Abstract
Codon usage can provide insights into the nature of the genes in a genome. Genes that are “native” to a genome (have not been recently acquired by horizontal transfer) range in codon usage from a low-bias “typical” usage to a more biased “high-expression” usage characteristic of genes encoding abundant proteins. Genes that differ from these native codon usages are candidates for foreign genes that have been recently acquired by horizontal gene transfer. In this study, we present a method for characterizing the codon usages of native genes—both typical and highly expressed—within a genome. Each gene is evaluated relative to a half line (or axis) in a 59D space of codon usage. The axis begins at the modal codon usage, the usage that matches the largest number of genes in the genome, and it passes through a point representing the codon usage of a set of genes with expression-related bias. A gene whose codon usage matches (does not significantly differ from) a point on this axis is a candidate native gene, and the location of its projection onto the axis provides a general estimate of its expression level. A gene that differs significantly from all points on the axis is a candidate foreign gene. This automated approach offers significant improvements over existing methods. We illustrate this by analyzing the genomes of Pseudomonas aeruginosa PAO1 and Bacillus anthracis A0248, which can be difficult to analyze with commonly used methods due to their biased base compositions. Finally, we use this approach to measure the proportion of candidate foreign genes in 923 bacterial and archaeal genomes. The organisms with the most homogeneous genomes (containing the fewest candidate foreign genes) are mostly endosymbionts and parasites, though with exceptions that include Pelagibacter ubique and Beutenbergia cavernae. The organisms with the most heterogeneous genomes (containing the most candidate foreign genes) include members of the genera Bacteroides, Corynebacterium, Desulfotalea, Neisseria, Xylella, and Thermobaculum.
Keywords: horizontal gene transfer, foreign genes, codon adaptation index, factorial correspondence analysis
Introduction
Most genomes are heterogeneous in codon usage due to the presence of highly expressed genes (encoding abundant protein products) and/or foreign genes (acquired by the genome via horizontal gene transfer) (e.g., Grantham et al. 1981; Médigue et al. 1991). The native genes of a genome span a continuum of codon usages ranging from that of weakly biased “typical” genes (e.g., Grantham, Gautier, and Gouy 1980; Grantham, Gautier, Gouy, Mercier, et al. 1980) to that of highly biased “high-expression” genes (e.g., Post et al. 1979; Grantham et al. 1981; Ikemura 1981a, 1981b; Médigue et al. 1991). This expression-related bias is a characteristic of most genomes, and many methods of codon usage analysis have been devised to characterize genes based on their adherence to this trend (e.g., Bennetzen and Hall 1982; Gribskov et al. 1984; McLachlan et al. 1984; Sharp and Li 1987).
The codon adaptation index (CAI) (Sharp and Li 1987) is the most commonly used method for characterizing the codon usages of genes in a genome. In the CAI, the codon usage of each gene is compared with an “optimal” codon usage, which is inferred from a hand-selected high-expression gene set. The more closely the codon usage of a gene matches this optimal codon usage profile, the higher its CAI value. The appeal of the method is that it is straightforward, and it provides a ranking of genes from those that have the most bias (look most highly expressed) to those that have the least. However, because this characterization is 1D, the method has limited ability to distinguish native genes with low CAI values from foreign genes.
Karlin and Mrázek (2000) devised a method that circumvents this problem. They define the typical codon usage of a genome as the average codon usage of its genes. They also define three categories of high-expression codon usage based on the average usages for each of three sets of genes that include ribosomal proteins, transcriptional and translation processing proteins, and chaperones. If a gene is sufficiently similar to the genome-wide average and sufficiently different from the high-expression usages, then it is called a typical gene. If it is sufficiently similar to two of the three categories of high-expression genes and sufficiently different from the average of the genome, then it is called a high-expression gene. All other genes are considered to be foreign. Despite its clear advantage in distinguishing typical and foreign genes, the Karlin and Mrázek method is far less used than the CAI for at least two reasons. First, it provides only a binary estimate of the expression level: low or high. Second, for each genome, the user must identify the genes belonging to each of the three high-expression gene sets prior to the analysis.
In a previous study, we defined the modal codon usage of a genome as the codon usage that matches (is not significantly different from) the largest number of genes (Davis and Olsen 2010). In this study, we describe a method for integrating this with the codon usages of more highly expressed genes. The method defines “native” codon usage as a continuum of potential codon usages that starts at the modal codon usage of the genome and extends through (and beyond) the modal codon usage of a set of (candidate) highly expressed genes. Unlike current methods, the identification of candidate highly expressed genes in a new genome is fully automated and does not rely on genome annotations. The subsequent characterization of each individual gene in the genome is based on whether it is statistically similar to any native codon usage and, if so, which level of expression best matches its codon usage. We describe the method using the well-characterized genome of Escherichia coli K-12 and then apply the analysis to Pseudomonas aeruginosa PAO1 and Bacillus anthracis A0248 to demonstrate its effectiveness in genomes with biased base compositions.
Materials and Methods
Gene and Protein Sequences
Unless otherwise indicated, all coding sequences, protein sequences, and protein annotations are taken from the National Center for Biotechnology Information (NCBI) bacterial genome ftp site (ftp://ftp.ncbi.nih.gov/genomes/Bacteria/).
Modal Codon Usage
The following is a brief outline of the concepts and methods; details are provided in Davis and Olsen (2010). The modal codon usage of a set of genes is defined as the expected codon usage that matches the largest number of genes. To minimize the influence of amino acid composition, codon usage frequencies are normalized for each amino acid (a form of relative codon usage). A gene is said to match a codon usage if its observed codon usage is not significantly different (P ≥ 0.1) in a chi-square test (41 degrees of freedom, unless the gene lacks some amino acids). To estimate the modal codon usage of a set of genes, we use a continuous approximation of the number of genes matching a set of expected codon usage frequencies and a simplex search in the 59D space codon usage frequencies (61 sense codons minus the codon frequencies for Met and Trp, which are always 1). The use of a chi-square test comes with two potential caveats. First, short genes, due to their lack of codons, are noisy and have a tendency to match a broad range of expected codon usages. This can cause short genes to match the native codon usage, even if they are of foreign origin. There are also concerns about the applicability of a chi-square test when the expected counts are low, though our previous work suggests that this is a minimal problem in the range of P values used here (Davis and Olsen 2010). Although our software to find native codon usage (below) and to identify genes that match it does not restrict the length of genes analyzed, it is a simple matter to prescreen the input sequences. Second, long genes contain so many codons that they must very precisely match an expected codon usage. We provide a mechanism by which a user can limit the effective length of genes in calculating P values from chi-square values (see below). Unless otherwise indicated, we include all genes regardless of length, and we do not limit the observed codon usages in the chi-square tests in our analyses.
