Abstract
The genotypic signature of spatially varying selection is ubiquitous across the Drosophila melanogaster genome. Spatially structured adaptive phenotypic differences are also commonly found, particularly along New World and Australian latitudinal gradients. However, investigation of gene expression variation in one or multiple environments across these well-studied populations is surprisingly limited. Here, we report genome-wide transcript levels of tropical and temperate eastern Australian populations reared at two temperatures. As expected, a large number of genes exhibit geographic origin-dependent expression plasticity. Less expected was evidence for an enrichment of down-regulated genes in both temperate and tropical populations when lines were reared at the temperature less commonly encountered in the native range; that is, evidence for significant differences in a “directionality” of plasticity across these two climatic regions. We also report evidence of small scale “neighborhood effects” around those genes significant for geographic origin-dependent plasticity, a result consistent with the evolution of high level, likely chromatin based gene regulation during range expansion in D. melanogaster populations.
Keywords: clinal variation, expression plasticity, chromatin, temperature, Drosophila
Introduction
Population genetic processes such as mutation–selection balance, genetic drift, and balancing selection contribute to the abundant genetic variation observed in natural populations. One particularly pervasive form of balancing selection, spatially varying selection, can maintain variation across heterogeneous landscapes if fitness variation is sufficiently great to overcome the homogenizing effects of gene flow (Felsenstein 1976; Endler 1977). The resultant adaptive phenotypic differences across populations often exhibit plasticity in response to different environmental conditions (Robinson and Partridge 2001; Gilchrist and Huey 2004; Swindell et al. 2007; Liefting et al. 2009).
Genetic variation for phenotypic plasticity, formally referred to as genotype × environment interactions (GEI), is widespread (DeWitt and Scheiner 2004). In Drosophila pseudoobscura, for example, as rearing temperature shifts from 14 to 21 °C, abdominal bristle number increases in some genotypes but decreases in others (Gupta and Lewontin 1982). In principle, both neutral and nonneutral evolutionary forces could maintain variants exhibiting GEI within and between natural populations. However, clinal variation for GEI, particularly along environmental gradients where gene flow is high, represents a strong case for selection contributing to geographically structured genetic variation for phenotypic plasticity.
New World and Australian latitudinal gradients have emerged as classic ecological contexts for exploring clinal variation, especially in the model system D. melanogaster (e.g., Stalker 1976; Oakeshott et al. 1982; Berry and Kreitman 1993; Verrelli and Eanes 2001; Turner et al. 2008). Microsatellite loci show virtually no spatial structure (Gockel et al. 2001; Kennington et al. 2003), yet many single nucleotide polymorphisms (Verrelli and Eanes 2001; Sezgin et al. 2004; Turner et al. 2008), inversions (Stalker 1976; Knibb et al. 1981; Umina et al. 2005), and phenotypes (e.g., Gockel et al. 2001; Hoffmann et al. 2002; Schmidt et al. 2005) vary clinally across latitude, implicating the action of spatially varying selection. Moreover, several clinal phenotypes exhibit genetic variation for plasticity (Robinson and Partridge 2001; Gilchrist and Huey 2004; Liefting et al. 2009).
Although there are several examples of clinal gene expression variation (Whitehead and Crawford 2006; Swindell et al. 2007), there are no published investigations of clinal variation for expression plasticity across D. melanogaster populations derived from these well-studied geographic regions, on a single gene or a whole-genome scale (i.e., clinal GEI, where “genotype” refers more precisely to “genotype of geographic origin”). Given that transcript abundance often exhibits plasticity across environmental conditions (Causton et al. 2001; Kreps et al. 2002; Bochdanovits et al. 2003), one would expect many genes to exhibit expression plasticity correlated with locally adapted genotypes.
If such variation exists, it may be attributable to variation at local regulatory sequence or at regulatory proteins, such as transcription factors (Li et al. 2006; Smith and Kruglyak 2008). Genes regulated by chromatin remodeling factors may harbor a distinct transcriptional signature, namely a “neighborhood effect” of correlated expression change (Sproul et al. 2005; Batada et al. 2007). The physical scale of this effect (generally greater than five genes) renders unlikely a role for variation at local features, such as bidirectional promoters or read-through transcription (Drosophila: Boutanaev et al. 2002; Spellman and Rubin 2002; Kalmykova et al. 2005; Blanco et al. 2008; Mezey et al. 2008; humans: Caron et al. 2001; zebrafish: Ng et al. 2009; mouse: Li et al. 2005). Moreover, several recent studies have documented an association between chromatin structure and physical coexpression. For example, genomic regions harboring elevated nucleosome occupancy are associated with elevated coexpression in yeast cells (Batada et al. 2007). In fruit flies, the genomic region surrounding a cluster of nonhomologous, male-germline coexpressed genes is sensitive to DNase I in germ cells but not sensitive in somatic cells (Kalmykova et al. 2005). Recent evidence that chromatin remodeling factors show clinal variation (Harr et al. 2002; Levine and Begun 2008) suggests that chromatin regulation may contribute to adaptive expression variation along latitudinal gradients. If so, clinal expression plasticity may span physically clustered gene neighborhoods.
