Abstract
The genetic basis of organisms’ adaptation to different environments is a central issue of molecular evolution. The budding yeast Saccharomyces cerevisiae and its relatives predominantly ferment glucose into ethanol even in the presence of oxygen. This was suggested to be an adaptation to glucose-rich habitats, but the underlying genetic basis of the evolution of aerobic fermentation remains unclear. In S. cerevisiae, the first step of glucose metabolism is transporting glucose across the plasma membrane, which is carried out by hexose transporter proteins. Although several studies have recognized that the rate of glucose uptake can affect how glucose is metabolized, the role of HXT genes in the evolution of aerobic fermentation has not been fully explored. In this study, we identified all members of the HXT gene family in 23 fully sequenced fungal genomes, reconstructed their evolutionary history to pinpoint gene gain and loss events, and evaluated their adaptive significance in the evolution of aerobic fermentation. We found that the HXT genes have been extensively amplified in the two fungal lineages that have independently evolved aerobic fermentation. In contrast, reduction of the number of HXT genes has occurred in aerobic respiratory species. Our study reveals a strong positive correlation between the copy number of HXT genes and the strength of aerobic fermentation, suggesting that HXT gene expansion has facilitated the evolution of aerobic fermentation.
Keywords: aerobic fermentation, HXT, glucose metabolism, glucose transport, adaptive evolution
Introduction
The genetic basis of organisms' adaptation to their environments is a central issue in evolution. Aerobic fermentation is a glucose metabolic pathway that predominantly ferments glucose to ethanol even in the presence of oxygen. It has been proposed that aerobic fermentation in yeasts evolved as an adaptation to glucose-rich habitats (Thomson et al. 2005; Piskur et al. 2006; Conant and Wolfe 2007; Merico et al. 2007). The hemiascomycete yeasts that prefer aerobic fermentation are called Crabtree-positive yeasts (De Deken 1966); these include Saccharomyces cerevisiae and its relatives. In contrast, the Crabtree-negative yeasts, such as the dairy yeast Kluyveromyces lactis and the filamentous fungus Ashbya gossypii, prefer to completely oxidize glucose to CO2 through the mitochondrial respiration pathway for maximum energy and biomass gain (Merico et al. 2007). The fission yeast Schizosaccharomyces pombe, which diverged from the hemiascomycete lineage around 330–420 Ma (Wood et al. 2002), also prefers aerobic fermentation (Fiechter et al. 1981). Therefore, it is believed that aerobic fermentation has evolved independently in the S. cerevisiae lineage and in the S. pombe lineage. The aerobic fermentation lifestyle enabled the ancestral Crabtree-positive yeasts to consume surrounding glucose rapidly by transforming it into ethanol. Because most of them can efficiently use ethanol as carbon and energy sources after glucose depletion, it was speculated that this metabolism pathway provides a selective advantage for Crabtree-positive yeasts in glucose-rich environments (Thomson et al. 2005).
Many studies have shed light on the underlying genetic basis for the origin and evolution of aerobic fermentation, but different explanations were proposed (Wolfe and Shields 1997; Kellis et al. 2004; Ihmels et al. 2005; Thomson et al. 2005; Conant and Wolfe 2007; Field et al. 2009). Several studies have shown that a large proportion of glucose metabolism is controlled by glucose uptake, which is the first and obligatory step of glucose metabolism (Gancedo and Serrano 1989; Diderich et al. 1999; Ye et al. 1999; Elbing et al. 2004; Otterstedt et al. 2004; Conant and Wolfe 2007). Glucose uptake in S. cerevisiae is carried out by a group of hexose transporters (Hxt), which belong to a superfamily of monosaccharide facilitators with 12 transmembrane domains (Boles and Hollenberg 1997; Ozcan and Johnston 1999). A total number of 20 putative HXT genes have been identified in S. cerevisiae: HXT1–HXT17, GAL2, SNF3, and RGT2 (fig. 1) (Boles and Hollenberg 1997). Among the 20 putative S. cerevisiae HXT genes, 17 encodes glucose transporters (Hxt1–Hxt17), but under normal conditions, the glucose uptake is mainly mediated by only 6 of them (Hxt1 and Hxt3–Hxt7). Two of the HXT genes (SNF3 and RGT2) encode glucose sensors and one (GAL2) encodes transporters that mediate D-galactose uptake. The homologues of S. cerevisiae HXT genes have been identified in other species. For example, a gene encodes a low-affinity glucose transporter (Rag1) has been characterized as a HXT homologue in K. lactis (Goffrini et al. 1990), and eight HXT homologues (GHT1–8) were described in S. pombe (Heiland et al. 2000; Wood et al. 2002). When the HXT1–HXT17 genes in S. cerevisiae were replaced by a chimera HXT gene, decreased ability of ethanol production due to reduced glucose consumption rates or even a switch to fully respiratory metabolism was observed (Elbing et al. 2004; Otterstedt et al. 2004). Quantitative analyses in another aerobic fermentation yeast Saccharomyces bayanus revealed that a high proportion of glycolytic flux relies on the level of glucose transport (Diderich et al. 1999). In addition to experimental data, Pritchard and Kell (2002) used a mathematical model to investigate flux control patterns of S. cerevisiae glycolysis and drew the same conclusion that hexose transport is the major rate-limiting step in yeast glycolysis. Moreover, after introducing S. cerevisiae HXT genes (GAL2 or HXT4) to K. lactis, the modified cells can grow on galactose and raffinose through fermentation (Goffrini et al. 2002). All these results suggest that glucose uptake by Hxt proteins plays an influential role in controlling glucose metabolism activities.
FIG. 1.
Genomic locations of the 20 Saccharomyces cerevisiae HXT genes. The six HXT genes that play a major role in glucose uptake form two 3-gene clusters (Italic). Eight HXT genes (underlined) are located within or near subtelomeric regions. This figure is drawn in scale.
Based on a study in 7 hemiascomycete yeasts, Conant and Wolfe (2007) have noticed that all the 4 aerobic fermentation species have at least twice as many HXT genes as the 3 aerobic respiration yeasts. However, in-depth investigation of the evolutionary history of HXT genes and their role in the evolution of aerobic fermentation in yeasts has not been reported so far. In view of the fact that glucose uptake is a key step in glucose metabolism, analysis of the evolutionary dynamics of the HXT gene family will likely provide new insights into the evolution of yeast aerobic fermentation. Such a study has become possible because genome sequences are now available for many fungal species. In this study, we identified all HXT homologues in 23 fungal species and comprehensively studied the births and deaths of HXT genes through phylogenetic analyses and comparative genomics. Our study provides evidence that the evolution of aerobic fermentation in yeasts has been facilitated by expansion in the copy number of HXT genes.