Native Codon Usage
We use the term native codon usage to describe the typical and high-expression codon usages of a genome. These genes range from low to high levels of expression-linked bias. In our 59-dimensional space of codon usage frequencies, we represent native codon usage as a half line beginning at the modal codon usage of the genome (f0) and extending through a point representing a high-expression codon usage (f1). In a parametric representation of this line, which is related to that of Kloster and Tang (2008), we specify the expected codon usage frequencies f(x) as a function of “expression level” x (−∞ ≤ x ≤ ∞). Letting fi(x) be the expected frequency of codon i at x, we define
| (1) |
| (2) |
| (3) |
where si is the set of codons for the amino acid encoded by i (i.e., the set of synonymous codons that includes i), f0i is the frequency of codon i in the modal codon usage, and f1i is the frequency of codon i in a high-expression codon usage. The vector w(x) is the unnormalized preference for each codon as a function of expression level. Equation (1) normalizes these values for the codons of each amino acid, so that 0 ≤ fi(x) ≤ 1, and the codon frequencies of each amino acid sum to 1. Equation 2 causes the relative preference for codon i to vary (up or down) exponentially with expression level and sets the preference for codon i at x = 0 to f0i (so f(0) = f0). Kloster and Tang (2008) point out that equations 1 and 2 constitute a partition function in which each codon responds to a pressure (in our case, expression level) with its own characteristic sensitivity ki. Finally, equation 3 defines this response of each codon in terms of the ratio of its frequency in the highly expressed genes to its frequency in typical genes, so that f(1) = f1. There is no absolute scale to x; the scale depends on the set of genes used to define f1.
To evaluate whether a gene matches the native codon usage, we use a combination of grid search and divide and conquer strategies to find the value of x and corresponding expected codon frequencies f(x) from which the gene differs least significantly. Although the mathematical domain of x includes negative values, we define native codon usages as the values of f(x) for x ≥ 0. Thus, if the optimal value of x is negative, the value is reset to 0, and the match to the codon usage frequencies is reevaluated. The genes that do not differ significantly (P ≥ 0.1 or other P value, when appropriate) from their best matching point on the line are classified as matching native codon usage.
Projecting Highly Expressed Proteins to a New Genome
For a given reference genome (or set of diverse reference genomes), all the “high-expression” protein sequences are included in a perl module (our default organisms for this are E. coli K-12 substr. MG1655 and Methanococcus maripaludis S2). The module also includes a list of the identifiers of proteins considered to be highly expressed (see Results). Given the coding sequences of a new genome, we seek the orthologs of the reference highly expressed proteins in two steps. First, TBlastN is used to find the best matching gene of each highly expressed protein in the new genome (Altschul et al. 1997). Then, each of these best matches is used as a BlastX query against the full protein set of the reference genome (Altschul et al. 1997). Those cases in which the best match is the same as the original query are bidirectional best hits and likely orthologs. By default, the following constraints are applied to the basic local alignment search tool matches: E value ≤10−5, coverage of query and subject sequences ≥70%, fraction sequence identity ≥20%, and fraction positive-scoring aligned residues ≥30%. Because the first search includes only the highly expressed proteins in the reference genome(s), all the resulting bidirectional best hits are candidate highly expressed genes in the new genome. Although bidirectional best hits are an imperfect tool for identifying orthologous genes, the combination of this criterion with E value, sequence coverage, and sequence similarity tests results in a very clean set of candidate genes (certainly more precise than that defined by annotations). When the analysis is carried out on DNA sequences that do not represent a genome or are a very small fraction of a genome, there will be few if any bidirectional best hits, and the analysis terminates without estimating high-expression codon usage (below).
Inferring High-Expression Codon Usage from Candidate Highly Expressed Genes
Because the goal is to improve the discrimination between the modal usage of the genome and the high-expression codon usage, before calculating the modal codon usage of the candidate highly expressed genes, we identify and remove genes whose codon usage matches (is not significantly different from, P ≥ 0.1) the modal codon usage of the genome. For the same reason that we use the modal codon usage (rather than the average codon usage) to characterize the typical genes in a genome (f0 above), we use the modal codon usage of the candidate highly expressed genes to minimize sensitivity to the outlying genes in estimating high-expression codon usage (f1 above). This approach ensures that even if the candidate high-expression gene set is imperfect, there will be minimal influence on the outcome. With these values, we can classify all genes based on their similarity to native codon usage (i.e., native gene vs. foreign gene) and their x position (expression level) along the axis from typical to high expression.
Iterative Refinement of Candidate Highly Expressed Genes
Because the estimation of high-expression codon usage is based on a (potentially small) fraction of the genes that are highly expressed in a given genome, we are interested in the behavior of iterative approaches to refining the estimate. In outline, we find the modal codon usage of the candidate high-expression genes (above) at a relaxed P value (P ≥ 0.05). The idea is that if the original estimate of high-expression genes is slightly in error, the new estimate may draw in genes that had been previously missed. This new set of genes is then filtered for those whose x value is ≥0.5. That is, we screen for genes that are closer to the current high-expression estimate than to the modal codon usage of the genome. Finally, the list of candidate genes is limited to 10% of the total number of genes in the genome being evaluated, sorted from highest to lowest value of x. These last two filters are both based on the value of x (the first on the absolute value, and the latter on the relative rank), so in any given case, only one of them can be limiting. The resulting set becomes the new candidate highly expressed genes for the next estimate of high-expression codon usage.
This approach works well for most genomes, but if a genome has little or no expression-related codon bias, then the candidate highly expressed genes will also match to the overall genomic (modal) codon usage. Given the candidate highly expressed genes from a genome, three situations are distinguished and handled differently. First, if there are <20 candidate highly expressed genes, the analysis terminates without an estimate of high-expression codon usage. Generally, this only occurs when the data being analyzed are not a complete archaeal or bacterial genome. Second, when the candidate highly expressed genes are compared with the modal codon usage, if ≥20% of them differ significantly from the genome modal usage and the number of these differing genes is ≥20, then the high-expression codon usage estimate is the modal usage of the candidates that differ from the genome mode. Finally, if <20% of the candidates (or <20 genes) differ significantly from the mode, it is concluded that there is little or no high-expression bias, so the modal codon usage of all candidates is reported as the high-expression usage, and the analysis terminates.
If the number of original high-expression candidate genes declines between iterations by more than 20%, the iteration is cancelled, and the original high-expression mode is used to define the axis. Here, the rationale is that the high-expression genes were either too weakly biased or their numbers were too small to prevent the axis from drifting away from the genes with expression-related codon usage. These iterations end after a specified number of cycles (the default is 2).
Factorial Correspondence Analysis
Factorial correspondence analysis (FCA) of the codon usages of all the genes in a genome was analyzed using the CODONW program (Peden 1999). In all illustrations, the projection shown is that defined by the first two axes.
