Abstract
The FoxP gene subfamily of transcription factors is defined by its characteristic 110 amino acid long DNA-binding forkhead domain and plays essential roles in vertebrate biology. Its four members, FoxP1–P4, have been extensively characterized functionally. FoxP1, FoxP2, and FoxP4 are involved in lung, heart, gut, and central nervous system (CNS) development. FoxP3 is necessary and sufficient for the specification of regulatory T cells (Tregs) of the adaptive immune system.
In Drosophila melanogaster, in silico predictions identify one unique FoxP subfamily gene member (CG16899) with no described function. We characterized this gene and established that it generates by alternative splicing two isoforms that differ in the forkhead DNA-binding domain. In D. melanogaster, both isoforms are expressed in the embryonic CNS, but in hemocytes, only isoform A is expressed, hinting to a putative modulation through alternative splicing of FoxP1 function in immunity and/or other hemocyte-dependent processes. Furthermore, we show that in vertebrates, this novel alternative splicing pattern is conserved for FoxP1. In mice, this new FoxP1 isoform is expressed in brain, liver, heart, testes, thymus, and macrophages (equivalent in function to hemocytes). This alternative splicing pattern has arisen at the base of the Bilateria, probably through exon tandem duplication. Moreover, our phylogenetic analysis suggests that in vertebrates, FoxP1 is more related to the FoxP gene ancestral form and the other three paralogues, originated through serial duplications, which only retained one of the alternative exons. Also, the newly described isoform differs from the other in amino acids critical for DNA-binding specificity. The integrity of its fold is maintained, but the molecule has lost the direct hydrogen bonding to DNA bases leading to a putatively lower specificity and possibly affinity toward DNA.
With the present comparative study, through the integration of experimental and in silico studies of the FoxP gene subfamily across the animal kingdom, we establish a new model for the FoxP gene in invertebrates and for the vertebrate FoxP1 paralogue. Furthermore, we present a scenario for the structural evolution of this gene class and reveal new previously unsuspected levels of regulation for FoxP1 in the vertebrate system.
Keywords: evolution, FoxP, gene duplication, alternative splicing
Introduction
The evolutionarily conserved family of Fox genes encompasses a large number of transcription factors involved in many developmental and differentiation processes (Mazet et al. 2003; Banerjee-Basu and Baxevanis 2004). Its many members share a conserved DNA-binding domain of 110 amino acids termed winged helix or forkhead domain but can differ markedly in the rest of the molecule. The forkhead domain was defined in 1990 by homology in the DNA-binding domains of the mouse hepatocyte nuclear factor 3 (HFN3) and Drosophila melanogaster forkhead gene (fkh) (Weigel et al. 1989; Lai et al. 1990). The functional importance of the FOX proteins is revealed by the multitude of diseases caused by spontaneous mutations in these genes, both in mice and in humans (Hannenhalli and Kaestner 2009). This family has undergone expansion and diversification during evolution through various steps of duplication (tandem and whole genome), being further divided into 19 subfamilies—from A to S—according to the position of particular conserved amino acids within the forkhead domain (Wijchers et al. 2006; Hannenhalli and Kaestner 2009).
Within these families further duplications occurred, for instance, in the FoxP subfamily whose members play an essential role in development, physiology, and immunity of vertebrates. Data from human and mouse models suggest that in vertebrates, this subfamily comprises four genes (Hannenhalli and Kaestner 2009). Distinguishing features of the FoxP subfamily include a carboxy-terminal DNA-binding domain (leucine zipper) and a zinc finger motif located at the N terminus of this domain (Li et al. 2004).
FoxP1, the member that was first isolated, is an important regulator of mouse lung, heart, brain, testis, kidney, and gut development (Ferland et al. 2003; Tamura et al. 2003; Schon et al. 2006; Shu et al. 2007). Moreover, FoxP1 has been found to be an essential transcriptional regulator of B cell and macrophage development (Shi et al. 2004; Hu et al. 2006) and to act as a Hox gene cofactor in specifying motor neuron identity and connectivity throughout the anteroposterior axis of the developing central nervous system (CNS) (Dasen et al. 2008; Rousso et al. 2008). FOXP2 mutations are associated with language disorders in humans (Lai et al. 2001) and with bird song defects in zebra finches (Haesler et al. 2004), suggesting a role in CNS development. Furthermore, FoxP2 plays a role in lung, heart, and gut development (Shu et al. 2007). FoxP3, the seemingly less pleiotropic gene of this group, is involved in regulatory T-cell specification and plays a central role in adaptive immunity (Brunkow et al. 2001; Yagi et al. 2004). Finally, FoxP4 is expressed in the developing lung and gut (Lu et al. 2002), and in humans, its downregulation has been correlated with kidney tumorigenesis (Teufel et al. 2003).