Materials and Methods
Data Search
The amino acid sequences of the 20 S. cerevisiae HXT genes were retrieved from the Yeast Genome Browser (http://www.yeastgenome.org/). We used each of the 20 S. cerevisiae Hxt protein sequences as a query in BlastP searches against the following species with available annotated protein data: Aspergillus fumigatus, S. pombe, A. fumigatus, Magnaporthe grisea, Phaeosphaeria nodorum, Gibberella zeae, Ustilago maydis, S. cerevisiae, Candida glabrata, K. polysporus (Vanderwaltozyma polyspora), Zygosaccharomyces rouxii, K. thermotolerans (Lachancea thermotolerans), K. lactis, A. gossypii, Pichia stipitis, Debaryomyces hansenii, Candida albicans, Yarrowia lipolytica, and two outgroup genomes human and Arabidopsis thaliana. TBlastN searches were performed against genomic sequences for the following species without genome-wide proteomic data: Saccharomyces paradoxus, Saccharomyces mikatae, S. bayanus, Saccharomyces castellii, and K. waltii. Each of the TBlastN-hit sequences was extended in both directions along the genome sequence, and the longest coding sequence from the ATG to the stop codon was extracted. If an open reading frame was truncated by the end of genomic contig, we further searched for overlapping contigs by BlastN and assembled overlapping contigs into a large contig using CAP3 (Huang and Madan 1999).
Identification of HXT Members
Because the HXT gene family belongs to the major facilitator superfamily (MFS), many hits in our Blast searches may belong to other MFS families instead of the HXT family. To determine if a hit belongs to the HXT family, we used the Blast search hits with an E value < 10−10 from each of target species and the 20 S. cerevisiae HXT genes to construct a Neighbor-Joining (NJ) tree using MEGA 4.0 (Tamura et al. 2007) (data not shown). Those sequences that are grouped with the S. cerevisiae HXT genes were taken as HXT members and used in subsequent analyses. A complete list of the HXT genes from the 23 fungal species is included in supplementary table S1 (Supplementary Material online).
Phylogenetic Analyses of the HXT Gene Family and Inference of Gene Gains and Losses
Preliminary multiple sequence alignments of all Hxt protein sequences under study were carried out using MUSCLE version 3.52 with default parameter settings (Edgar 2004). These alignments were manually inspected and corrected using GeneDoc version 2.6.002 (Nicholas et al. 1997). We constructed phylogenetic trees using both the maximum likelihood (ML) and NJ methods to ensure the robustness of our analysis. We used ProtTest 1.4 (Abascal et al. 2005) to identify the most appropriate model and parameters (WAG + I + G + F model) for the Hxt protein alignment. These model and parameters were used in our ML tree reconstruction using Phyml 2.4 (Guindon and Gascuel 2003) with 500 bootstrap replicates. The proportion of invariable sites and the α-parameter of β-law distribution were optimized according to the data. NJ trees were constructed using MEGA 4.0 (Tamura et al. 2007). The confidence of internal branches of NJ trees was assessed with 1000 bootstrap pseudoreplicates using “pairwise deletion option” of amino acid sequences with Poisson correction and the Jones-Taylor-Thornton model.
We primarily used the reconstructed phylogenetic trees to determine the orthologous relationships of the HXT genes. However, due to potentially frequent gene conversion and unequal evolutionary rates among lineages, an orthologous gene sometimes cannot be determined merely from phylogenetic analyses (Lynch and Conery 2000). Under this condition, we further compared their synteny structures for the presence of other homologs using the Yeast Gene Order Browser (Byrne and Wolfe 2005). The phylogeny and synteny methods worked effectively to determine the orthologous groups for almost all HXT genes. In a few cases, reciprocal best hits were used to assign orthologs among a set of homologs if they cannot be resolved from phylogenetic analyses and synteny conservation.
The reconciled tree method has been commonly used to estimate the numbers of gene gains and losses (Slowinski et al. 1997; Nam and Nei 2005). This method estimates the number of genes in the most recent common ancestor (MRCA) on the basis of tree topology. If the phylogenetic tree is not well resolved, the results become less reliable. Because a large number of sequences were used in this study, the phylogenetic tree is not completely resolved. To obtain more reliable results, based on our gene classification results (subgroups I–VIII, see Results), we constructed a phylogenetic tree for each subgroup and estimated the gene gain and loss events separately using the reconciled tree method. We then combined the results from all subgroups to obtain the overall numbers of gain and loss events.
Results
The HXT Family Consists of Two Distinct Subfamilies: Transporters and Sensors
To study the evolution of the HXT family, we constructed a phylogenetic tree using the homologous sequences from human, Arabidopsis, and 7 fungal species representing 4 major fungal lineages (see Materials and Methods). The 20 S. cerevisiae HXT genes are found in a well-supported monophyletic clade in the tree of the MFS superfamily (supplementary fig. S1, Supplementary Material online). The HXT clade also includes members from other major fungal lineages, such as A. fumigatus, U. maydis, and S. pombe, but does not contain any human and Arabidopsis member, suggesting that the HXT family originated after the separation of the fungal lineage from the animal and plant lineages or became lost in the animals and plants. The HXT family consists of two well-supported groups (supplementary fig. S1, Supplementary Material online), and each group contains members from multiple fungal lineages, suggesting that the two groups were formed before the divergence of major fungal lineages. One group contains 18 S. cerevisiae HXT genes (HXT1–17 and GAL2) and their homologs in other species from all 4 examined fungal lineages. We named this group the Transporter subfamily because most of these gene products in S. cerevisiae are directly involved in transporting glucose across cell membrane (Boles and Hollenberg 1997). The other group consists of the S. cerevisiae SNF3 and RGT2 and their homologs from 3 of the 4 fungal lineages examined. Because SNF3 and RGT2 encode sensors that recognize the concentration of extracellular glucose in S. cerevisiae (Boles and Hollenberg 1997; Ozcan and Johnston 1999), we called this group the Sensor subfamily.