Here, using natural populations of D. melanogaster from the tips of the well-studied eastern Australia latitudinal cline, we investigate clinal expression plasticity by testing for an interaction between genotype and rearing temperature, where genotype refers to either tropical or temperate origin. We also characterize genotypic variation for expression plasticity at physically clustered unrelated genes to test for evidence of adaptive chromatin domain organization.
Materials and Methods
D. melanogaster isofemale lines were collected from temperate Australia (Cygnet, Southern Tasmania, latitude 43°09′12″) and tropical Australia (Innisfail, Queensland 17°31′20″) during the summer of 2008 (provided by A. Hoffmann). The long-term monthly averages for the temperate region range from 11.9 to 21.9 °C and the equivalent averages for the tropical region range from 24.1 to 30.8 °C (www.weatherzone.com.au).
Fifteen isofemale lines from each population were reared at 25 °C on standard medium for more than five generations before two replicate single pair matings per isofemale line were aspirated into two separate vials and placed immediately into either an 18 °C or a 30 °C incubator with 12 h day/night cycles. We therefore had four treatment combinations represented by 15 single pair matings per treatment: tropical at 18 °C, temperate at 18 °C, tropical at 30°, and temperate at 30 °C. The choice of temperatures was based on both the native range of temperatures experienced by these populations in nature (see above) and the known fertility limits of natural populations of D. melanogaster (Chakir et al. 2002).
Three pools of 15 males were generated per treatment combination by collecting three F1 adult males from each vial (using light CO2) 24–48 h posteclosion, and adding each male to one of three pools. These pools therefore served as both technical and biological replication (the isofemales lines are highly heterozygous). Prior to freezing down each pool of 15 males in liquid nitrogen, flies were allowed to acclimate at room temperature for 120 min to enrich for (though not rigorously account for) expression plasticity specifically associated with the temperature at which development occurred.
Total RNA was extracted using Trizol (Invitrogen), followed by cDNA synthesis, amplification, and labeling using the Affymetrix 3′IVT Express Kit (Affymetrix). Three replicate Affymetrix GeneChip 2.0 expression arrays were used for each population sample at each of two temperatures for a total of 12 arrays. Arrays were probed, stained, washed, and scanned at the UC Davis Microarray Core Facility following Affymetrix guidelines.
Probesets called absent by the Affy GCOS software on at least 6 of the 12 arrays were removed from the analysis, leaving 12,560 of the original 18,952 probesets. All chips were normalized and background corrected using rma from the Bioconductor affy package (Gentleman et al. 2004) followed by calculation of the mean log2 expression intensity. The genefilter package was used to remove all probesets below an interquartile range of 0.5 across arrays (Scholtens and von Heydebreck 2005), leaving 1,760 probesets for the analysis (see supplementary fig. 1a–c, Supplementary Material online for quality assessment figures). These 1,760 probesets therefore represent only those exhibiting substantial variation across arrays. Using the limma package, three a priori contrasts were set up: 1) tropical at 18 °C − tropical 30 °C; 2) temperate at 18 °C − temperate at 30 °C; and 3) (temperate at 18 °C − temperate at 30 °C) − (tropical at 18 °C − tropical at 30 °C). The first two contrasts test for plasticity across the two temperatures for each genotype pool separately, whereas the third contrast effectively tests for the interaction of temperature and geographic origin (“GEI contrast” hereafter). Although traditional GEI cannot be tested for directly using pooled samples (rather than individual genotypes), plasticity differences between the tropical and temperate genotype pools should reflect, on average, differences between these pools that are greater than differences within pools. Note that all of our contrasts use within-region comparisons and so are not confounded by intensity differences that may result from sequence differentiation between populations (due to differential hybridization of alternative alleles). False discovery rates (FDR) were generated following (Benjamini and Hochberg 1995). Gene ontology analyses were conducted using GeneMerge, which uses hypergeometric distributions to generate probabilities associated with overrepresentation in particular categories (Castillo-Davis and Hartl 2003). We report the Bonferroni corrected P values (“e-values”) generated by GeneMerge.