FoxP-member expression, in vertebrates, is highly regulated by tissue-specific alternative splicing of functionally important domains, suggesting that the same FoxP protein can perform different functions in different cells and tissues. In mice, FoxP1 has 11 described isoforms, 8 for FoxP2, 4 for FoxP3, and 5 for FoxP4 (Ensembl data). These findings suggest that in this gene subfamily, alternative splicing is an important mechanism to create variation from a single locus. It is no wonder then that the functional and expression complexity of this family have generated much interest regarding its evolutionary history. Gene duplication and alternative splicing are processes that diversify the protein repertoire with significant impact on both structural and functional levels (Chothia et al. 2003). Interestingly, recent comparative genomics studies have shown these two processes to be inversely correlated (Kopelman et al. 2005; Su et al. 2006). Studying the function of FoxP in invertebrates can unravel some important aspects concerning the evolution of the FoxP family of vertebrates.
FoxP expression and function have been only sporadically approached in invertebrates, with reports of expression patterns in sponge (Suberites domuncula) (Adell and Muller 2004), honeybee (Apis mellifera) (Kiya et al. 2008), Drosophila (D. melanogaster) (Lee and Frasch 2004), and sea urchin (Strongylocentrotus purpuratus) (Tu et al. 2006). In D. melanogaster, only one gene of the FoxP family has been predicted (CG16899). In this study, we report that DmFoxP produces two alternative splicing products, which differ in their forkhead domain. This fact suggests that DNA-binding FoxP protein diversity could be achieved in vertebrates also by alternative splicing. Indeed, alternative splicing producing different Forkhead domains has never been observed in any FoxP gene, which raises questions about the extent of its conservation throughout the animal kingdom.
In the current study, with the integration of molecular and in silico studies, we present a scenario for the molecular evolution of FoxP genes from invertebrates to vertebrates revealing new unsuspected levels of functional regulation and diversification.
Materials and Methods
RNA Extraction and Reverse Transcriptase–Polymerase Chain Reaction in Drosophila
RNA extracted from each developmental stage of D. melanogaster by using the TRIzol reagent (Invitrogen) was used for cDNA synthesis with an Oligo(dT) primer using RevertAid H Minus First Strand cDNA Synthesis Kit (Fermentas). To distinguish between the two possible forkhead isoforms, we design the specific primers. In embryo, larvae, and adults (fig.1D), we used FoxPConstantForward (5′-CTGAATACGGAACATGGTTT-3′), FoxPIsoAReverse (5′-CTATTTGAGACCCACATACC-3′), or CG32937IsoBReverse (5′-TTATCGATTGTGCTCATTG–3′). For hemocytes (fig. 1F), we amplified isoA with commonFwd1DmFox (5′-GAGTCCGCTTACCGTAAATA-3′) and PartMelFoxfwd (5′-TTTCAGCTATATGCACGATG-3′) and isoB with commonFwd2DmFox (5′-GATAATGAGGTGTGCAACAA-3′) and Alt2RevDmFox (5′-GTGCTCATTGGCACTACTCT-3′). The PCR conditions were 20 s at 95 °C, 30 s at 52 °C, and 1.5 min at 72 °C for 35 cycles.