Low Copy Number Variation in the Sensor Subfamily
To achieve a better understanding of the evolutionary changes of the HXT family, we conducted a detailed phylogenetic analysis for each subfamily. We identified the members of the Sensor subfamily from 23 fungal species with complete genomes and reconstructed their evolutionary history (supplementary fig. S2, Supplementary Material online). Except for S. pombe, all species have 1–4 Sensor genes and 14 species each contain 2 copies, indicating that these fungal species have maintained largely a constant number of Sensor genes throughout their evolution (supplementary fig. S2 and table S1, Supplementary Material online). All post–whole-genome duplication (WGD) species, except for K. polysporus, contain two Sensor genes (SNF3 and RGT2) and they form two separate clades (supplementary fig. S2, Supplementary Material online), suggesting that they were produced by the WGD. This is further supported by the fact that the two genes share similar synteny structures in the post-WGD species (data not shown). One of the two Sensor genes is lost in K. polysporus, and the sequence of the remaining copy has significantly diverged (supplementary fig. S2, Supplementary Material online). Many pre-WGD yeasts such as C. albicans and P. stipitis also contain two Sensor genes, which were annotated as SNF3 and RGT2. As inferred above, the S. cerevisiae SNF3 and RGT2 were likely produced by the WGD, which occurred after the S. cerevisiae lineage diverged from the C. albicans lineage. However, the two C. albicans Sensor genes were produced before the divergence of S. cerevisiae and C. albicans (supplementary fig. S2, Supplementary Material online). The common ancestor of S. cerevisiae SNF3 and RGT2 seems to have originated from the C. albicans SNF3 lineage. Functional study of the C. albicans SNF3 gene (HGT4) supports its role as a glucose sensor (Brown et al. 2006). Therefore, we named this lineage of Sensor genes the EuSensor group. Accordingly, the other Sensor genes were named as the ProSensor genes (supplementary fig. S2, Supplementary Material online). The orthologues of ProSensor genes are not present in any of the post-WGD yeasts, and their proteins lack the extra C-terminal domain, which is present in the EuSensor proteins. The C-terminal domain has been shown to be important for sensing the availability of extracellular glucose (Ozcan et al. 1998). The C. albicans RGT2 (HGT12) protein is probably just a transporter (Brown et al. 2006). Combining our phylogenetic study and previous functional characterizations, we postulate that the EuSensor proteins acquired the sensor function through acquiring the extra C-terminal domain after they diverged from ancestral transporters during the early evolution of hemiascomycete yeasts.
Amplification of the Transporter Subfamily in Two Aerobic Fermentation Lineages
In contrast to the Sensor subfamily, the numbers of Transporter subfamily genes vary extensively among yeasts (figs. 2 and 3). Our phylogenetic analysis of nine representative fungal species shows that the Transporter genes originated from a single gene in their MRCA but have undergone many amplification events in two different lineages: S. pombe (from 1 to 8 copies) and S. cerevisiae (from 1 to 18 copies). Notably, both these two lineages prefer aerobic fermentation over aerobic respiration. This correlation suggests that the increase of Transporter gene number has facilitated the evolution of aerobic fermentation.
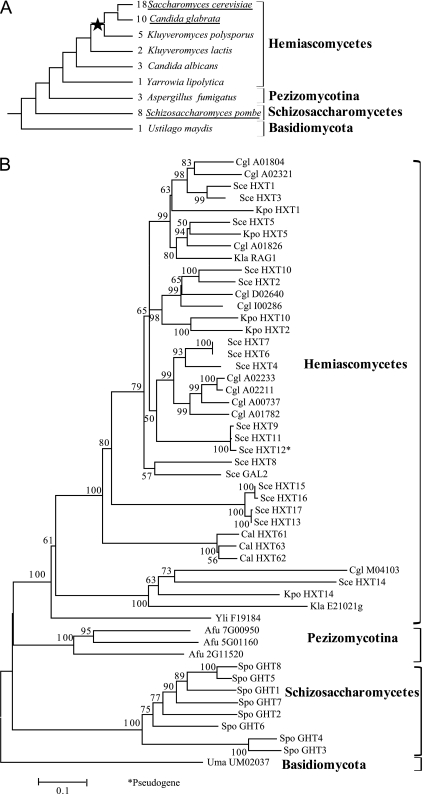
FIG. 2.
The Transporter genes were greatly amplified in two aerobic fermentative yeast lineages. (A) The evolutionary change of copy numbers of Transporter genes in nine fungal species. The evolutionary relationships of the nine species were derived from the trees in Hedges (2002) and Souciet et al. (2009). The aerobic fermentative species are underlined. The star indicates the WGD event. The number in front of each species indicates the copy number of Transporter genes in the genome. (B) The ML tree of the Transporter subfamily genes for the nine fungal species. This tree shows that the Transporter genes originated from a single gene in the common ancestor of the fungi under study and have experienced gene expansions in the two lineages with aerobic fermentation. Species names are abbreviated as follows: Sce, Saccharomyces cerevisiae; Cgl, Candida glabrata; Kpo, Kluyveromyces polysporus; Kla, Kluyveromyces lactis; Cal, Candida albicans; Yli, Yarrowia lipolytica; Afu, Aspergillus fumigatus; Spo, Schizosaccharomyces pombe; and Uma, Ustilago maydis.
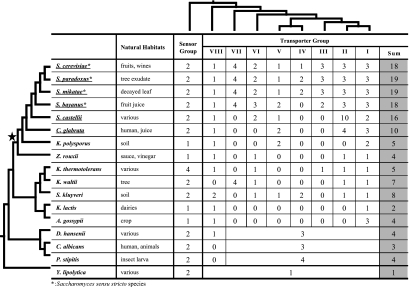
FIG. 3.
Copy number variation of HXT genes in 17 hemiascomycete yeasts. The evolutionary relationships of the 17 hemiascomycete yeasts were modified from Souciet et al. (2009). The WGD is indicated by a star on the species tree. The names of aerobic fermentative species were underlined. Information of yeast natural habitats is obtained from Barnett et al. (2000). The phylogenetic relationships among subgroups were inferred using the 20 Saccharomyces cerevisiae HXT genes. The 18 S. cerevisiae Transporter genes are distributed into eight groups: I (HXT1, HXT3, and HXT5); II (HXT4, HXT6, and HXT17), III (HXT9, HXT11, and HXT12), IV (HXT8), V (GAL2), VI (HXT2 and HXT10), VII (HXT13, HXT15, HXT16, and HXT17), and VIII (HXT14). The “Sum” column shows the total number of Transporter subfamily genes in each species.
Many genomes in the hemiascomycete class, which includes S. cerevisiae, have been sequenced, and they can be used to define orthologous genes based on conservation of their syntenic structures in addition to the phylogenetic analyses. Furthermore, these yeasts display different glucose metabolism lifestyles. Therefore, it offered us a chance to systematically study how the Transporter genes were amplified and whether amplification was correlated with the evolution of aerobic fermentation. As shown in figure 3, the copy number of Transporter genes had continually increased through the evolution of the S. cerevisiae lineage, starting from a single copy in Y. lipolytica, which is most distantly related to S. cerevisiae in hemiascomycetes, to 2–8 copies in K. lactis and Saccharomyces kluyveri, and to 18–19 copies in the Saccharomyces sensu stricto species (fig. 3). Interestingly, the number increases match the levels of aerobic fermentation of these species: Y. lipolytica is exclusively respiratory; K. lactis and S. kluyveri prefer aerobic respiration with limited aerobic fermentation; and Saccharomyces sensu stricto species much prefer aerobic fermentation (Wieczorke et al. 1999).