To investigate whether geographic origin-dependent expression plasticity (the GEI contrast) is manifest at the level of chromosomal domain, we asked whether GEI in the neighborhood of significant genes is significantly different from randomly assembly neighborhoods. We positioned windows of varying sizes (7, 11, 15, … ,61 genes) centered on each of the top 50 genes (FDR < 0.09) from the GEI contrast and then calculated the median t-statistic from the “unfiltered genes,” excluding the significant gene. Windows containing a potential paralog of the significant gene were removed from the analysis. We inferred paralogy if a BLAST analysis (Altschul et al. 1990) using all coding sequences of significant genes as both the query and the database returned an e-value of 1e−5 or less (a very conservative cutoff) and overlapped >25% of coding sequence of the significant gene. The distribution of median t-statistics for each window size was compared with an empirically derived null distribution generated in two different ways. In the first, we randomly picked genes (there were no observed chromosome-specific effects) in numbers corresponding to those used to calculate observed medians (6, 10, 14,… genes) for windows centered on the top 50 genes. This was repeated 1,000 times to generate P values. In the second, we randomly picked 50 genes, generated local windows of sizes corresponding to those used in the analysis of the actual data and calculated the median plasticity of these randomly placed windows. This was repeated 1,000 times. The results of the two approaches were qualitatively equivalent, so only results from the former approach are presented. All statistical analyses were conducted in R.
Results
Within-Region Expression Differences across Temperatures
At a FDR of 0.10, 11 genes were expressed differently at 18° versus 30° for the temperate population sample and 44 genes for the tropical population sample (supplementary table S1a and S1b, Supplementary Material online). At an FDR of 0.125, the corresponding numbers of significant genes for the temperate and tropical population samples were 48 and 341, respectively. The associated gene lists with log-fold change, t-statistic, probability value, FDR, gene name, and location can be found in supplementary table S1a and S1b, Supplementary Material online.
We observed a strong statistical signature of “directionality” in the plasticity data, with a consistent excess of significant genes characterized by higher transcript abundance in the rearing temperature more commonly encountered in the geographic origin of the population pool. Our a priori contrast design predicts that if such “home” versus “away” dynamics hold, we would observe an excess of positive t-statistics in the temperate population (contrast with home italicized: temperate at 18 °C − temperate at 30 °C) and an excess of negative t-statistics in the tropical population sample (tropical at 18 °C − tropical at 30 °C). We indeed observed a significant excess of positive t-statistics in the temperate population sample (genes with higher 18 °C expression) and a significant excess of negative t-statistics in the tropical population sample (genes with higher expression at 30 °C) for both the 25 and the 50 most significant genes (fig. 1; supplementary table S1a and S1b, Supplementary Material online). The next 50 most significant genes exhibited the same pattern, although only the tropical region was significantly enriched (fig. 1). These results suggest that for a gene exhibiting highly significant expression plasticity in our experiment, transcript abundance tends to be higher at the rearing temperature most frequently occurring in the population of origin (see Methods).
FIG. 1.
t-statistics generated from a priori contrasts comparing 18 °C and 30 °C transcript abundance plotted against rank order based on significance. Across the top of the figure, the number of positive and negative t-statistics is reported for nonoverlapping, 50 gene segments of the rank order list. The segment containing the 50 most significant genes also reports these data for the 25 most significant genes in each pool. Asterisks refer to binomial probability associated with enrichment of positive (temperate) or negative (tropical) t-statistics (***P < 0.001, **P < 0.01.
Gene Ontology analyses for the 50 most significant genes exhibiting plasticity across temperatures in the temperate pool (FDR ∼ 0.13) revealed that categories “galactose binding” (e = 0.019), “monophenol monooxygenase” (e = 0.003), “oxygen transport” (e = 0.040), and “defense response” (e = 0.008) are significantly overrepresented, whereas in the tropical pool (FDR ∼ 0.11), “lipase activity” (e = 0.002), “steriod metabolism” (e = 0.022), and “chitin metabolism” (e = 0.044) are overrepresented. In the temperate zone only, multiple odorant-binding protein genes exhibited plasticity (Obp19a, Obp8a), whereas in the tropical zone, many more metabolic genes emerged as particularly plastic (e.g., CG6776, CG6012, and CG13325). Both lipase- and carbohydrate metabolism are known to show clinal variation (reviewed in Eanes 1999; de Jong and Bochdanovits 2003), and several odorant-binding proteins are known to vary clinally along the Australian cline (Turner et al. 2008), though not the two described here. Frost, a gene associated with cold shock recovery (Sinclair et al. 2007), was expressed at significantly lower levels in the flies raised at 18° (supplementary table S1b, Supplementary Mateiral online) in the tropical pool. Finally, eight genes significant in this analysis (FDR < 0.125) are also significant in a recently published data set of up/down regulation across the D. melanogaster genome in response to artificial selection for increased resistance to chill coma (Telonis-Scott et al. 2009; supplemental table S2, Supplementary Material online). These experimental lines were derived from the same region of Australia as the tropical pool sampled in this analysis, and seven of the eight are significant for expression plasticity only in the tropical pool.