Software Options
The software that was developed in this study has several user-defined options. For example, it is possible to seed the search for high-expression genes by using a user-defined high-expression gene set. It is also possible to dictate the chi-square P-value cutoff for genes matching (or not matching) the axis. As noted above, it is also possible to limit the precision with which long genes must match an expected codon usage by modifying the calculation of the chi-square P value for genes longer than a user-defined threshold (see also Davis and Olsen 2010). Additional options are described in the readme files of the distribution (supplementary material, Supplementary Material online).
Software Availability
Our method for assessing the native codon usage of a genome is implemented in the program native_codon_usage. This and other programs used to perform this work are written in perl and C. Several of the analysis steps utilize multiple processors, if available. They have been tested on PPC and i386 Macintosh computers, under OS X 10.4 and 10.5, but should work in any Unix environment. They require that the formatdb and blastall (Altschul et al. 1997) programs be installed. The versions of our programs that were current at the time of submission are deposited as supplemental information on the journal World Wide Web site, and current versions are available through links at http://www.life.illinois.edu/gary/programs.html (Supplementary Material online).
Results and Discussion
Our Concept of Native Codon Usage and the Algorithm for Finding It
We have previously described our concept of modal codon usage as the codon usage that represents that largest number of genes in a genome (Davis and Olsen 2010). In many genomes, genes for abundant proteins (high-expression genes) have a distinct codon usage. In practice, there is a continuum of genes with codon usages ranging from typical to the most highly expressed. We model this by a half line (or axis) in a 59-dimensional codon usage space (Materials and Methods). The line begins at the modal codon usage, and it extends through the codon usage of a set of highly expressed genes. Due to the mathematical properties of codon usage, we use a parametric representation of the line, similar to that of Kloster and Tang (2008) (Materials and Methods), so the line is not straight. Thus, we define a function f(x) whose value is the expected codon usage at position x along the axis. The function is defined so that x = 0 at the beginning of the half line [thus, f(0) is the modal codon usage] and x = 1 at the codon usage of the set of highly expressed genes.
Given this framework, for each gene, we ask which position along the axis (at which value of x, constrained to x ≥ 0) does the expected codon usage best match (least significantly differ from) the codon usage of the gene. If the gene is not significantly different from the codon usage at that x value, we say it matches the native codon usage, and the value of x provides a measure of how much it looks like a highly expressed gene. If the gene is significantly different from all codon usages along the axis, then it is a candidate for a foreign gene.
In general, the procedure we use for inferring the native codon usages of a genome can be summarized as follows: 1) find the modal codon usage of the genes in the genome; 2) identify a set of candidate highly expressed genes by finding the bidirectional best hits to highly expressed proteins from one or more reference genomes; 3) remove from the candidate high-expression genes, those that are too similar to the genome mode, and find the modal usage of the remaining genes; 4) terminate if a stopping condition has been reached; and 5) otherwise, produce a new set of candidate highly expressed genes and go back to step 3. Calculation of the modal codon usage has been described previously (Davis and Olsen 2010). The identification and refinement of candidate highly expressed genes are covered in more detail in the following sections.
Defining a Set of Highly Expressed Genes in E. coli K-12
Our overall strategy for analyzing a new genome includes the projection (by bidirectional best hits) of highly expressed genes from a reference genome to the new genome. Because it is so well characterized, we started with E. coli as the default reference. The ribosomal protein genes provide good examples of high-expression codon usage in E. coli and are among the genes most commonly used for representing high expression in current methodologies (e.g., Sharp and Li 1987; Karlin and Mrázek 2000). However, ribosomal proteins tend to be small (hence, they provide a noisy sample of codon usage) and are limited in number. In table 1, we consider several alternative gene sets for representing the codon usage of highly expressed genes. In each case, we use the gene set to provide an estimate of a high-expression codon usage and use this along with the genomic modal codon usage to define a native codon usage axis. In each case, we report the total number of genes matching the corresponding axis (limited to x ≥ 0). The number of matching genes ranges from 2,402 to 2,565. Encouragingly, 2,306 of the matching genes are common to all five sets (table 1, column 3), though most of these also match the genome mode. To focus on the highly expressed genes, we compare the 415 genes (10% of the genes in the genome) whose projections on the axis have the highest value of x. Of these, 225 are held in common. These data indicate that different starting codon usages provide similar, but not identical, predictions of high-expression genes.
Table 1.
Iterative Development of a High-Expression Gene Set in E. coli K-12.
| Number of genes | Matching genesa | Iterations |
|||
| 1 | 2 | 3 | |||
| Initial high-expression codon usage estimate | |||||
| Ribosomal protein genes (mode) | 55 | 2,402 | 2,533 | 2,571 | 2,588 |
| Ribosomal protein genes (average) | 55 | 2,476 | 2,552 | 2,582 | 2,598 |
| CAI genes (average)b | 27 | 2,512 | 2,563 | 2,586 | 2,600 |
| aa-transfer RNA synthetase genes (average) | 22 | 2,565 | 2,593 | 2,600 | 2,597 |
| rpoB gene | 1 | 2,436 | 2,597 | 2,600 | 2,598 |
| Comparison of matching gene setsc | |||||
| Genes in any set (union) | 2,655 | 2,658 | 2,639 | 2,622 | |
| Genes in at least three of five sets | 2,479 | 2,562 | 2,584 | 2,599 | |
| Genes in all sets (intersection) | 2,306 | 2,477 | 2,533 | 2,563 | |
| Comparison of 415 matching genes with highest x valued | |||||
| Genes in any set (union) | 658 | 499 | 469 | 444 | |
| Genes in at least three of five sets | 394 | 416 | 414 | 417 | |
| Genes in all sets (intersection) | 225 | 327 | 362 | 381 | |
Genes matching the axis that intersects the mode of the genome and the original high-expression codon usage from the first column. Genes that do not match the mode and have negative x values are excluded.
Genes used to define optimal codons in CAI analysis (Sharp and Li 1986).
Generated by combining the native genes in each column.
Top 10% of the genes in the genome (415 genes) with highest x values for each column.
We were interested in exploring strategies that might converge upon a common estimate, in spite of the diverse starting points. The basic strategy was an iterative refinement procedure. The idea was to generate a new set of candidate high-expression genes that are close to the current axis and then take the modal codon usage of these. We first selected the set of genes that match the axis at a reduced stringency, P ≥ 0.05 (rather than our usual P ≥ 0.1). This allows us to gather additional highly expressed genes that we may have missed in our previous estimate. We then reduced this to the 415 genes (10% of the genome) with the highest x values. We use the modal codon usage of this gene set as our new representation of highly expressed genes, thereby reorienting our native codon usage axis. In the case of the E. coli genome, this increases the number of matching genes and makes the matching gene sets more uniform between alternative starting points. After three iterations, the native gene sets are nearly identical—between 2,588 and 2,600 genes match the native set in each case, and 2,563 of these genes are held in common in all five sets (table 1, last column). After the third iteration, 381 (of 415) of the highest expression genes are shared among all five sets. These results suggest that in an organism with a large amount of expression-linked codon bias, our approach to refining the estimate of high-expression codon bias is robust.