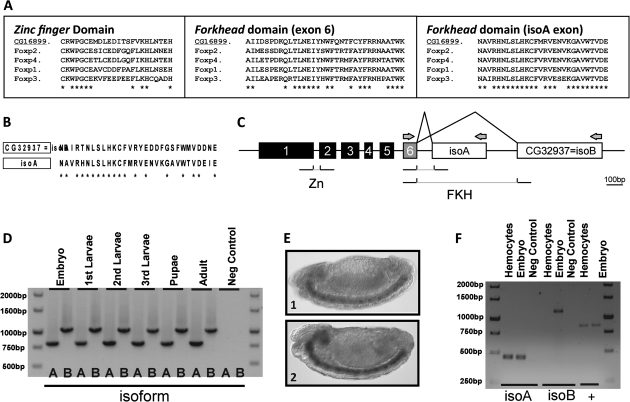
FIG. 1.
(A) Alignment of the protein sequences from the conserved domains of M. musculus FoxP subfamily and D. melanogaster FoxP (CG16899). (B) Alignment of the first 30 amino acids from the protein sequence of the putative alternative exons. The asterisks represent amino acid conservation between the two sequences confirming that CG32937 is a forkhead domain–encoding sequence. (C) Schematic representation of the predicted gene structure of D. melanogaster FoxP (DmFoxP–CG16899). The described forkhead domain is encoded by exons 6 and isoA. The remaining numbered boxes (1–5) represent the 5′ exons from DmFoxP. The zinc finger domain (Zn) is coded by exons 1 and 2. In silico prediction showed that CG32937 may be an extra exon of DmFoxP and that the forkhead domain (FKH) may have two different isoforms (using exons 6 + isoA or exons 6 + isoB). Arrows show the region for which specific primers were designed that would discriminate between the putative isoforms. (D) RT-PCR results of DmFoxP expression at all the life stages. The negative control used no RNA as the PCR template. Expression of both isoforms of DmFoxP was detected in all stages, confirming the existence of an alternatively spliced isoform only differing in the second part of the forkhead domain. (E) DmFoxP expression pattern in D. melanogaster embryos detected by in situ hybridization. Lateral view of stage 14 embryos stained for isoform A (1) and isoform B (2) showing expression ventrally across the whole CNS. Anterior is to the left and ventral to the bottom of the image. (F) RT-PCR results of DmFoxP expression in hemocytes. Lanes 3 and 6 are negative controls and lanes 7 and 8 are positive controls for a gene (Heme Oxygenase) expressed in this cell type. Only DmFoxP IsoA expression was detected in hemocytes.
The same procedure was performed with the RNA extracted from D. melanogaster hemocytes (see below).
RNA Extraction and Reverse Transcriptase–Polymerase Chain Reaction in Mouse
RNA was extracted and cDNA was prepared as described above from isolated tissues taken from perfused animals.
The two isoforms were distinguished using specific primer pairs as follows:
Isoform 1, 102Fwd (5′-TCTCCAGAAAAGCAGCTAAC-3′) and A1Rev (5′-GGTTACCACTGATCTTTTGT-3′) and isoform 2, Mm_Alt2Fwd (5′-GCCATTCGCACCAACCTC-3`) and CommonRev2 (5′-GTCAAAATCTGGACTGTGGT-3′).
Hemocyte Extraction
Hemocytes were collected by rupturing abdominal larval cuticle in ice-cooled Schneider’s medium. For each Fluorescence-activated cell sorting (FACS) analysis, 150 larvae were bled in 800 μl of medium. Subsequently, hemocytes were stained with a modified protocol from Tirouvanziam et al. (2004). Two hundred microliters of Schneider’s medium with 100 μM monochlorobimane was added to 800 μl hemocyte suspension and incubated at 25 °C for 20 min. The reaction was stopped by adding 3 ml of ice-cooled Schneider’s medium. Hemocytes were pelleted by centrifugation at 430 g for 5 min at 4 °C and ressuspended in 400 μl Schneider’s medium with 2 μg/ml propidium iodide just before cell sorting.
In Situ Hybridization
We used the cDNA amplified with the previous combinations of primers to generate sense and antisense RNA probes for each isoform with a Digoxigenin (DIG) labeling kit (Roche Applied Science). In situ hybridization was performed as described in (O'Neill and Bier 1994).
Data Mining of FoxP Sequences
tBlastn searches were performed using the sequence from FoxP forkhead domain from D. melanogaster in ENSEMBL and JGI databases. Species names, gene names, and accession numbers are listed in supplementary table S1, Supplementary Material online.