Geographic Origin-Dependent Plasticity
Our interest in GEI motivated a test for geographic origin-dependent plasticity differences (see Methods for our definition of “GEI” using pooled samples) by evaluating the contrast: (temperate at 18 °C − temperate at 30 °C) − (tropical at 18 °C − tropical at 30 °C). At an FDR of 0.10, 56 genes showed significant GEI, whereas at an FDR 0.125, 64 genes were significant. The associated gene list with log-fold change, t-statistic, probability value, and FDR can be found in supplementary table S1c, Supplementary Material online.
The directional biases observed for within-tropical and within-temperate plasticity contrasts predict the possibility of directional bias in this GEI contrast, but only if the same genes exhibit opposing, geographic origin-specific, directional biases. In other words, if a given gene exhibits a positive t-statistic in the temperate data (higher 18° expression) and/or a negative t-statistic in the tropical data (higher 30° expression), then we predict a positive t-statistic for the combined contrast (home expression italicized: [temperate at 18° − temperate at 30°] − [tropical at 18°–tropical at 30°]). We indeed observed a significant excess of positive t-statistics for the GEI contrast. In the top 25 genes (FDR < 0.06), there were 17 positive t-statistics (binomial probability = 0.05) and in the top 50 (FDR < 0.0.09), 34 positive t-statistics (binomial probability = 0.007; see supplementary table S1c, Supplementary Mateiral online). The six most significant GEI genes, five of which exhibit this home versus away pattern, provide a striking illustration of this enrichment (fig. 2).
FIG. 2.
Mean probeset intensity of the six most significant genes from the GEI contrast across the two rearing temperature treatments. Note that home in these figures refers to the temperate population reared at 18 °C and the tropical population reared at 30 °C.
Gene Ontology analysis of the 50 most significant GEI genes (FDR < 0.09) from this analysis suggests enrichment in the categories galactose binding (e = 0.045), “triacylglycerol lipase activity” (e = 0.018), “oxidoreductase activity” (e = 0.014), “monooxygenase activity” (e = 0.019), and “heme binding” (e = 0.042). Interestingly, 20 of these top 50 genes overlap the 50 genes most significantly diverged for transcript level in a study comparing African and European populations (Hutter et al. 2008; supplementary table S2, Supplementary Material online). Finally, two of these 50 most significant “GEI” genes are also found in regions significant for sequence differentiation in Australia (Cyp6a17, CG1304) (Turner et al. 2008).
In summary, a considerable number of D. melanogaster genes exhibit evidence of transcript abundance plasticity in samples collected from the endpoints of the Australian latitudinal gradient, particularly in the tropical genotype pool. There appears to be geographic structure to expression plasticity variation, with tropical genotypes raised at 30 °C and temperate genotypes raised at 18 °C exhibiting higher transcript levels than the corresponding genotypes raised in opposite conditions consistent with higher expression at home and lower expression away.
Chromatin Organization and Geographic Origin-Dependent Expression Plasticity
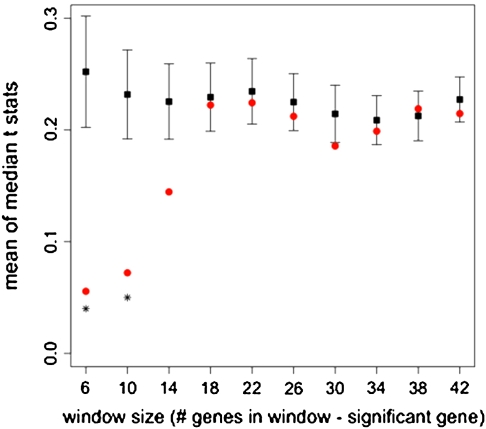
If spatially varying selection at chromatin remodeling proteins underlies a portion of the expression variation described above, expression plasticity may also vary across genotype pools at a scale greater than individual genes. We tested for such neighborhood effects by testing for elevated GEI in the genes located near the most significant genes in our GEI analysis (see Methods). We found such neighborhood effects for geographic origin-dependent expression plasticity at the scale of 6–10 genes but not at larger physical scales (fig. 3). Interestingly, the distribution of medians around significant genes was shifted toward zero relative to the genome-wide average, suggesting that more small values occur around these significant genes. Any one (or more) of the four components of the GEI contrast may underlie this observation.