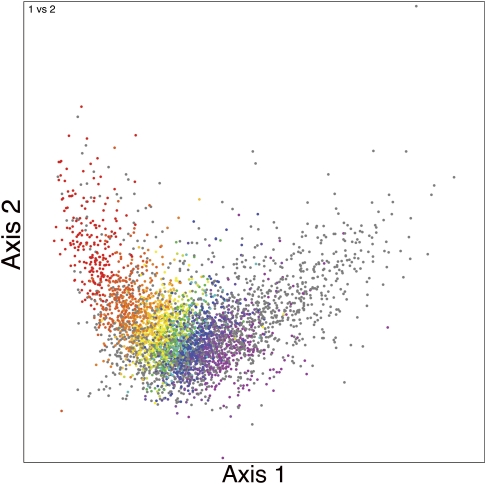
This analysis indicates that that ∼2,600 E. coli K-12 genes (63% of the genes in the genome) have a native codon usage, and ∼1,549 genes (37% of the genes in the genome) have a foreign codon usage. These data closely correspond with recent large-scale comparative analyses of E. coli genomes that estimate the core E. coli genome size to be approximately 2,200–2,900 genes (Fukiya et al. 2004; Chen et al. 2006; Rasko et al. 2008). The net result of this analysis is shown in figure 1. The figure displays all the genes in the E. coli K-12 genome separated by their positions on the first two axes of a FCA plot of relative codon usage (Materials and Methods). Each gene that matches the native codon usage axis is assigned a color of the visible spectrum according to the position at which it projects on the native codon usage axis, from violet (x = 0) to red (x > 1). Genes that do not match the native codon usage are colored light gray. The figure clearly distinguishes the major features previously noted in similar analyses of E. coli codon usage (Médigue et al. 1991; Badger 1999): a “rabbit’s head” shape with the head representing typical native genes (green through violet), an ear composed primarily of highly expressed genes (orange and red), and an ear composed of foreign genes (gray). For each gene, the axis position and P value of its match to the axis are reported in supplementary table S1 (Supplementary Material online).
FIG. 1.
FCA plot of E. coli K-12. Each plot point shows the location of a gene in the first two axes of the analysis. Genes are colored according to their axis position (x value) based upon the colors of the visible spectrum, with red genes indicating the highest expression-related codon usage bias and violet genes indicating the least. Genes that differ significantly from all points on the native codon usage axis (likely to be foreign) are colored gray and are drawn behind the colored genes. Each gene’s position along the first axis of the plot also corresponds with its G + C content (from left to right: high G + C to low G + C) (see also Médigue et al. 1991).
Characterizing Native Codon Usage in Other Organisms
The above results indicate that we have a robust and relatively impartial method for selecting a native gene set in E. coli and assessing the expected expression level. We want to expand this analysis to study other genomes. The first step, using modal codon usage to represent the typical genes, is straightforward. The second step, picking candidate genes to represent high-expression codon usage, is more subtle. Previous studies have done so on the basis of annotation (e.g., Sharp and Li 1987; Karlin and Mrázek 2000). This relies upon having a list of protein names (functions) that are likely to be highly expressed, having accurate annotations for the genes in the new genome, finding the annotations in the new genome that correspond to those in the highly expressed list, and only then analyzing these genes for codon usage. We bypass all the annotation-based steps. Instead, we use sequence similarity searches (bidirectional best hits) between the high-expression genes of a reference genome(s) and the genome of interest in order to identify candidate highly expressed genes. The modal codon usage of these candidate high-expression orthologs is used as our initial estimate of high-expression codon usage. In the previous section, we offered a detailed portrait of developing a high-expression gene set for E. coli and we have found that this provides a good reference for all bacterial genomes that we have tested. We have performed a similar analysis of the M. maripaludis S2 genome, and we have found that this provides a good reference for all archaeal genomes that we have tested (data not shown). Our default reference “genome” is a concatenation of these two data sets.
In the analysis of E. coli highly expressed genes (above), we explored the utility of iterative refinement of the estimate of high-expression codon usage. To do this, we must recognize those genomes in which the high-expression usage is sufficiently distinct from that of other genes that the process converges on a common set of genes in spite of variations in the starting gene set. By default, our program attempts to do this for each new genome in which ≥20% of the candidate highly expressed genes differ from the modal codon usage, subject to several tests to minimize the chance that the process drifts from high-expression bias to some other bias that is common in the genome (see Materials and Methods for more details).
Native Codon Usage in Genomes with Base Compositional Bias
Previous studies have also recognized the problems associated with hand-selecting potential high-expression genes, and methods have been devised that iterate the CAI as a hands-free approach in order to find the most highly expressed genes in a genome (Carbone et al. 2003; Puigbò et al. 2007); however, in genomes that are A + T or G + C rich, unsupervised application of the CAI can fail. A good example of this effect comes from studies of P. aeruginosa, which has a genomic G + C content of 66% (Gupta and Ghosh 2001; Grocock and Sharp 2002).
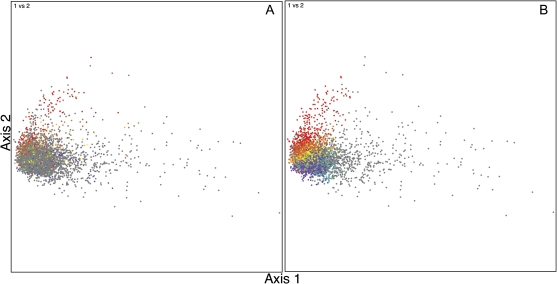
To test the robustness of our approach when analyzing genomes with strong compositional bias, we calculate the native codon usage in P. aeruginosa. We start by illustrating the correspondence between P. aeruginosa genes and their orthologs in E. coli K-12. Figure 2A displays all the genes in P. aeruginosa separated by their positions on the first two axes of an FCA plot. Each gene with a bidirectional best hit to an E. coli gene is colored the same as the corresponding E. coli gene in figure 1; genes without putative orthologs are colored gray. Grocock and Sharp (2002) observed that P. aeruginosa genes separate on the first axis of the correspondence analysis plot by G + C content and on the second axis by expression bias. In figure 2A, we see that the P. aeruginosa genes with E. coli orthologs are distributed along the second axis and that second axis position is well correlated with the position of the corresponding E. coli gene axis position, as reflected in gene colors (e.g., red genes group with red, orange with orange, etc.). This is true even though the high-expression codons differ between E. coli and P. aeruginosa.
FIG. 2.
FCA plot of P. aeruginosa PAO1. (A) Genes that are orthologous to those in E. coli K-12 are colored based upon E. coli axis position (x value) from figure 1. The nonorthologous genes are colored gray. (B) Genes are colored according to P. aeruginosa axis position (x value) based on the colors of the visible spectrum, with red genes having the highest expression-related codon usage bias and violet genes having the least. Genes that differ significantly from all points on the native codon usage axis (likely to be foreign) are colored gray. In both panels, gray genes are drawn behind the colored genes. Genes in the right portion of the first axis have low G + C contents (see also Grocock and Sharp 2002).