The 18 S. cerevisiae Transporter genes are distributed into 8 well-supported clades, denoted as Groups I–VIII (fig. 3 and supplementary fig. S3, Supplementary Material online). Based on the species composition in each group and the phylogenetic tree, the approximate time when these groups arose can be inferred. The Group VIII (HXT14) gene is present in 13 of the 17 hemiascomycete yeasts under study; in these yeasts, the most distant relative to S. cerevisiae is D. hansenii, so Group VIII likely arose before the divergence of the D. hansenii and S. cerevisiae lineages. All other groups, except for Group II (HXT4, HXT6, and HXT7), contain members from at least one of the following pre-WGD species: K. thermotolerans, K. waltii, S. kluyveri, K. lactis, and A. gossypii. A small number of Transporter genes of these pre-WGD species, such as S. kluyveri G07678 and K. thermotolerans E02772, are not grouped with any of the eight groups on the phylogenetic tree, but they are located at the orthologous locus of Group II genes based on syntenic structure comparison, implying they are likely members of Group II (fig. 4). Therefore, all the eight groups arose before the separation of the S. cerevisiae lineage from the A. gossypii lineage (Wolfe and Shields 1997; Dietrich et al. 2004; Kellis et al. 2004).
FIG. 4.
Expansion of Transporter genes by WGD, tandem duplication, and telomeric gene recombination. (A) A schematic illustration for the gene syntenic structure and the evolutionary history of six major Transporter genes. The star indicates the WGD event. The names of aerobic fermentative species were underlined. The arrows indicate gene orientations. White arrows represent Group I genes, and black arrows represent Group II genes. The missing orthologous genes are shown as dashed white arrows. The KHT2 gene is present only in some Kluyveromyces lactis strains. The Saccharomyces kluyveri G07678 and K. thermotolerans E02772 were assigned as orthologous genes of HXT6/7 on the basis of synteny structure. This figure was not drawn to scale. (B) The Saccharomyces cerevisiae subtelomeric Transporter genes are found in large newly duplicated regions. The syntenic structures of 30-kb region are drawn in scale, and each chromosome end is labeled by a dot. The arrows represent the gene-coding regions and their orientation. The numbers indicate percentage of identical nucleotide sequences in shaded regions.
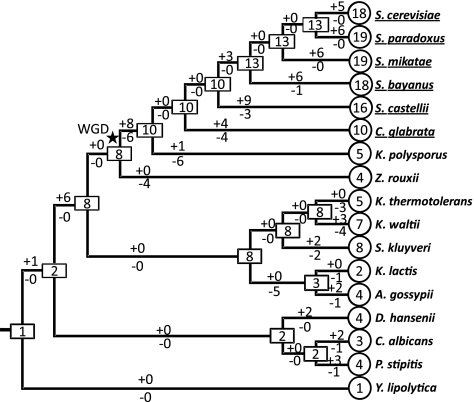
We used the reconciled tree method to estimate the number of Transporter genes in each ancestral node or species and then infer the numbers of gains and losses in each branch of the tree of 17 hemiascomycete yeasts (fig. 5, see Materials and Methods). The overall evolutionary history of Transporter genes of hemiascomycete yeasts can be summarized as follows: 1) The Transporter genes very likely originated from a single gene in the MRCA of hemiascomycete yeasts because the most basal species in this lineage Y. lipolytica contains only one Transporter gene and the duplication event that produced the HXT14 and the common ancestor of the other Transporter genes occurred after divergence of Y. lipolytica from other yeasts (fig. 3 and supplementary fig. S3, Supplementary Material online). However, we cannot rule out the possibility that the ancestral Y. lipolytica contained two Transporter genes and later lost one of them. 2) The Transporter genes have experienced multiple duplication events during the evolution of hemiascomycetes. The first duplication event might have occurred before the divergence of the C. albicans lineage from the S. cerevisiae lineage and produced Group VIII and the common ancestor of the other seven groups. The second major expansion occurred before the divergence of the S. cerevisiae lineage from the A. gossypii lineage and generated the other seven groups (Groups I–VII). Therefore, the MRCA of S. cerevisiae and A. gossypii contained at least eight Transporter genes. We observed distinct evolutionary fates of the eight groups of Transporter genes between aerobic fermentation and respiratory species. Among the 13 species that in principle contain the 8 groups of Transporter genes, 6 of them are aerobic fermentative species and all of them have increased the number of Transporter genes, even up to 19 copies. In contrast, the seven aerobic respiration yeasts show a reduction in Transporter genes (fig. 3). For example, the four Saccharomyces sensu stricto yeasts have gained at least 10 more Transporter genes, but the dairy yeast K. lactis has lost six Transporter genes. Note that K. polysporus, which is the species most distantly related to S. cerevisiae among the post-WGD yeasts (Kurtzman and Robnett 2003), retains only five Transporter genes. Unlike other post-WGD yeasts, K. polysporus cannot carry out efficient fermentation and is considered an intermediate yeast between respiratory and aerobic fermentation yeasts (Fekete et al. 2007; Chen et al. 2008; Jiang et al. 2008). To evaluate the correlation between the number of Transporter genes and the levels of aerobic fermentation in yeasts, we calculated the Spearman's rank correlation coefficients using the ethanol production rate data from two independent studies (Blank et al. 2005; Merico et al. 2007). The coefficient is ρ = 0.79 using data from eight species with ethanol production data in the study of Blank et al. (2005) and ρ = 0.88 using data from six species in the study of Merico et al. (2007), both suggesting a strong positive correlation (supplementary table S2, Supplementary Material online). This and the above observations strongly suggest that gains and losses of Transporter genes have significant effects on the glucose metabolic style.
FIG. 5.
Evolutionary change in the copy number of Transporter genes in 17 hemiascomycete yeasts. The numbers in rectangles represent the numbers of Transporter genes in the extant or ancestral species. The numbers with plus and minus signs indicate the numbers of gene gains and losses, respectively, for a branch. The star indicates the WGD event, and names of aerobic fermentative species were underlined.
Amplification of the Transporter Genes by WGD and Tandem Duplication
The six S. cerevisiae Transporter genes (HXT1 and HXT3–7) that play a major role in glucose uptake form two 3-gene clusters (HXT5-1-4 and HXT3-6-7) on chromosomes IV and VIII, respectively (fig. 1). By comparing the gene neighbor contexts of the two clusters, we found that they share similar syntenic structures (fig. 4A), indicating that the two gene clusters were produced either by segmental duplication or by the WGD. The two duplicate segments are present in all post-WGD yeast genomes, whereas pre-WGD yeasts contain only a single segment, suggesting that the two segments were produced by the WGD (fig. 4A). The number of Transporter genes varies in each cluster among these post-WGD yeasts. The Saccharomyces sensu stricto yeasts (e.g., S. cerevisiae and S. bayanus) contain three genes, S. castellii contains only two, and K. polysporus contains only one Transporter gene in each locus. In the pre-WGD yeasts, S. kluyveri, K. waltii, K. thermotolerans, and A. gossypii contain two Transporter genes, and Z. rouxii and K. lactis each contain only one gene in this locus. One possibility is that there was only one gene at this locus before the WGD, and tandem duplication events have occurred independently in both duplicate segments to produce up to three Transporter genes in each cluster. An alternative hypothesis is that the two neighboring Transporter genes were already present before the divergence of the S. cerevisiae lineage from the A. gossypii lineage. The second hypothesis is more parsimonious, as it requires fewer duplication and deletion events than the first one. Further tandem duplication events on the each cluster have occurred in the MRCA of Saccharomyces sensu stricto yeasts, leading to three genes in each cluster (fig. 4A). In the other aerobic fermentation lineage, S. pombe contains 8 Transporter genes (GTH1–8), 4 of which are tandemly arrayed on chromosome III, suggesting that they were produced by a series of tandem duplication events (data not shown). These results indicate that tandem duplication has contributed to the expansion of Transporter genes in aerobic fermentation yeasts.