FIG. 3.
Means of the distributions of median t-statistics for a range of window sizes. Black squares refer to windows of randomized gene order and red circles refer to windows around significant genes. Asterisks indicate significant difference (at P = 0.05) of the means generated from the resampling analysis. Note that the randomized median GEI t-statistics span 0.2–0.4, a range of nonzero, positive values that likely result from the observed genome-wide difference between the contrast components (temperate plasticity t-statistic mean ≈ 0.0 and tropical plasticity t-statistic mean ≈ −0.3).
Discussion
We have reported genome-wide rearing temperature-dependent transcript levels of temperate and tropical D. melanogaster population samples from the endpoints of a well-studied cline. Although the relationship between transcript abundance and fitness remains unclear (Feder and Walser 2005), genetic differences maintained despite the homogenizing force of gene flow across eastern Australia (Gockel et al. 2001; Kennington et al. 2003) suggest that the gene expression phenotypes documented here may be adaptive. Moreover, our data suggest that a component of the expression variation differences between populations may be explained by changes in chromatin conformation at the scale of 6–10 genes. The fact that we detected significant geographic origin-dependent plasticity for several individual genes and a genomic signature of chromatin regulation in our relatively crude experiment (two temperatures, whole animal transcript abundance) suggests that more detailed analyses will reveal abundant genetic variation for environment-dependent gene expression.
We observed a strong trend toward many more genes showing expression plasticity in the tropical genotype pool than in the temperate genotype pool. This difference could simply be a function of the two particular rearing temperatures used in our experiment, that is, an artifact. For example, temperate genotypes could be similarly (or more) plastic across rearing temperatures than tropical ones, but the 18 °C treatment could have been be more stressful for the tropical lines than the 30 °C for the temperate lines, thereby inducing a more dramatic plasticity response. Alternatively, if our results reflect a biologically relevant phenomenon, they suggest that temperate genotypes may be more buffered against temperature variation occurring during development. Further experiments are required to distinguish between these two possibilities.
Our results suggest a general pattern of lower transcript abundance in away environments (i.e., temperate genotypes at 30° and tropical genotypes at 18°). Environmental stress (oxidative stress, ultraviolet, heat, etc.) in Saccharomyces cerevisiae induces dramatic genome-wide down-regulation of vast numbers of genes that results in suspended basal cellular processes until the stress is removed (Causton et al. 2001). Thus, the directional plasticity found in our data may reflect a passive stress response in the away treatment rather than adaptive plasticity. Future studies may reveal that down-regulation (of genes unrelated to stress response induction) is generally observed when genotype “mismatches” environment.
Recent evidence of spatially varying selection at several chromatin remodeling factors raises the possibility of high-level regulation of adaptive gene expression variation. The neighborhood effects described here support the idea that chromatin regulation contributes to temperature-dependent genotypic variation in transcript abundance. The observed scale of effect is consistent with findings from other physical coexpression papers in Drosophila (Spellman and Rubin 2002; Mezey et al. 2008). If chromatin remodeling factors are at least in part responsible for elevated buffering found in the temperate pool, we might expect neighborhood effects to be more pronounced in the tropical pool. We tested this post hoc hypothesis by performing a similar neighborhood analysis using the absolute values of the t-statistics from the within-genotype plasticity data. We observed no neighborhood effect for temperate genotype plasticity at any window size but a significant neighborhood effect at six gene-windows for the tropical genotypes (supplementary fig. S2a and S2b, Supplementary Material onine). These data are consistent with the idea that temperate genotypes at chromatin factors may buffer environmental variation more effectively than tropical ones or alternatively (again), an artifact. Intriguingly, the population genetic signature of recent strong selection at clinal chromatin remodeling factors in temperate populations (Harr et al. 2002; Levine and Begun 2008) also supports this idea. Moreover, five chromatin remodeling factors putatively targeted by spatially varying selection (Harr et al. 2002; Levine and Begun 2008) are associated with either the Polycomb or the Trithorax group proteins—a class of chromatin remodeling factors that maintain gene expression programs through mitosis at target loci regulating developmental programs and stem cell maintenance (Schwartz and Pirrotta 2007).