Next, we calculate the P. aeruginosa native codon usage axis by finding the modal codon usage of the genome, finding the mode of the genes that are orthologous to the E. coli highly expressed genes, and iteratively refining the high-expression estimate. After two iterations, very little change is observed in the number of genes matching the axis. Overall, 4,178 genes (75% of the genes in the genome) match the native codon usage axis and 1,388 genes (25% of the genes in the genome) do not. Data on each gene are presented in supplementary table S2 (Supplementary Material online). Unlike our results with E. coli, the number of candidate native genes is smaller than the reported core genome of P. aeruginosa genes, which is said to be 85–90% of the genes in the genome (Nelson et al. 2002; Lee et al. 2006). However, it is difficult to correlate data from comparative studies with this type of codon usage analysis because the divergence between (and hence diversity of) the strains of a species is not consistent across taxa. The results of this analysis are shown on a correspondence analysis plot by coloring the genes that match the axis by their axis position and showing those that do not match the axis in gray (fig. 2B). A continuum of genes is observed, and very few low G + C genes in the extreme right end of the first axis of the FCA plot match the native codon usage axis. The most highly expressed genes appear in a hook-shaped pattern—a consequence of the nonlinear response in codon usages and the projection used in FCA. As expected, we are able to accommodate the high G + C content of P. aeruginosa that has confounded CAI-based methods (e.g., Sharp and Li 1987; Carbone et al. 2003).
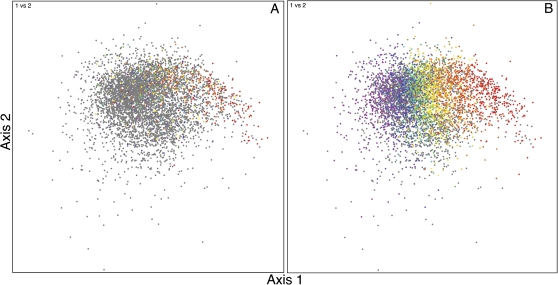
As a final example, we apply our analysis to B. anthracis A0248. Bacillus anthracis has an A + T-rich genome (65% A + T), and has relatively little codon usage variation between genes (as exhibited by its amorphous correspondence analysis pattern). Most of the genes that are orthologs between B. anthracis and our E. coli reference genome are classified as highly expressed genes in E. coli (red through yellow plot symbols in fig. 3A), and most of these are clearly displaced to the right relative to the major codon usage cluster. Thus, we see the broad phylogenetic conservation of our candidate highly expressed genes and that presumably because they retain the property of being highly expressed, most remain distinctive in codon usage even though the codon usages responsible for the distinction are wildly different in E. coli and B. anthracis.
FIG. 3.
FCA plot of B. anthracis A0248. (A) Genes that are orthologous to those in E. coli K-12 are colored based upon E. coli axis position (x value) from figure 1. (B) Genes are colored based upon B. anthracis axis position (x value) based on the colors of the visible spectrum, with red genes having the highest expression-related codon usage bias and violet genes having the least. Genes that differ significantly from all points on the native codon usage axis (likely to be foreign) are colored gray. In both panels, gray genes are drawn behind the colored genes. Each gene’s position along the first axis of the plot also roughly corresponds with its G + C content (from left to right: low G + C to high G + C).
As in the P. aeruginosa example, we next calculate the B. anthracis native codon usage axis and compare each gene with that axis (supplementary table S3, Supplementary Material online). In figure 3B, we show each gene colored by its position along the expression axis or in gray for those that do not match the axis. Overall, 3,788 (72%) of the (candidate native) genes in the genome match the axis, meaning that 1,409 (28%) of the (candidate foreign) genes in the figure do not. These are colored gray. Several of the genes that fail to match are obviously native genes (supplementary table S3, Supplementary Material online). With the P-value threshold of 0.1, we expect a 10% false negative rate, but this still leaves about 20% of the genes in the genome as nonnative. However, unlike the FCA plots of E. coli and P. aeruginosa, there is no obvious clustering of foreign genes in the FCA plot for B. anthracis. As with P. aeruginosa, it is difficult to correlate the number of candidate native genes from this analysis with the number of core B. anthracis genes. However, this small fraction of candidate foreign genes observed in B. anthracis is consistent with pangenome studies of B. anthracis and its relatives (Tettelin et al. 2005; Nelson et al. 2010) in which a relatively small number of novel genes were discovered with each newly sequenced strain. Even though B. anthracis has a biased base composition and its foreign genes do not share a distinct codon usage, this method is effective for characterizing the genes in this genome.
Genomes With Very Little Expression-Related Codon Usage Bias
The method presented accurately detects and characterizes the native (typical and high expression) codon usages in genomes with sufficient expression-related bias. However, some genomes, particularly those of endosymbionts and parasites, are nearly uniform in codon usage (e.g., Andersson and Sharp 1996; Wernegreen and Moran 1999; Herbeck et al. 2003; Rispe et al. 2004; Banerjee and Ghosh 2006). We have assessed the relative magnitude of expression-related bias for 923 “effectively complete” bacterial and archaeal genomes from the SEED database (Overbeek et al. 2005) by computing the distance (Davis and Olsen 2010) between the modal codon usage of the genome and that of the candidate high-expression gene set. In all cases, the high-expression set was more distant from the genome modal usage than was the average distance to randomly chosen gene sets (equal in size to the high-expression set). For 881 of the 923 genomes (>95%), the distance to the high-expression gene mode was greater than that of the random set by ≥2 standard deviations of the sampling variation.
Although this suggests that nearly all genomes have significant expression-related codon bias (see also Kloster and Tang 2008), the magnitude is often too small to reliably assess the status of individual genes; that is, there is a large overlap in the codon usages of the more highly expressed genes and the typical genes. If all gene codon usages in a genome were random samples of a uniform underlying codon usage, 7–10% of the genes are expected to not match the modal usage (Davis and Olsen 2010). Based on this, the default behavior of our program differs depending of the fraction of candidate high-expression genes matching the mode (Materials and Methods). If >20% of the candidates do not match the mode, then this subset is used to estimate high-expression usage. If ≥80% match the mode, then the genome is judged to have too little bias for reliable gene-by-gene assessment, and all of the candidates are used to make a zeroth-order estimate of the high-expression usage.