Phylogenetic Analyses
To understand the origins of the evolutionary history of alternative splicing in the FoxP family, we performed several phylogenetic studies. Multiple sequence alignments were prepared using ClustalX (v. 2.0) x2 (Thompson et al. 1994). Maximum likelihood (ML) trees were obtained with PAUP (v. 4.0b10) (Swofford 2001), and Bayesian trees were obtained with MrBayes (v. 3.1.2) (Ronquist and Huelsenbeck 2003). In the former, we used four Markov chain Monte Carlo that run for 2 × 106 iterations, and trees were sampled at each 100 generations. In the ML method, the search for the best tree was performed with a heuristic search using the tree bisection and reconnection algorithm and the resampling was performed by 100 bootstrap replicates. To determine the evolutionary model that adjusts best to our data, we used MODELTEST (v. 3.06) (Posada and Crandall 1998).
Modeling the 3D Structure of DmFoxP and MmFoxP1 Isoforms
Protein-only models of the both isoforms of Drosophila and mouse alternative splice variants were constructed using SWISS-MODEL(Arnold et al. 2006) and based on the FoxP2 structure (PDB: 2A07) chain K. A model incorporating DNA was constructed by alignment of the model with 2A07 chain K (in PYMOL) and rotamer search for T554 in coot (Emsley and Cowtan 2004). Models of the domain-swapped form were created using PHYRE (Kelley and Sternberg 2009). Alignments were performed with ClustalW (Thompson et al. 1994).
Results
The Single Copy of D. melanogaster FoxP Gene (DmFoxP) Is Alternatively Spliced
Only one FoxP gene is reported in D. melanogaster, CG16899, and our own analysis did not uncover other genes that would fill the criteria regarding the forkhead domain structure characteristic of Fox P subfamily (data not shown). CG16899 has a coding sequence composed of seven exons, of which the last two encode the forkhead domain. Comparison of the protein sequences from the conserved domains (zinc finger and Forkhead domains) of FoxP1/2/3/4 from Mus musculus and CG16899 from D. melanogaster reveal an overall 62% of amino acid identity (fig. 1A).
Our in silico analysis suggested that CG16899 could include an extra exon at its 3′ extremity, to date reported as a separate gene (CG32937). This prediction stems from the nature of the predicted protein encoded by CG32937 that showed forkhead domain–like features. The alignment of the predicted protein from CG32937 to exon 7 of CG16899 revealed a 60% amino acid identity (fig. 1B). These results suggest that the predicted locus CG32937 could be instead an additional exon of CG16899 that would be alternatively spliced, joining with exon 6 to form two different, yet recognizable, forkhead domains (fig. 1C).
To confirm the above prediction, we performed reverse transcriptase–polymerase chain reaction (RT-PCR) at different life cycle stages of Drosophila. Different combinations of primers were used to distinguish between the two putative alternative isoforms (locations indicated by arrows in fig. 1C). Amplification of both isoforms was observed at every life stage of D. melanogaster (fig. 1D). The subsequent sequencing of the RT-PCR products confirmed that CG16899 and CG32937 constitute a single Open Reading Frame with alternative splicing. Hereafter, we will refer to this locus as DmFoxP. Together, these results establish that DmFoxP has one more exon than the predicted model and that this locus produces two alternative isoforms differing at the forkhead domain, DmFoxP-A, and DmFoxP-B.
FoxP Is Transcribed in the Embryonic CNS and Differentially Expressed in Hemocytes
In previous expression studies FoxP had been detected in honeybee adult brain (Kiya et al. 2008) and in D. melanogaster embryonic CNS (Lee and Frasch 2004). We performed in situ hybridization on all embryonic developmental stages, using two DIG-labeled riboprobes that could distinguish DmFoxP-A from DmFoxP-B. Both isoforms show an onset of expression in the CNS at embryonic stage 8, which persists throughout the rest of the embryonic development (fig. 1D and E). Interestingly, CNS expression is also consistent with the vertebrate data showing that three of the four FoxP genes (FoxP3 excluded) participate in the development and physiology of the CNS.