The “countergradient variation” model (Levins 1968; Conover and Schultz 1995; Grether 2005) describes the distribution of variation across environmentally heterogeneous habitats such that the effects of the environment “oppose” the effects of selection on the focal phenotype. The mean trait values are consequently more similar across natural populations experiencing different (native) environments than those raised in a single, common environment. Stabilizing selection on phenotypes that are environmentally plastic may result in underlying adaptive evolution across populations, even though mean phenotypes are the same across populations in their native habitat.
Although we have not tested directly for stabilizing selection on transcript level, our data fit the countergradient variation/genetic compensation model well. At a disproportionate number of loci, we observed relatively high levels of gene expression for home temperatures compared with the frequent down-regulation observed for away temperatures. Thus, the expression at a disproportionate number of genes was more similar across home environments than within a common environment. The contrasting responses to the same temperature shift at common loci suggest geographically differentiated genetic variation for plasticity, the regulation of which may be at least partially chromatin based.
These findings are consistent with the idea that many genes are evolving adaptively to maintain expression levels associated with fundamental developmental processes across variable environments. Integration of investigations of clinal developmental phenotypes, chromatin organization, and gene expression may reveal novel chromatin-based mechanisms of adaptive evolution.
Supplementary Material
Supplementary tables S1a–S1c, S2 and figures S1, S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
The authors are grateful to A. Hoffmann for generously providing the Australian isofemales lines used in this study. The authors also thank M. Rolston at the UC Davis Microarray facility for sample preparation, hybridization, and scanning, and G. Coop and J. Haloin for extremely useful discussions about the neighborhood analyses. M. Hahn, G. Lee, and A. Hoffmann read earlier drafts of the manuscript and provided helpful feedback. The manuscript was significantly improved by the critical comments of two anonymous reviewers. This research was supported by a National Science Foundation Doctoral Dissertation Improvement Grant to M.T.L. and D.J.B. and the National Institute of Health grant GM084056 to D.J.B. M.T.L. was supported by a UC Davis Dissertation Year Fellowship.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Batada NN, Urrutia AO, Hurst LD. Chromatin remodelling is a major source of coexpression of linked genes in yeast. Trends Genet. 2007;23:480–484. doi: 10.1016/j.tig.2007.08.003. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Series B. 1995;57:289–300. [Google Scholar]
- Berry A, Kreitman M. Molecular analysis of an allozyme cline: alcohol dehydrogenase in Drosophila melanogaster on the east coast of North America. Genetics. 1993;134:869–893. doi: 10.1093/genetics/134.3.869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco E, Pignatelli M, Beltran S, Punset A, Perez-Lluch S, Serras F, Guigo R, Corominas M. Conserved chromosomal clustering of genes governed by chromatin regulators in Drosophila. Genome Biol. 2008;9:R134. doi: 10.1186/gb-2008-9-9-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochdanovits Z, van der Klis H, de Jong G. Covariation of larval gene expression and adult body size in natural populations of Drosophila melanogaster. Mol Biol Evol. 2003;20:1760–1766. doi: 10.1093/molbev/msg179. [DOI] [PubMed] [Google Scholar]
- Boutanaev AM, Kalmykova AI, Shevelyov YY, Nurminsky DI. Large clusters of co-expressed genes in the Drosophila genome. Nature. 2002;420:666–669. doi: 10.1038/nature01216. [DOI] [PubMed] [Google Scholar]
- Caron H, van Schaik B, van der Mee M, et al. (13 co-authors) The human transcriptome map: clustering of highly expressed genes in chromosomal domains. Science. 2001;291:1289–1292. doi: 10.1126/science.1056794. [DOI] [PubMed] [Google Scholar]
- Castillo-Davis CI, Hartl DL. GeneMerge—post-genomic analysis, data mining, and hypothesis testing. Bioinformatics. 2003;19:891–892. doi: 10.1093/bioinformatics/btg114. [DOI] [PubMed] [Google Scholar]