Native Codon Usage in Eukaryotes
Many eukaryotes have been documented to have expression-related codon usage bias (e.g., Bennetzen and Hall 1982; Shields et al. 1988; Sharp and Devine 1989; Sharp and Cowe 1991; Duret and Mouchiroud 1999; Kanaya et al. 2001; Hiraoka et al. 2009). However, there are potential pitfalls for automated codon usage analysis in eukaryotes. For instance, eukaryotic genomes often encode multiple copies of the same genes as well as multiple protein isoforms, which may influence the outcome of an automated analysis. In higher level eukaryotes, codon usage can be related to isochore base composition (and codon usage) rather than gene expression (Bernardi et al. 1985; Bernardi 1989). Codon usage can also be influenced by CpG methylation (Bernardi et al. 1985; Bernardi 1989), tissue-specific gene expression (Plotkin et al. 2004; Sémon et al. 2006), and intragenic variations relating to translational efficiency (Tuller et al. 2010).
Although these factors may complicate our approach, we attempted a codon usage analysis of several eukaryotic genomes to assess the utility of this algorithm for studying eukaryotes. We started by creating a eukaryotic high-expression reference gene set from the Schizosaccharomyces pombe genome. We seeded the search for S. pombe high-expression genes by using the S. pombe ribosomal proteins, which have been recently shown to have codon usage bias directly related to gene expression in microarray data (Hiraoka et al. 2009). We were able to compute a native codon usage axis in this organism, and we used the top 10% of the S. pombe genes with the highest x values as our reference gene set to search for native codon usage in other organisms (as above). Due to the longer gene lengths found in eukaryotes, which may cause a given gene to fail the chi-square test, we also limited the observed codon usage in the chi-square tests to 300 codons.
We were able to generate a native codon usage axis for the genomes of Caenorhabditis elegans, Dictyostelium discoideum, Drosophila melanogaster, and Saccharomyces cerevisiae. Consistent with our observations above, biased base composition does not appear to have a strong influence on this measure because the D. discoideum genome is extremely biased (27% G + C in protein-encoding genes). When we analyzed Arabidopsis thaliana, the software reverted to the zeroth-order high-expression estimate because the axis drifted away from the high-expression genes (see above), indicating a considerable amount of codon usage homogeneity between A. thaliana genes. Finally, in the case of the Oryza sativa genome, iterations for improving the native codon usage axis also failed. However, unlike A. thaliana, which failed due to homogeneity, the analysis of O. sativa may have failed because the genes of the O. sativa genome form a bimodal distribution based on codon usage (Wang and Hickey 2007).
In summary, this method of codon usage analysis is useful for studying eukaryotes with distinct expression-related codon usage bias but may require careful scrutiny in organisms with more complex genomes. Because horizontal gene transfer can be more restricted in eukaryotes, our designation of the genes not matching the axis as being “foreign” may also require additional scrutiny.
The Proportion of Foreign Genes in Bacterial and Archaeal Genomes
One of the main goals of this study was to create an automated framework for characterizing horizontal gene transfer, particularly in cases where close relatives are not sequenced, or for when potentially foreign genes lack the homologs necessary for sequence comparisons. Because our characterization of native genes is automated, it is possible to efficiently perform a large-scale assessment of foreign gene content over many genomes. We measured the nonnative gene content of 923 bacterial and archaeal genomes from the SEED database (Overbeek et al. 2005). In table 2, we report the genomes with the lowest and highest fraction of nonnative genes (P ≥ 0.1, which is consistent with the other analyses in this work).
Table 2.
Percentage of Candidate Foreign Genes in Each Genome for the Ten Bacterial and Archaeal Species With the Most and Least Homogenous Genomesa.
| Organism | CDSb | %G+Cc | % Foreign |
| Most homogeneous | |||
| Wigglesworthia glossinidia | 639 | 23.6 | 3.8 |
| Borrelia turicatae 91E135 | 838 | 29.4 | 7.8 |
| B. garinii PBi | 933 | 28.5 | 7.8 |
| B. hermsii DAH | 855 | 30.1 | 8.3 |
| Blochmannia floridanus | 584 | 28.9 | 8.6 |
| Buchnera aphidicola str. 5A | 591 | 27.3 | 8.8 |
| Nanoarchaeum equitans Kin4-M | 543 | 31.2 | 10.1 |
| Pelagibacter ubique HTCC1062 | 1,355 | 29.8 | 10.3 |
| Beutenbergia cavernae DSM 12333 | 4,278 | 73.2 | 10.5 |
| Rickettsia rickettsii | 1,292 | 32.9 | 11.0 |
| Least homogeneous | |||
| Bacteroides vulgatus ATCC 8482 | 3,900 | 43.2 | 65.0 |
| Ba. thetaiotaomicron VPI-5482 | 4,832 | 43.9 | 64.5 |
| Ba. fragilis ATCC 25285 | 4,233 | 44.1 | 64.1 |
| Corynebacterium diphtheriae NCTC 13129 | 2,343 | 54.1 | 61.6 |
| Desulfotalea psychrophila LSv54 | 3,240 | 47.5 | 59.1 |
| Neisseria meningitidis MC58 | 2,243 | 52.9 | 58.9 |
| C. glutamicum ATCC 13032 | 2,994 | 54.8 | 57.5 |
| Xylella fastidiosa 9a5c | 2,917 | 53.8 | 54.9 |
| N. gonorrhoeae FA 1090 | 2,023 | 54.0 | 53.8 |
| Thermobaculum terrenum ATCC BAA-798 | 3,048 | 53.8 | 52.4 |
When multiple strains of a species are available, only the most extreme is included.
Number of coding sequences in the genome (all replicons combined).
In coding sequences.
The most homogeneous genomes, with the smallest fraction of foreign genes, are very well characterized (e.g., Andersson and Sharp 1996; Wernegreen and Moran 1999; Herbeck et al. 2003; Rispe et al. 2004). In most cases, these are endosymbionts and parasites. They have been historically easy to identify because they usually live in isolation with limited access to foreign genes, and their genomes are usually reduced and have extreme base compositions (and thus limited codon usage variation between genes). Our results recapitulate these previous studies (table 2, top). The genome of the Wigglesworthia glossinidia, an endosymbiont of Glossina brevipalpis, is the most homogenous observed. Only 4% of the genes in this genome have a foreign codon usage, which is even less than random expectation for a homogeneous genome. All the other homogeneous genomes reported in table 2 are either endosymbionts or parasites with the exceptions of the free-living Pelagibacter ubique, a marine bacterium that has one of the most streamlined genomes for any free-living organism (Giovannoni et al. 2005), and the free-living cave isolate Beutenbergia cavernae (Groth et al. 1999). The high degree of homogeneity in the B. cavernae genome is somewhat surprising due to its large size (4,278 protein-encoding genes); however, the genome has an extremely biased base composition (73% G + C for protein-encoding genes), which is consistent with extreme base composition being a hallmark of homogeneous genomes.
Unlike the most homogeneous genomes, the organisms with the most heterogeneous genomes (having the largest fraction of foreign genes) are not well characterized. They are typically free living, and their genomic base compositions tend to be moderate. Many previous studies have searched genomes for genes with aberrant base compositions and codon usages (e.g., Médigue et al. 1991; Lawrence and Ochman 1997; Daubin and Ochman 2004); however, the vast majority of the studies of this kind have focused on individual genomes. For this reason, the current analysis may provide broader insights in this area.