In vertebrates, the FoxP subfamily also plays an essential role in immunity, inspiring us to test for FoxP expression in D. melanogaster hemocytes. Hemocytes are the blood cells and immunocytes from arthropods, constituting the cellular component of the lymph in charge of phagocytosis (amongst other processes), a role undertaken in vertebrates by macrophages. To test for FoxP expression in this cell type, we took advantage of a D. melanogaster strain with a hemocyte-specific Green Fluorescent Protein (GFP) expression (Hemolectin-Δ-GAL4 UAS-GFP, kindly provided by A. Jacinto) and isolated positive cells from the hemolymph by FACS analysis and sorting. We then extracted the RNA from the hemocytes and performed an RT-PCR with the set of primers described above. Drosophila hemocytes only express FoxP isoform A (fig 1F).
These results suggest that alternative splicing in DmFoxP may play relevant functional roles in embryogenesis, metamorphosis, or/and Drosophila immunity, the main stages of hemocyte function.
FoxP Alternative Splicing Is a Bilateria Feature
To further define the extent of conservation of this alternative splicing pattern, we next examined (in silico) the structure of the forkhead domain in other organisms. To determine if this pattern was conserved throughout evolution or if it was a derived character of Drosophila, we searched the available genomes (invertebrates and vertebrates) for this extra alternative exon. BLAST of DmFoxP resulted in no orthologues in fungi, plants, bacteria, and protozoa, dating the emergence of the FoxP subfamily to the animal lineage.
In invertebrates, all species for which the genome is available have only one FoxP subfamily gene member. The search for orthologues in invertebrates belonging to the Parazoa and Radiata group (Porifera, Cnidaria, and Placozoa) shows that these organisms only possess one isoform in their genome, which means that probably there is no alternative splicing mechanism generating forkhead domain variability in this group. An exception concerning the conservation of the alternative splicing pattern in Bilateria is the Nematoda phylum. Nevertheless, this absence of alternative splicing must represent a derived state for such feature is present in all other observed Bilateria groups. Taken together, these data suggest that alternative splicing in the forkhead domain appeared in the Bilateria (fig. 2, arrow 1) possibly by tandem exon duplication (Kondrashov and Koonin 2001).
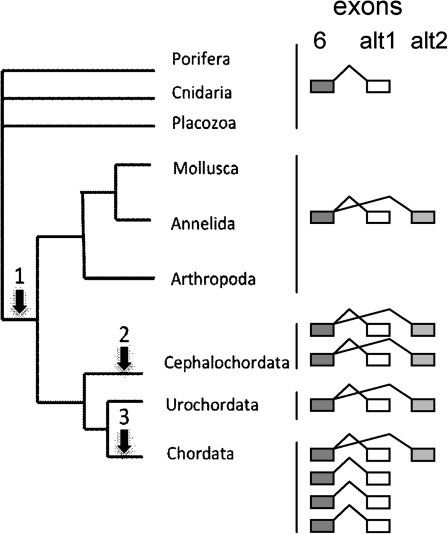
FIG. 2.
Main events of FoxP gene structure evolution in Animals. The arrows show the main duplication events: arrow 1, tandem duplication of the exon coding for the second part of the forkhead domain (AS origin); arrow 2, FoxP duplication in Cephalochordata clade; arrow 3, two duplication events of the FoxP family in vertebrates (with an extra one in Teleosts; Amores et al. 1998). The diagram on the right depicts the number and structure of alternative splicing for the FoxP genes in the different groups. Only exon 6 (containing the first half of the forkhead domain) and (alternative) exon(s) 7 (containing the second half) are shown.
In silico predictions of the FoxP gene structure in other invertebrates belonging to the phyla, Arthropod, Annelida, and Mollusca also reveal the presence of two alternative isoforms with different forkhead domains (supplementary table S1, Supplementary Material online). Within chordates, the amphioxus (Branchiostoma floridae, Cephalochordata) genome contains two copies of the gene (Yu et al. 2008), both exhibiting the same predicted alternative splicing pattern as DmFoxP. On the other hand, Urochordata (Ciona savignyi) only possess one copy of FoxP with alternative splicing. The lack of a complete urochordate genome project makes it hard to date the first duplication event at the base of chordates or as two independent events that occurred in the Cephalochordata and Vertebrate lineages. We tend to favor the independent duplication alternative, given that there is evidence that Cephalochordata does not represent an intermediate stage between vertebrates and invertebrates in what pertains to the 2R hypothesis (fig. 2, arrow 2). Indeed, amphioxus does not seem to have been through a complete genome duplication as revealed by its single Hox cluster (de Rosa et al. 1999).