- Causton HC, Ren B, Koh SS, Harbison CT, Kanin E, Jennings EG, Lee TI, True HL, Lander ES, Young RA. Remodeling of yeast genome expression in response to environmental changes. Mol Biol Cell. 2001;12:323–337. doi: 10.1091/mbc.12.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakir M, Chafik A, Moreteau B, Gibert P, David JR. Male sterility thermal thresholds in Drosophila: D. simulans appears more cold-adapted than its sibling D. melanogaster. Genetica. 2002;114:195–205. doi: 10.1023/a:1015154329762. [DOI] [PubMed] [Google Scholar]
- Conover DO, Schultz ET. Phenotypic similarity and the evolutionary significance of countergradient variation. Tree. 1995;10:248–252. doi: 10.1016/S0169-5347(00)89081-3. [DOI] [PubMed] [Google Scholar]
- de Jong G, Bochdanovits Z. Latitudinal clines in Drosophila melanogaster: body size, allozyme frequencies, inversion frequencies, and the insulin-signalling pathway. J Genet. 2003;82:207–223. doi: 10.1007/BF02715819. [DOI] [PubMed] [Google Scholar]
- DeWitt TJ, Scheiner SM. Phenotypic plasticity: functional and conceptual approaches. New York: Oxford University Press; 2004. p. 247. [Google Scholar]
- Eanes WF. Analysis of selection on enzyme polymorphism. Annu Rev Ecol Syst. 1999;30:301–326. [Google Scholar]
- Endler JA. Geographic variation, speciation and clines. Princeton (NJ): Princeton University Press; 1977. [PubMed] [Google Scholar]
- Feder ME, Walser JC. The biological limitations of transcriptomics in elucidating stress and stress responses. J Evol Biol. 2005;18:901–910. doi: 10.1111/j.1420-9101.2005.00921.x. [DOI] [PubMed] [Google Scholar]
- Felsenstein J. The theoretical population genetics of variable selection and migration. Annu Rev Genet. 1976;10:253–280. doi: 10.1146/annurev.ge.10.120176.001345. [DOI] [PubMed] [Google Scholar]
- Gentleman RC, Carey VJ, Bates DM, et al. (25 co-authors) Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrist GW, Huey RB. Plastic and genetic variation in wing loading as a function of temperature within and among parallel clines in Drosophila subobscura. Integr Comp Biol. 2004;44:461–470. doi: 10.1093/icb/44.6.461. [DOI] [PubMed] [Google Scholar]
- Gockel J, Kennington WJ, Hoffmann A, Goldstein DB, Partridge L. Nonclinality of molecular variation implicates selection in maintaining a morphological cline of Drosophila melanogaster. Genetics. 2001;158:319–323. doi: 10.1093/genetics/158.1.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grether GF. Environmental change, phenotypic plasticity, and genetic compensation. Am Nat. 2005;166:E115–E123. doi: 10.1086/432023. [DOI] [PubMed] [Google Scholar]
- Gupta AP, Lewontin RC. A study of reaction norms in natural populations of Drosophila pseudoobscura. Evolution. 1982;36:934–948. doi: 10.1111/j.1558-5646.1982.tb05464.x. [DOI] [PubMed] [Google Scholar]
- Harr B, Kauer M, Schlotterer C. Hitchhiking mapping: a population-based fine-mapping strategy for adaptive mutations in Drosophila melanogaster. Proc Natl Acad Sci U S A. 2002;99:12949–12954. doi: 10.1073/pnas.202336899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Anderson A, Hallas R. Opposing clines for high and low temperature resistance in Drosophila melanogaster. Ecol Lett. 2002;5:614–618. [Google Scholar]
- Hutter S, Saminadin-Peter SS, Stephan W, Parsch J. Gene expression variation in African and European populations of Drosophila melanogaster. Genome Biol. 2008;9:R12. doi: 10.1186/gb-2008-9-1-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmykova AI, Nurminsky DI, Ryzhov DV, Shevelyov YY. Regulated chromatin domain comprising cluster of co-expressed genes in Drosophila melanogaster. Nucleic Acids Res. 2005;33:1435–1444. doi: 10.1093/nar/gki281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennington WJ, Gockel J, Partridge L. Testing for asymmetrical gene flow in a Drosophila melanogaster body-size cline. Genetics. 2003;165:667–673. doi: 10.1093/genetics/165.2.667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knibb WR, Oakeshott JG, Gibson JB. Chromosome inversion polymorphisms in Drosophila melanogaster. I. Latitudinal clines and associations between inversions in Australasian populations. Genetics. 1981;98:833–847. doi: 10.1093/genetics/98.4.833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreps JA, Hur-Song C, Zhu T, Wang X, Harper JF. Transcirptomic changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol. 2002;130:2129–2141. doi: 10.1104/pp.008532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine MT, Begun DJ. Evidence of spatially varying selection acting on four chromatin-remodeling loci in Drosophila melanogaster. Genetics. 2008;179:475–485. doi: 10.1534/genetics.107.085423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levins R. Evolution in changing environments. Princeton (NJ): Princeton University Press; 1968. [Google Scholar]