The most heterogeneous genome observed was that of Bacteroides vulgatus, with 65% of the genes in the genome having a nonnative codon usage (table 2, bottom). Other heterogeneous genomes include members of the genera Corynebacterium, Desulfotalea, Neisseria, Xylella, and Thermobaculum. The base compositions of these genomes are moderate (43–55% G + C in protein-encoding genes). Surprisingly, the genome sizes of the most heterogeneous genomes are also moderate, ranging from 2,023 to 4,832 genes. Because all replicons were considered in this analysis, it is tempting to attribute genomic heterogeneity to the existence of extrachromosomal elements; however, five of the heterogeneous genomes in table 2 do not contain a plasmid or secondary chromosome. Clearly, horizontal gene transfer is strongly contributing to the evolution of these genomes.
Codon usage homogeneity is not always constant within a phylogenetic group. In one interesting example, Prochlorococcus marinus str. MIT 9301 and Pr. marinus subsp. pastoris, are free-living organisms with very homogeneous genomes (only 11% of the genes in these genomes do not match native codon usage), whereas Pr. marinus MIT 9313 has one of the least homogeneous genomes with 44% of the genes in this genome differing from native codon usage.
Conclusion
In this study, we have provided a robust automated method for characterizing the native codon usages of a genome. In defining the CAI, Sharp and Li (1987) provided a continuous measure of how optimal (or fully adapted) the codon usage of a gene is to an ideal for highly expressed genes in the given genome. In place of this match to an ideal codon usage, we use the projected position of each gene along an axis of native codon usage, which is a half line starting at “typical usage” and extending through a codon usage representing highly expressed genes. The concept of an axis is implicit in the method of Karlin and Mrázek (2000) in that they represent high-expression usage by three separate point estimates, whereas using the average codon usage to represent “typical” genes. Yet in the end, they classify genes into three categories (highly expressed, typical native, and if neither of these, then foreign), without providing an estimate of relative expression levels of the various high-expression genes. In addition, their analysis is based on a distance measure rather than a statistical assessment of the fit of a gene to an expected codon usage, which forced them to limit their analyses to genes greater than an arbitrary minimum length. A final way in which our method differs from all others is the convenience of a fully automated identification of candidate high-expression genes, entirely bypassing the use of annotations. In addition to being simpler, this eliminates common errors in gene annotation and the subjective interpretation of the annotations.
Supplementary Material
Supplementary tables S1, S2, and S3, supplementary material; codon_usage_software.tgz (the software for computing the native codon usage axis); and Davis_and_Olsen_examples.tgz (a sample analysis) are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/), and current versions are available through links at http://www.life.illinois.edu/gary/programs.html.
Acknowledgments
We wish to thank Claudia Reich and the anonymous reviewers for their helpful comments and suggestions on the manuscript. We are also grateful to Carl Woese for his encouragement and support of the project. Portions of this material are based upon work supported by the National Aeronautics and Space Administration under Grant NAG 5-12334 issued through the Exobiology Program, by the Department of Energy under Grant FG02-01ER63146, and by the National Institutes of Health under Contract HHSN266200400042C via subcontract from the University of Chicago to the University of Illinois.
References
- Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson SG, Sharp PM. Codon usage and base composition in Rickettsia prowazekii. J Mol Evol. 1996;42:525–536. doi: 10.1007/BF02352282. [DOI] [PubMed] [Google Scholar]
- Badger JH. Exploration of microbial genomic sequences via comparative analysis [PhD dissertation] 1999 [Urbana (IL)]: University of Illinois at Urbana-Champaign. p. 45–92. [Google Scholar]
- Banerjee T, Ghosh TC. Gene expression level shapes the amino acid usages in Prochlorococcus marinus MED4. J Biomol Struct Dyn. 2006;23:547–553. doi: 10.1080/07391102.2006.10507079. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Hall BD. Codon selection in yeast. J Biol Chem. 1982;257:3026–3031. [PubMed] [Google Scholar]
- Bernardi G. The isochore organization of the human genome. Annu Rev Genet. 1989;23:637–661. doi: 10.1146/annurev.ge.23.120189.003225. [DOI] [PubMed] [Google Scholar]
- Bernardi G, Olofsson B, Filipski J, Zerial M, Salinas J, Cuny G, Meunier-Rotival M, Rodier F. The mosaic genome of warm-blooded vertebrates. Science. 1985;228:953–958. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- Carbone A, Zinovyev A, Képès A. Codon adaptation index as a measure of dominating codon bias. Bioinformatics. 2003;19:2005–2015. doi: 10.1093/bioinformatics/btg272. [DOI] [PubMed] [Google Scholar]
- Chen SL, Hung CS, Xu J, et al. (18 co-authors) Identification of genes subject to positive selection in uropathogenic strains of Escherichia coli: a comparative genomics approach. Proc Natl Acad Sci U S A. 2006;103:5977–5982. doi: 10.1073/pnas.0600938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daubin V, Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis JJ, Olsen GJ. Modal codon usage: assessing the typical codon usage of a genome. Mol Biol Evol. 2010;27:800–810. doi: 10.1093/molbev/msp281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:4482–4487. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukiya S, Mizoguchi H, Tobe T, Mori H. Extensive genomic diversity in pathogenic Escherichia coli and Shigella strains revealed by comparative genomic hybridization microarray. J Bacteriol. 2004;186:3911–3921. doi: 10.1128/JB.186.12.3911-3921.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni SJ, Tripp HJ, Givan S, et al. (14 co-authors) Genome streamlining in a cosmopolitan oceanic bacterium. Science. 2005;309:1242–1245. doi: 10.1126/science.1114057. [DOI] [PubMed] [Google Scholar]
- Grantham R, Gautier C, Gouy M. Codon frequencies in 119 individual genes confirm consistent choices of degenerate bases according to genome type. Nucleic Acids Res. 1980;8:1893–1912. doi: 10.1093/nar/8.9.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R, Gautier C, Gouy M, Mercier R, Pave A. Codon catalog usage and the genome hypothesis. Nucleic Acids Res. 1980;8:r49–r62. doi: 10.1093/nar/8.1.197-c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grantham R, Gautier C, Gouy M, Jacobzone M, Mercier R. Codon catalog usage is a genome strategy modulated for gene expressivity. Nucleic Acids Res. 1981;9:r43–r74. doi: 10.1093/nar/9.1.213-b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M, Devereux J, Burgess RR. The codon preference plot: graphic analysis of protein coding sequences and prediction of gene expression. Nucleic Acids Res. 1984;12:539–549. doi: 10.1093/nar/12.1part2.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grocock RJ, Sharp PM. Synonymous codon usage in Pseudomonas aeruginosa PAO1. Gene. 2002;289:131–139. doi: 10.1016/s0378-1119(02)00503-6. [DOI] [PubMed] [Google Scholar]