In vertebrates, we witness the appearance of four FoxP copies in accordance with the postulated two rounds of whole-genome duplication (fig. 2, arrow 3). How has the alternative splicing pattern of FoxP evolved upon gene duplication in the vertebrate lineage?
Evolutionary Conservation of Alternative Splicing in the Vertebrate FoxP1 Gene
The C terminus of FoxP2 and FoxP4 are similar, with two exons that code for the forkhead domain followed by three more exons (fig. 3A, gray boxes). FoxP3 protein is more similar to the invertebrate gene structure, although without a described alternative splicing. We have not found evidence for the existence of alternative splicing involving changes within the forkhead domain–coding region for any of these three paralogues, FoxP2, P3, and P4. As for FoxP1, in silico translation of the mouse genomic region revealed a putative nonannotated exon, immediately downstream of the exon that codes for the second part of the forkhead domain (fig. 3A). The resulting protein is similar to the one obtained for isoform B of D. melanogaster FoxP (fig. 3B). Moreover, the splicing acceptor and donor sites in the junctions of this new exon are also conserved. The search for this undescribed exon in other vertebrate genomes (Danio rerio and Homo sapiens) revealed that such DNA sequence is present in a form also consistent with the production of a viable protein corresponding to a partial forkhead domain. We hypothesized that this FoxP1 alternative splicing pattern is conserved in vertebrates and that it encodes a yet undescribed isoform containing a forkhead domain similar to DmFoxP-B.
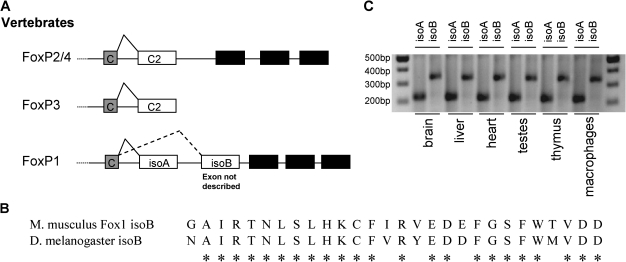
FIG. 3.
C terminus structure and splicing of vertebrate FoxP genes. (A) Boxes represent exons starting with the forkhead domain–encoding constant exon (C) orthologous to exon 6 of Drosophila. The new alternative splicing pattern was only predicted for FoxP1, the exon coding for isoform B had not been described but the translation of the genomic sequence revealed a putative coding sequence for an alternative forkhead domain in FoxP1. (B) Alignment of the first 28 amino acids of the protein sequence of the predicted new FoxP1 exon and the exon IsoB from D. melanogaster. The asterisks represent conservation between the two sequences. (C) RT-PCR results of FoxP1 expression in M. musculus. Expression of both isoforms of FoxP1 was detected in brain, liver, heart, testes, thymus, and macrophages confirming the expression of the predicted new alternatively spliced isoform of FoxP1 in vertebrates.
To test this hypothesis, we performed PCR on cDNA from various murine cell types/tissues/organs, with specific primers that can distinguish between the two putative isoforms. Consistent with our prediction, expression of both isoforms was detected in brain, liver, heart, testis, thymus, and macrophages (fig. 3C). The two isoforms fragments were sequenced confirming that at this gross level of analysis both isoforms are simultaneously expressed. Further analysis is needed to determine whether in this vertebrate model each isoform may have time- and space-specific expressions patterns. With this result, we show the conservation of this alternative splicing pattern in M. musculus and we describe a new isoform for FoxP1. These results suggest that FoxP1 from vertebrates is more closely related to FoxP from invertebrates than any other vertebrate FoxP, at least at the gene structure level. Furthermore, they raise the question of which of the alternatively spliced exons has been lost in FoxP2-4, the vertebrate paralogues of FoxP1.