- Li Q, Lee BT, Zhang L. Genome-scale analysis of positional clustering of mouse testis-specific genes. BMC Genomics. 2005;6:7. doi: 10.1186/1471-2164-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Alvarez OA, Gutteling EW, et al. (12 co-authors) Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLoS Genet. 2006;2:e222. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liefting M, Hoffmann AA, Ellers J. Plasticity versus environmental canalization: population differences in thermal responses along a latitudinal gradient in Drosophila serrata. Evolution. 2009;63:1954–1963. doi: 10.1111/j.1558-5646.2009.00683.x. [DOI] [PubMed] [Google Scholar]
- Mezey JG, Nuzhdin SV, Ye F, Jones CD. Coordinated evolution of co-expressed gene clusters in the Drosophila transcriptome. BMC Evol Biol. 2008;8:2. doi: 10.1186/1471-2148-8-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng YK, Wu W, Zhang L. Positive correlation between gene coexpression and positional clustering in the zebrafish genome. BMC Genomics. 2009;10:42. doi: 10.1186/1471-2164-10-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakeshott JG, Gibson JB, Anderson PR, Knibb WR, Anderson DG, Chambers GK. Alcohol dehydrogenase and glycerol-3-phosphate dehydrogenase clines in Drosohpila melanogaster on different continents. Evolution. 1982;36:86–96. doi: 10.1111/j.1558-5646.1982.tb05013.x. [DOI] [PubMed] [Google Scholar]
- Robinson SJW, Partridge L. Temperature and clinal variation in larval growth efficiency in Drosophila melanogaster. J Evol Biol. 2001;14:14–21. doi: 10.1046/j.1420-9101.2001.00259.x. [DOI] [PubMed] [Google Scholar]
- Schmidt PS, Matzkin L, Ippolito M, Eanes WF. Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution. 2005;59:1721–1732. [PubMed] [Google Scholar]
- Scholtens D, von Heydebreck A. Analysis of differential gene expression studies in R.C. Gentleman. In: Huber W, Carey VJ, Irizarry R, Dudoit S, editors. Bioinformatics and computational biology solutions using R and Bioconductor. New York: Springer; 2005. [Google Scholar]
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- Sezgin E, Duvernell DD, Matzkin LM, Duan Y, Zhu CT, Verrelli BC, Eanes WF. Single-locus latitudinal clines and their relationship to temperate adaptation in metabolic genes and derived alleles in Drosophila melanogaster. Genetics. 2004;168:923–931. doi: 10.1534/genetics.104.027649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair BJ, Gibbs AG, Roberts SP. Gene transcription during exposure to, and recovery from, cold and desiccation stress in Drosophila melanogaster. Insect Mol Biol. 2007;16:435–443. doi: 10.1111/j.1365-2583.2007.00739.x. [DOI] [PubMed] [Google Scholar]
- Smith EN, Kruglyak L. Gene-environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. doi: 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spellman PT, Rubin GM. Evidence for large domains of similarly expressed genes in the Drosophila genome. J Biol. 2002;1:5. doi: 10.1186/1475-4924-1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul D, Gilbert N, Bickmore WA. The role of chromatin structure in regulating the expression of clustered genes. Nat Rev Genet. 2005;6:775–781. doi: 10.1038/nrg1688. [DOI] [PubMed] [Google Scholar]
- Stalker HD. Chromosome studies in wild populations of D. melanogaster. Genetics. 1976;82:323–347. doi: 10.1093/genetics/82.2.323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swindell WR, Huebner M, Weber AP. Plastic and adaptive gene expression patterns associated with temperature stress in Arabidopsis thaliana. Heredity. 2007;99:143–150. doi: 10.1038/sj.hdy.6800975. [DOI] [PubMed] [Google Scholar]
- Telonis-Scott M, Hallas R, McKechnie SW, Wee CW, Hoffmann AA. Selection for cold resistance alters gene transcript levels in Drosophila melanogaster. J Insect Physiol. 2009;55:549–555. doi: 10.1016/j.jinsphys.2009.01.010. [DOI] [PubMed] [Google Scholar]
- Turner TL, Levine MT, Eckert ML, Begun DJ. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics. 2008;179:455–473. doi: 10.1534/genetics.107.083659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umina PA, Weeks AR, Kearney MR, McKechnie SW, Hoffmann AA. A rapid shift in a classic clinal pattern in Drosophila reflecting climate change. Science. 2005;308:691–693. doi: 10.1126/science.1109523. [DOI] [PubMed] [Google Scholar]
- Verrelli BC, Eanes WF. Clinal variation for amino acid polymorphisms at the Pgm locus in Drosophila melanogaster. Genetics. 2001;157:1649–1663. doi: 10.1093/genetics/157.4.1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proc Natl Acad Sci U S A. 2006;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.