- Groth I, Schumann P, Schuetze B, Augsten K, Kramer I, Stackebrandt E. Beutenbergia cavernae gen. nov., sp. nov., an L-lysine-containing actinomycete isolated from a cave. Int J Syst Bacteriol. 1999;49:1733–1740. doi: 10.1099/00207713-49-4-1733. [DOI] [PubMed] [Google Scholar]
- Gupta S, Ghosh T. Gene expressivity is the main factor in dictating the codon usage variation among the genes in Pseudomonas aeruginosa. Gene. 2001;273:63–70. doi: 10.1016/s0378-1119(01)00576-5. [DOI] [PubMed] [Google Scholar]
- Herbeck JT, Wall DP, Wernegreen JJ. Gene expression level influences amino acid usage, but not codon usage, in the tsetse fly endosymbiont Wigglesworthia. Microbiology. 2003;149:2585–2596. doi: 10.1099/mic.0.26381-0. [DOI] [PubMed] [Google Scholar]
- Hiraoka Y, Kenichi K, Haraguchi T, Chikashige Y. Codon usage bias is correlated with gene expression levels in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2009;14:499–509. doi: 10.1111/j.1365-2443.2009.01284.x. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes. J Mol Biol. 1981a;146:1–21. doi: 10.1016/0022-2836(81)90363-6. [DOI] [PubMed] [Google Scholar]
- Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981b;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol. 2001;53:290–298. doi: 10.1007/s002390010219. [DOI] [PubMed] [Google Scholar]
- Karlin S, Mrázek J. Predicted highly expressed genes of diverse prokaryotic genomes. J Bacteriol. 2000;182:5238–5250. doi: 10.1128/jb.182.18.5238-5250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloster M, Tang C. SCUMBLE: a method for systematic and accurate detection of codon usage bias by maximum likelihood estimation. Nucleic Acids Res. 2008;36:3819–3827. doi: 10.1093/nar/gkn288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence JG, Ochman H. Amelioration of bacterial genomes: rates of change and exchange. J Mol Evol. 1997;44:383–397. doi: 10.1007/pl00006158. [DOI] [PubMed] [Google Scholar]
- Lee DG, Urbach JM, Wu G, et al. (17 co-authors) Genomic analysis reveals that Pseudomonas aeruginosa virulence is combinatorial. Genome Biol. 2006;7:R90. doi: 10.1186/gb-2006-7-10-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLachlan AD, Staden R, Boswell DR. A method for measuring the non-random bias of a codon usage table. Nucleic Acids Res. 1984;12:9567–9575. doi: 10.1093/nar/12.24.9567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médigue C, Rouxel T, Vigier P, Hénaut A, Danchin A. Evidence for horizontal gene transfer in Escherichia coli speciation. J Mol Biol. 1991;222:851–856. doi: 10.1016/0022-2836(91)90575-q. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Weinel C, Paulsen IT, et al. (43 co-authors) Complete genome sequence and comparative analysis of the metabolically versatile Pseudomonas putida KT2440. Environ Microbiol. 2002;4:799–808. doi: 10.1046/j.1462-2920.2002.00366.x. [DOI] [PubMed] [Google Scholar]
- Nelson KE, Weinstock GM, Highlander SK, et al. (83 co-authors) A catalog of reference genomes from the human microbiome. Science. 2010;328:994–999. doi: 10.1126/science.1183605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, et al. (40 co-authors) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peden J. Analysis of codon usage [PhD dissertation] [Nottingham (UK)]: University of Nottingham; 1999. pp. 50–102. [Google Scholar]
- Plotkin JB, Robins H, Levine AJ. Tissue-specific codon usage and the expression of human genes. Proc Natl Acad Sci U S A. 2004;101:12588–12591. doi: 10.1073/pnas.0404957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Post LE, Strycharz GD, Nomura M, Lewis H, Dennis PP. Nucleotide sequence of the ribosomal protein gene cluster adjacent to the gene for RNA polymerase subunit β in E. coli. Proc Natl Acad Sci U S A. 1979;76:1697–1701. doi: 10.1073/pnas.76.4.1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puigbò P, Guzmán E, Romeu A, Garcia-Vallvé S. OPTIMIZER: a web server for optimizing the codon usage of DNA sequences. Nucleic Acids Res. 2007;35:W126–W131. doi: 10.1093/nar/gkm219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasko DA, Rosovitz MJ, Myers GSA, et al. (13 co authors) The pangenome structure of Escherichia coli: comparative genomic analysis of E. coli commensal and pathogenic isolates. J Bacteriol. 2008;190:6881–6893. doi: 10.1128/JB.00619-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rispe C, Delmotte F, van Ham R, Moya A. Mutational and selective pressures on codon and amino acid usage in Buchnera, endosymbiotic bacteria of aphids. Genome Res. 2004;14:44–53. doi: 10.1101/gr.1358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M, Lobry JR, Druet L. No evidence for tissue-specific adaptation of synonymous codon usage in humans. Mol Biol Evol. 2006;23:523–529. doi: 10.1093/molbev/msj053. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Cowe E. Synonymous codon usage in Saccharomyces cerevisiae. Yeast. 1991;7:657–678. doi: 10.1002/yea.320070702. [DOI] [PubMed] [Google Scholar]
- Sharp PM, Devine KM. Codon usage and gene expression level in Dictyostelium discoideum: highly expressed genes do 'prefer' optimal codons. Nucleic Acids Res. 1989;17:5029–5039. doi: 10.1093/nar/17.13.5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li W. Codon usage in regulatory genes in Escherichia coli does not reflect selection for 'rare' codons. Nucleic Acids Res. 1986;14:7737–7749. doi: 10.1093/nar/14.19.7737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp PM, Li W. The codon adaptation index—a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;29:1281–1295. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shields DC, Sharp PM, Higgins DG, Wright F. “Silent” sites in Drosophila genes are not neutral: evidence of selection among synonymous codons. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, et al. (46 co-authors) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome”. Proc Natl Acad Sci U S A. 2005;102:13950–13955. doi: 10.1073/pnas.0506758102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller T, Carmi A, Vestsigian K, Navon S, Dorfan Y, Zaborske J, Pan T, Dahan O, Furman I, Pilpel Y. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–354. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- Wang HC, Hickey DA. Rapid divergence of codon usage patterns within the rice genome. BMC Evol Biol. 2007;7(Suppl 1):S6. doi: 10.1186/1471-2148-7-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernegreen JJ, Moran NA. Evidence for genetic drift in endosymbionts (Buchnera): analyses of protein-coding genes. Mol Biol Evol. 1999;16:83–97. doi: 10.1093/oxfordjournals.molbev.a026040. [DOI] [PubMed] [Google Scholar]