Subtelomeric Transporter Genes Have Experienced Frequent Duplication and Deletion Events
Eight S. cerevisiae Transporter genes are located within 30 kb of the ends of several chromosomes (fig. 1). The 8 genes are distributed into 3 Transporter groups: III (HXT9, HXT11, and HXT12), IV (HXT8), and VII (HXT13, HXT15, HXT16, and HXT17) (fig. 3 and supplementary fig. S3, Supplementary Material online). From our phylogenetic analysis, we found that the three groups share very similar evolutionary patterns. First, members of the three groups were lost in many species since their appearance. According to our phylogenetic analysis, the three groups were produced prior to the divergence between the S. cerevisiae and A. gossypii lineages (fig. 3). Among the 13 species that potentially contain these genes, 7 of them lost the Group III genes, 9 lost the Group IV genes, and 8 lost the Group VII genes, indicating that subtelomeric Transporter genes are prone to gene deletion events. Meanwhile, these genes have been frequently duplicated. For example, Group VII genes are only found in 5 species, but all these 5 species contain 4 highly similar copies. Groups II and IV also contain duplicate genes in several species. Furthermore, the duplicate genes in each group are more similar to each other than to the orthologous genes from other species (supplementary fig. S3, Supplementary Material online), indicating that they were produced by species-specific duplication events or have been homogenized by gene conversion events. Therefore, the Transporter genes in subtelomeric regions have experienced frequent duplication and deletion events.
We then compared the syntenic structures for each group of subtelomeric Transporter genes to learn more about how these genes have evolved. We found that large fragments of neighboring sequences of some pairs of subtelomeric Transporter genes are highly similar. Specifically, HXT9 is located near the left end of chromosome IX and HXT12 is located near the left end of chromosome X, but strikingly, the first 21-kb sequences of the two telomeric regions are 99% identical to each other (fig. 4B). Indeed, only 4 nt differences are detected in the first 19-kb nucleotides between the two segments. In this pair of segments, the sequence conservation level is similar in the coding and intergenic regions, suggesting that the two segments were produced by a very recent duplication event. Similar patterns are also observed in other gene pairs: HXT11/HXT12, HXT13/HXT17, and HXT15/HXT16 (fig. 4B). Considering that gene conversion events in yeast usually homogenized a tract of sequence <2 kb (Borts and Haber 1989), it is unlikely that such long highly similar fragments were produced by gene conversion events. The loss of telomere DNA may occur during cell proliferation because DNA polymerases are unable to replicate the end of a linear DNA molecule. It is well known that an important mechanism of telomere length maintenance is interchromosome recombination: the 3′ end of one chromosome invades to a second chromosome and uses it as a template for telomere elongation (Kass-Eisler and Greider 2000). As a consequence, this process leads to highly repetitive and identical subtelomeric sequences. Therefore, the telomere shortening and elongation processes may have contributed to the frequent birth and death of the highly identical Transporter genes in the subtelomeric regions.
Discussion
Gains and Losses of Transporter Genes May Reflect the Adaptation of Yeasts to Their Habitats
Our study revealed that the Transporter and Sensor subfamilies differed greatly in the evolution of gene copy number. The Transporter genes have experienced significant amplifications in both the two lineages that have evolved predominant aerobic fermentation: the S. pombe and S. cerevisiae lineages. Most of the enzymes involved in glycolysis, the tricarboxylic acid cycle, and ethanol fermentation have been conserved between the two lineages, implying that similar glucose metabolic pathways operate in the two yeasts (Wood et al. 2002). Therefore, increase in copy number of Transporter genes might have been used as a common strategy by the two aerobic fermentation lineages to adapt to their glucose-rich niches. In contrast, the copy number of Sensor genes is much less variable in all yeast lineages. The distinct patterns suggest that organisms are able to adapt to their environment by changing the dosage of specific genes.
An interesting observation is that the number of Transporter genes was expanded from 2 to 8 copies before the divergence of the aerobic fermentation lineage (S. cerevisiae) from the respiratory lineage (A. gossypii), which diverged about 100 Ma (Wolfe and Shields 1997; Dietrich et al. 2004; Kellis et al. 2004). Coincidently, the flowering plants (angiosperms), which produced abundance of glucose, became widespread around the same time (Bremer 2000; Moore et al. 2007). Therefore, the expansion of Transporter genes in the MRCA of S. cerevisiae and A. gossypii might be a response to rapid glucose consumption. The WGD that occurred after the divergence of the S. cerevisiae lineage from the A. gossypii lineage led to further expansion of Transporter genes and many other glucose metabolism genes, providing extra raw genetic materials for further development of the aerobic fermentation pathway. The natural habitats of many aerobic fermentation yeasts are rich in fruit juices, which may contain up to 1.5 M glucose, so it is important to have low-affinity glucose transporters to efficiently uptake glucose from environments to feed the need of aerobic fermentation. The expansion of Transporter genes allowed the evolution of low-affinity glucose transporters, such as Hxt1p and Hxt3p, providing a selective advantage for these aerobic fermentation yeasts. Meanwhile, aerobic respiratory yeasts had adapted to glucose-limited habitats and subsequently lost some, if not most, Transporter genes produced by the first major amplification process. For example, the respiratory yeast K. lactis is found mostly in dairy products, which are rich in lactose but limited in glucose. Kluyveromyces lactis has evolved a special ability to assimilate lactose, and thus, a high capacity of glucose transport became unnecessary in its natural habitats. Accordingly, K. lactis has retained only two Transporter genes (RAG1 and HXT14). Interestingly, the RAG1 locus in K. lactis is polymorphic. That is, some K. lactis strains contain two tandemly arrayed genes (KHT1 and KHT2) at the RAG1 locus. It was speculated that the RAG1 gene was produced by recombination of KHT1 and KHT2 (Weirich et al. 1997). Strains with KHT1 and KHT2 show a higher ethanol production rate compared with RAG1-containing strains, probably because strains with more copies of Transporter genes have a higher sugar uptake capacity (Weirich et al. 1997; Milkowski et al. 2001). Because RAG1 encodes a low-affinity glucose transporter and its expression is mainly induced by high concentration of glucose (Chen et al. 1992), uptake of glucose is mainly carried by a high-affinity glucose transporter gene HGT1 in K. lactis on low concentration of glucose (Billard et al. 1996). However, the K. lactis Hgt1p, sharing only a 27% sequence identity to Rag1p, is not a member of the HXT family (supplementary fig. S1, Supplementary Material online). The orthologous genes of K. lactis HGT1 are present in several fungal lineages, and they form a well-supported clade in the phylogenetic tree (supplementary fig. S1, Supplementary Material online), suggesting that the HGT genes have separated from the HXT family before divergence of fungi. Interestingly, the HGT gene is only present in nonaerobic fermentation species, suggesting that they were lost during evolution of both aerobic fermentation lineages. Therefore, it seems that two glucose uptake systems, high affinity (HGT) and low affinity (HXT), have been present in the common ancestor of fungi. The low-affinity system has been rapidly expanded in aerobic fermentation species to adapt to glucose-rich environments, whereas the HGT genes have been retained in respiratory yeast to adapt glucose-limited environments. Meanwhile, due to functional divergence on HXT duplicate genes, new high-affinity genes emerged, such as HXT6 and HXT7, have replaced the function of HGT in S. cerevisiae on low concentration of glucose. The copy number of S. cerevisiae HXT6 and HXT7 can be significantly increased by tandem duplication after incubating in a glucose-limited environment for 450 generations (Brown et al. 1998). In summary, all these studies suggest that gains and losses of Transporter genes in different yeasts reflect adaptation to different natural environments.
