Hum Evol. 1999;37:653–693. doi: 10.1006/jhev.1999.0336. [DOI] [PubMed] [Google Scholar]
- Delson E, Tattersall I, Van Couvering J, Brooks A. Encyclopedia of human evolution and prehistory. New York: Garland Press; 2000. [Google Scholar]
- Drummond AJ, Ho SYW, Phillips MJ, Rambaut A. Relaxed phylogenetics and dating with confidence. PloS Biol. 2006;4:699–710. doi: 10.1371/journal.pbio.0040088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Rambaut A, Shapiro B, Pybus OG. Bayesian coalescent inference of past population dynamics from molecular sequences. Mol Biol Evol. 2005;22:1185–1192. doi: 10.1093/molbev/msi103. [DOI] [PubMed] [Google Scholar]
- Edwards SV, Beerli P. Perspective: gene divergence, population divergence, and the variance in coalescence time in phylogeographic studies. Evolution. 2000;54:1839–1854. doi: 10.1111/j.0014-3820.2000.tb01231.x. [DOI] [PubMed] [Google Scholar]
- EPICA Community Members. Eight glacial cycles from an Antarctic ice core. Nature. 2004;429:623–628. doi: 10.1038/nature02599. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ. Characterizing regions of ambiguous alignment caused by the expansion and contraction of hairpin-stem loops in ribosomal RNA molecules. Mol Phylogenet Evol. 2004;33:936–943. doi: 10.1016/j.ympev.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Gillespie JJ, McKenna CH, Yoder MJ, Gutell RR, Johnston JS, Kathirithamby J, Cognato AI. Assessing the odd secondary structural properties of nuclear small subunit ribosomal RNA sequences (18S) of the twisted-wing parasites (Insecta: Strepsiptera) Insect Mol Biol. 2005;14:625–643. doi: 10.1111/j.1365-2583.2005.00591.x. [DOI] [PubMed] [Google Scholar]
- Gilligan I. The prehistoric development of clothing: archeological implications of a thermal model. J Archeol Method Theory. 2010;17:15–80. [Google Scholar]
- Hey J. On the number of New World founders: a population genetic portrait of the peopling of the Americas. PloS Biol. 2005;3:965–975. doi: 10.1371/journal.pbio.0030193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hey J, Nielsen R. Multilocus methods for estimating population sizes, migration rates and divergence time, with applications to the divergence of Drosophila pseudoobscura and D. persimilis. Genetics. 2004;167:747–760. doi: 10.1534/genetics.103.024182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittler R, Kayser M, Stoneking M. Molecular evolution of Pediculus humanus and the origin of clothing. Curr Biol. 2003;13:1414–1417. doi: 10.1016/s0960-9822(03)00507-4. [DOI] [PubMed] [Google Scholar]
- Kittler R, Kayser M, Stoneking M. Molecular evolution of Pediculus humanus and the origin of clothing. Curr Biol. 2004;14:2309. doi: 10.1016/s0960-9822(03)00507-4. [DOI] [PubMed] [Google Scholar]
- Krause J, Fu QM, Good JM, Viola B, Shunkov MV, Derevianko AP, Paabo S. The complete mitochondrial DNA genome of an unknown hominin from southern Siberia. Nature. 2010;464:894–897. doi: 10.1038/nature08976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Hedges SB. A molecular timescale for vertebrate evolution. Nature. 1998;392:917–920. doi: 10.1038/31927. [DOI] [PubMed] [Google Scholar]
- Leo NP, Hughes JM, Yang X, Poudel SKS, Brogdon WG, Barker SC. The head and body lice of humans are genetically distinct (Insecta: Phthiraptera, Pediculidae): evidence from double infestations. Heredity. 2005;95:34–40. doi: 10.1038/sj.hdy.6800663. [DOI] [PubMed] [Google Scholar]
- Levene H, Dobzhansky T. Possible genetic difference between the head louse and the body louse. Am Nat. 1959;873:347–353. [Google Scholar]
- Li WJ, Ortiz G, Fournier PE, Gimenez G, Reed DL, Pittendrigh B, Raoult D. Genotyping of human lice suggests multiple emergences of body lice from local head louse populations. PloS Negl Trop Dis. 2010;4(3):e641. doi: 10.1371/journal.pntd.0000641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light JE, Reed DL. Multigene analysis of phylogenetic relationships and divergence times of primate sucking lice (Phthiraptera: Anoplura) Mol Phylogenet Evol. 2009;50:376–390. doi: 10.1016/j.ympev.2008.10.023. [DOI] [PubMed] [Google Scholar]
- Light JE, Toups MA, Reed DL. What's in a name: the taxonomic status of human head and body lice. Mol Phylogenet Evol. 2008;47:1203–1216. doi: 10.1016/j.ympev.2008.03.014. [DOI] [PubMed] [Google Scholar]
- Nielsen R, Wakeley J. Distinguishing migration from isolation: a Markov chain Monte Carlo approach. Genetics. 2001;158:885–896. doi: 10.1093/genetics/158.2.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DL, Light JE, Allen JM, Kirchman JJ. Pair of lice lost or parasites regained: the evolutionary history of anthropoid primate lice. BMC Biol. 2007;5:7. doi: 10.1186/1741-7007-5-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed DL, Smith VS, Hammond SL, Rogers AR, Clayton DH. Genetic analysis of lice supports direct contact between modern and archaic humans. PloS Biol. 2004;2:e340. doi: 10.1371/journal.pbio.0020340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers AR, Iltis D, Wooding S. Genetic variation at the MC1R locus and the time since loss of human body hair. Curr Anthropol. 2004;45:105–108. [Google Scholar]
- Stringer C. Modern human origins: progress and prospects. Philos Trans R Soc B Biol Sci. 2002;357:563–579. doi: 10.1098/rstb.2001.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteman NK, Parker PG. Using parasites to infer host population history: a new rationale for parasite conservation. Anim Conserv. 2005;8:175–181. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.