Gains and losses of genes that are involved in metabolism of other sugars have been observed in previous studies (Naumov et al. 1996). The S. cerevisiae SUC genes encode β-fructosidase that controls sucrose fermentation. Copy number variation of SUC genes was found in different S. cerevisiae strains that were isolated from different environments. A single SUC2 gene was found in those S. cerevisiae strains isolated from wine fermentation. Grape juice used in wine fermentation contains glucose and fructose, but no sucrose. Two or three SUC genes were found in some other strains that were isolated from distillers and champagne fermentation where sucrose is added (Naumov et al. 1996). The α-galactosidase encoding MEL genes have also been amplified in some specific S. cerevisiae populations (Naumov et al. 1995). Frequent gains and losses of olfactory receptor genes, which are responsible for the detection of odor molecules, are observed in different vertebrate animals, and it is speculated that the copy number changes of this gene family reflect the adaptation of these animals to different environments (Nei and Rooney 2005).
Functional and Regulatory Divergence of Transporter Genes after Gene Duplication
We showed that the six major HXT genes in S. cerevisiae originated from a single gene in ancestral respiratory yeast (fig. 4A). These duplicate genes have subsequently experienced functional divergences and encode different types of glucose transporters (Boles and Hollenberg 1997; Ozcan and Johnston 1999). The HXT1 and HXT3 genes encode low-affinity transporters and play critical roles during aerobic fermentation (Ozcan and Johnston 1995). In contrast, HXT6 and HXT7 encode high-affinity transporters, which are more efficient to uptake glucose under low concentration (Reifenberger et al. 1995). From these observations, it seems that the duplication and functional divergence of HXT genes in aerobic fermentation yeasts have provided the flexibility to adapt to environments where availability of glucose varies enormously in time.
HXT1 and HXT3 are induced by high concentration of glucose and repressed by low concentration of glucose (DeRisi et al. 1997). Their expression levels increase up to 300-folds during active fermentation (Ozcan and Johnston 1995). HXT6 and HXT7 are induced by limited extracellular glucose but repressed by excess amount of environmental glucose (DeRisi et al. 1997). Similarly, high glucose concentration induces the expression of S. pombe HXT members GHT2, GHT5, and GHT6, whereas GHT3 and GHT4 are highly expressed at low glucose concentration (Flores et al. 2000; Leandro et al. 2009). It will be interesting to study the changes in the promoter regions that have contributed to their expression divergence after gene duplication.
The Dynamic Gene Number Change in Subtelomeric Transporter Genes: Adaptation or Drift?
We observed frequent duplication and deletion events on subtelomeric Transporter genes due to high frequency of recombination. Many other sugar metabolism genes, such as MEL, SUC, and MAL, are also located in subtelomeric regions and have similar evolutionary patterns (Carlson et al. 1985; Michels et al. 1992; Turakainen et al. 1993). The malaria parasite Plasmodium falciparum contains genes encoding surface antigen that are used to evade host immune attack, and many of these genes, such as VAR genes, are present in subtelomeric regions and have experienced frequent recombination (Freitas-Junior et al. 2000). These examples imply that expansion of gene families in subtelomeric regions may facilitate the adaptation of species to new environments. However, it is possible that the dynamic nature of subtelomeric Transporter genes might be a by-product of the maintenance of telomere length (Louis 1995; Kass-Eisler and Greider 2000). The olfactory receptor genes, which show frequent gains and losses during evolution, are located in subtelomeric regions in humans (Rouquier et al. 1998; Niimura and Nei 2003). Random variation was suggested to have substantially contributed to the extensive copy number variation of olfactory receptor genes (Nozawa et al. 2007). Therefore, genomic drift, a process of random gene duplication and deletion, has been considered as a common phenomenon in many multigene families (Nei et al. 2008). Considering the high plasticity of yeast subtelomeric regions, the number variation of subtelomeric Transporter genes might be in part due to genomic drift.
Conclusion
The switch from aerobic respiration to aerobic fermentation during the evolution of two yeast lineages was a complicated process, which should require modifications in many steps in this pathway. Amplification of HXT genes was unlikely the sole factor responsible for this metabolism switch. However, the observed strong positive correlation between the copy number of HXT genes and the strength of aerobic fermentation in different yeasts suggests that amplification of HXT genes has facilitated the evolution of aerobic fermentation. Moreover, the variation in HXT gene number might reflect different adaptation strategies of yeasts to their enormously diverse environments. Future studies to investigate what evolutionary changes were involved in the modification of gene regulation of the HXT genes as well as genes involved in respiration process will provide further insights into the origin of aerobic fermentation.
Supplementary Material
Supplementary figures S1–S3 and tables S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We would like to thank Hongzhi Kong, Yong Zhang, Chris Hittinger, and Barak Cohen for critical comments. We also gratefully thank three anonymous reviewers for valuable suggestions. This study was funded by National Institutes of Health grants GM30998.
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Barnett JA, Payne RW, Yarrow D. Cambridge: Cambridge University Press; 2000. Yeasts: Characteristics and Identification. [Google Scholar]
- Billard P, Menart S, Blaisonneau J, Bolotin-Fukuhara M, Fukuhara H, Wesolowski-Louvel M. Glucose uptake in Kluyveromyces lactis: role of the HGT1 gene in glucose transport. J Bacteriol. 1996;178:5860–5866. doi: 10.1128/jb.178.20.5860-5866.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blank LM, Lehmbeck F, Sauer U. Metabolic-flux and network analysis in fourteen hemiascomycetous yeasts. FEMS Yeast Res. 2005;5:545–558. doi: 10.1016/j.femsyr.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Boles E, Hollenberg CP. The molecular genetics of hexose transport in yeasts. FEMS Microbiol Rev. 1997;21:85–111. doi: 10.1111/j.1574-6976.1997.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Borts RH, Haber JE. Length and distribution of meiotic gene conversion tracts and crossovers in Saccharomyces cerevisiae. Genetics. 1989;123:69–80. doi: 10.1093/genetics/123.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bremer K. Early Cretaceous lineages of monocot flowering plants. Proc Natl Acad Sci U S A. 2000;97:4707–4711. doi: 10.1073/pnas.080421597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Todd KM, Rosenzweig RF. Multiple duplications of yeast hexose transport genes in response to selection in a glucose-limited environment. Mol Biol Evol. 1998;15:931–942. doi: 10.1093/oxfordjournals.molbev.a026009. [DOI] [PubMed] [Google Scholar]
- Brown V, Sexton JA, Johnston M. A glucose sensor in Candida albicans. Eukaryot Cell. 2006;5:1726–1737. doi: 10.1128/EC.00186-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne KP, Wolfe KH. The Yeast Gene Order Browser: combining curated homology and syntenic context reveals gene fate in polyploid species. Genome Res. 2005;15:1456–1461. doi: 10.1101/gr.3672305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson M, Celenza JL, Eng FJ. Evolution of the dispersed SUC gene family of Saccharomyces by rearrangements of chromosome telomeres. Mol Cell Biol. 1985;5:2894–2902. doi: 10.1128/mcb.5.11.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Xu L, Gu Z. Regulation dynamics of WGD genes during yeast metabolic oscillation. Mol Biol Evol. 2008;25:2513–2516. doi: 10.1093/molbev/msn212. [DOI] [PubMed] [Google Scholar]
- Chen XJ, Wesolowski-Louvel M, Fukuhara H. Glucose transport in the yeast Kluyveromyces lactis. II. Transcriptional regulation of the glucose transporter gene RAG1. Mol Gen Genet. 1992;233:97–105. doi: 10.1007/BF00587566. [DOI] [PubMed] [Google Scholar]
- Conant GC, Wolfe KH. Increased glycolytic flux as an outcome of whole-genome duplication in yeast. Mol Syst Biol. 2007;3:129. doi: 10.1038/msb4100170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Deken RH. The Crabtree effect: a regulatory system in yeast. J Gen Microbiol. 1966;44:149–156. doi: 10.1099/00221287-44-2-149. [DOI] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Diderich JA, Teusink B, Valkier J, Anjos J, Spencer-Martins I, van Dam K, Walsh MC. Strategies to determine the extent of control exerted by glucose transport on glycolytic flux in the yeast Saccharomyces bayanus. Microbiology. 1999;145(Pt 12):3447–3454. doi: 10.1099/00221287-145-12-3447. [DOI] [PubMed] [Google Scholar]
- Dietrich FS, Voegeli S, Brachat S, et al. (14 co-authors) The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbing K, Larsson C, Bill RM, Albers E, Snoep JL, Boles E, Hohmann S, Gustafsson L. Role of hexose transport in control of glycolytic flux in Saccharomyces cerevisiae. Appl Environ Microbiol. 2004;70:5323–5330. doi: 10.1128/AEM.70.9.5323-5330.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete V, Cierna M, Polakova S, Piskur J, Sulo P. Transition of the ability to generate petites in the Saccharomyces/Kluyveromyces complex. FEMS Yeast Res. 2007;7:1237–1247. doi: 10.1111/j.1567-1364.2007.00287.x. [DOI] [PubMed] [Google Scholar]
- Fiechter A, Fuhrmann GF, Kappeli O. Regulation of glucose metabolism in growing yeast cells. Adv Microb Physiol. 1981;22:123–183. doi: 10.1016/s0065-2911(08)60327-6. [DOI] [PubMed] [Google Scholar]
- Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flores CL, Rodriguez C, Petit T, Gancedo C. Carbohydrate and energy-yielding metabolism in non-conventional yeasts. FEMS Microbiol Rev. 2000;24:507–529. doi: 10.1111/j.1574-6976.2000.tb00553.x. [DOI] [PubMed] [Google Scholar]
- Freitas-Junior LH, Bottius E, Pirrit LA, Deitsch KW, Scheidig C, Guinet F, Nehrbass U, Wellems TE, Scherf A. Frequent ectopic recombination of virulence factor genes in telomeric chromosome clusters of P. falciparum. Nature. 2000;407:1018–1022. doi: 10.1038/35039531. [DOI] [PubMed] [Google Scholar]
- Gancedo C, Serrano R. Energy-yielding metabolism. London: Academic Press; 1989. [Google Scholar]
- Goffrini P, Ferrero I, Donnini C. Respiration-dependent utilization of sugars in yeasts: a determinant role for sugar transporters. J Bacteriol. 2002;184:427–432. doi: 10.1128/JB.184.2.427-432.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffrini P, Wesolowski-Louvel M, Ferrero I, Fukuhara H. RAG1 gene of the yeast Kluyveromyces lactis codes for a sugar transporter. Nucleic Acids Res. 1990;18:5294. doi: 10.1093/nar/18.17.5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- Heiland S, Radovanovic N, Hofer M, Winderickx J, Lichtenberg H. Multiple hexose transporters of Schizosaccharomyces pombe. J Bacteriol. 2000;182:2153–2162. doi: 10.1128/jb.182.8.2153-2162.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Madan A. CAP3: a DNA sequence assembly program. Genome Res. 1999;9:868–877. doi: 10.1101/gr.9.9.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Jiang H, Guan W, Pinney D, Wang W, Gu Z. Relaxation of yeast mitochondrial functions after whole-genome duplication. Genome Res. 2008;18:1466–1471. doi: 10.1101/gr.074674.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass-Eisler A, Greider CW. Recombination in telomere-length maintenance. Trends Biochem Sci. 2000;25:200–204. doi: 10.1016/s0968-0004(00)01557-7. [DOI] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature. 2004;428:617–624. doi: 10.1038/nature02424. [DOI] [PubMed] [Google Scholar]
- Kurtzman CP, Robnett CJ. Phylogenetic relationships among yeasts of the 'Saccharomyces complex' determined from multigene sequence analyses. FEMS Yeast Res. 2003;3:417–432. doi: 10.1016/S1567-1356(03)00012-6. [DOI] [PubMed] [Google Scholar]
- Leandro MJ, Fonseca C, Goncalves P. Hexose and pentose transport in ascomycetous yeasts: an overview. FEMS Yeast Res. 2009;9:511–525. doi: 10.1111/j.1567-1364.2009.00509.x. [DOI] [PubMed] [Google Scholar]
- Louis EJ. The chromosome ends of Saccharomyces cerevisiae. Yeast. 1995;11:1553–1573. doi: 10.1002/yea.320111604. [DOI] [PubMed] [Google Scholar]
- Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151–1155. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- Merico A, Sulo P, Piskur J, Compagno C. Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J. 2007;274:976–989. doi: 10.1111/j.1742-4658.2007.05645.x. [DOI] [PubMed] [Google Scholar]
- Michels CA, Read E, Nat K, Charron MJ. The telomere-associated MAL3 locus of Saccharomyces is a tandem array of repeated genes. Yeast. 1992;8:655–665. doi: 10.1002/yea.320080809. [DOI] [PubMed] [Google Scholar]
- Milkowski C, Krampe S, Weirich J, Hasse V, Boles E, Breunig KD. Feedback regulation of glucose transporter gene transcription in Kluyveromyces lactis by glucose uptake. J Bacteriol. 2001;183:5223–5229. doi: 10.1128/JB.183.18.5223-5229.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore MJ, Bell CD, Soltis PS, Soltis DE. Using plastid genome-scale data to resolve enigmatic relationships among basal angiosperms. Proc Natl Acad Sci U S A. 2007;104:19363–19368. doi: 10.1073/pnas.0708072104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J, Nei M. Evolutionary change of the numbers of homeobox genes in bilateral animals. Mol Biol Evol. 2005;22:2386–2394. doi: 10.1093/molbev/msi229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumov GI, Naumova ES, Korhola MP. Chromosomal polymorphism of MEL genes in some populations of Saccharomyces cerevisiae. FEMS Microbiol Lett. 1995;127:41–45. doi: 10.1111/j.1574-6968.1995.tb07447.x. [DOI] [PubMed] [Google Scholar]
- Naumov GI, Naumova ES, Sancho ED, Korhola MP. Polymeric SUC genes in natural populations of Saccharomyces cerevisiae. FEMS Microbiol Lett. 1996;135:31–35. doi: 10.1111/j.1574-6968.1996.tb07962.x. [DOI] [PubMed] [Google Scholar]
- Nei M, Niimura Y, Nozawa M. The evolution of animal chemosensory receptor gene repertoires: roles of chance and necessity. Nat Rev Genet. 2008;9:951–963. doi: 10.1038/nrg2480. [DOI] [PubMed] [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholas KB, Nicholas HB, Jr., Deerfield DW., 2nd GeneDoc: analysis and visualization of genetic variation. EMBNET News. 1997;4:1–4. [Google Scholar]
- Niimura Y, Nei M. Evolution of olfactory receptor genes in the human genome. Proc Natl Acad Sci U S A. 2003;100:12235–12240. doi: 10.1073/pnas.1635157100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nozawa M, Kawahara Y, Nei M. Genomic drift and copy number variation of sensory receptor genes in humans. Proc Natl Acad Sci U S A. 2007;104:20421–20426. doi: 10.1073/pnas.0709956104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otterstedt K, Larsson C, Bill RM, Stahlberg A, Boles E, Hohmann S, Gustafsson L. Switching the mode of metabolism in the yeast Saccharomyces cerevisiae. EMBO Rep. 2004;5:532–537. doi: 10.1038/sj.embor.7400132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Dover J, Johnston M. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 1998;17:2566–2573. doi: 10.1093/emboj/17.9.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Johnston M. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol Cell Biol. 1995;15:1564–1572. doi: 10.1128/mcb.15.3.1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozcan S, Johnston M. Function and regulation of yeast hexose transporters. Microbiol Mol Biol Rev. 1999;63:554–569. doi: 10.1128/mmbr.63.3.554-569.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Pritchard L, Kell DB. Schemes of flux control in a model of Saccharomyces cerevisiae glycolysis. Eur J Biochem. 2002;269:3894–3904. doi: 10.1046/j.1432-1033.2002.03055.x. [DOI] [PubMed] [Google Scholar]
- Reifenberger E, Freidel K, Ciriacy M. Identification of novel HXT genes in Saccharomyces cerevisiae reveals the impact of individual hexose transporters on glycolytic flux. Mol Microbiol. 1995;16:157–167. doi: 10.1111/j.1365-2958.1995.tb02400.x. [DOI] [PubMed] [Google Scholar]
- Rouquier S, Taviaux S, Trask BJ, Brand-Arpon V, van den Engh G, Demaille J, Giorgi D. Distribution of olfactory receptor genes in the human genome. Nat Genet. 1998;18:243–250. doi: 10.1038/ng0398-243. [DOI] [PubMed] [Google Scholar]
- Slowinski JB, Knight A, Rooney AP. Inferring species trees from gene trees: a phylogenetic analysis of the Elapidae (Serpentes) based on the amino acid sequences of venom proteins. Mol Phylogenet Evol. 1997;8:349–362. doi: 10.1006/mpev.1997.0434. [DOI] [PubMed] [Google Scholar]
- Souciet JL, Dujon B, Gaillardin C, et al. (54 co-authors) Comparative genomics of protoploid Saccharomycetaceae. Genome Res. 2009;19:1696–1709. doi: 10.1101/gr.091546.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4. Molecular Evolutionary Genetics Analysis (MEGA) Software Version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet. 2005;37:630–635. doi: 10.1038/ng1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turakainen H, Naumov G, Naumova E, Korhola M. Physical mapping of the MEL gene family in Saccharomyces cerevisiae. Curr Genet. 1993;24:461–464. doi: 10.1007/BF00351706. [DOI] [PubMed] [Google Scholar]
- Weirich J, Goffrini P, Kuger P, Ferrero I, Breunig KD. Influence of mutations in hexose-transporter genes on glucose repression in Kluyveromyces lactis. Eur J Biochem. 1997;249:248–257. doi: 10.1111/j.1432-1033.1997.t01-1-00248.x. [DOI] [PubMed] [Google Scholar]
- Wieczorke R, Krampe S, Weierstall T, Freidel K, Hollenberg CP, Boles E. Concurrent knock-out of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 1999;464:123–128. doi: 10.1016/s0014-5793(99)01698-1. [DOI] [PubMed] [Google Scholar]
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, et al. (134 co-authors) The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Ye L, Kruckeberg AL, Berden JA, van Dam K. Growth and glucose repression are controlled by glucose transport in Saccharomyces cerevisiae cells containing only one glucose transporter. J Bacteriol. 1999;181:4673–4675. doi: 10.1128/jb.181.15.4673-4675.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.