Isoform B Is More Recent and Has Been Lost in Paralogues 2, 3, and 4
We have generated a FoxP gene subfamily phylogeny to better dissect the evolutionary history of the FoxP subfamily and to infer the ancestral state of the FoxP gene before the duplication events. This knowledge is essential to generate hypotheses about the functional evolution of this gene subfamily. Forkhead domain–encoding genes have been widely conserved during evolution and are found in species as distant as yeast and humans, but the extent of sequence conservation among products from distant species is largely restricted to the forkhead and zinc finger domains (Hannenhalli and Kaestner 2009). Therefore, these two domains are the best sequences to construct a strong phylogeny.
In vertebrates, three of the genes (FoxP2/3/4) lost the forkhead alternative splicing described here and have only one exon coding for the second part of the forkhead domain. To construct a phylogeny, we must determine the homologous exons that constitute this second part of the domain. With this in mind, we constructed a phylogeny of the two alternative exons from FoxP found in invertebrates, the two alternative exons of FoxP1, and the exons that code for the second part of the forkhead domain of FoxP2, 3, and 4 (fig. 4A).
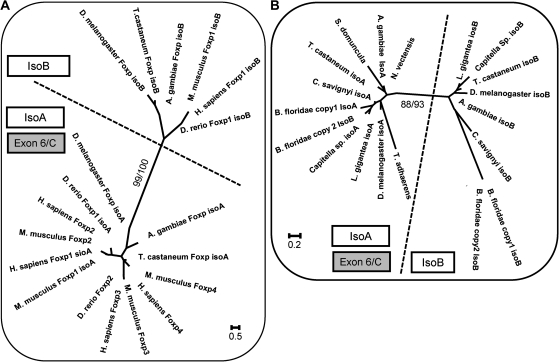
FIG. 4.
Evolution of FoxP alternative splicing in the forkhead domain. Unrooted ML phylogenetic trees generated with PAUP program with 100 replicates using the sequences from the exons coding for the second part of the forkhead domain. (A) vertebrates and arthropods. (B) Bilateria invertebrates and Cnidaria/Porifera/Placozoa organisms. In both phylogenies, the two alternatively spliced exons separate into two independent clusters. Isoform A always clusters with the ancestral (without alternative splicing) protein. The value before the slash is the bootstrap and after is the posterior probability determined with the Bayesian method, with n = 4 in 2 × 106 generations. (A) The distance matrix was determined with the generalized time reversible model assuming heterogeneity in nucleotide mutation rates with a = 0.2156 and I = 0.1316, with “a” being the gamma distribution rate variation among sites and “I” being the proportion of invariable sites in our data set. (B) The distance matrix was determined with the TrNef (Tamura–Nei) model assuming heterogeneity in nucleotide mutation rates with a = 0.3192.
In this phylogeny, there are two distinct groups: One composed of alternative exon B and another composed of alternative exon a clustering with the exons from FoxP2, P3, and P4 that encode for the second part of the forkhead domain. The analysis of this exon phylogeny, strongly supports the hypothesis that the ancestral state within Bilateria corresponds to the structure of the invertebrate gene with two alternatively spliced exon. This entails that alternative exon 2 has degenerated in FoxP2, P3, and P4 and that the forkhead domain from isoform A of FoxP1 is homologous to the forkhead domain present in FoxP2, P3, and P4. Thus, we can exclude the hypothesis that alternative splicing of FoxP1 arose independently in vertebrates.
The results from the exon structure comparison suggest that the ancestral condition of the FoxP gene before the split of bilaterians did not have alternative splicing and that this pattern appeared only in the Bilateria lineage before the diversification of all the other groups. Was the ancestral second part of the forkhead domain more similar to isoform A or B? To answer to this question, we performed a phylogenetic analysis in which we used only the nonvertebrate exon sequences from the forkhead domain (fig. 4B).
The phylogeny resulted in the division of two distinct groups, one constituted by isoform B and the other constituted by isoform A plus the other FoxPs belonging to the nonbilateria clade that do not have alternative splicing. This result suggests that the ancestral forkhead domain was more similar to isoform A and that the alternative exon B only appeared in the bilaterian lineage. This result reinforces our hypothesis that exon B appeared through tandem duplication of the exon corresponding to the second part of the forkhead domain. Upon duplication, the second isoform could evolve and acquire different properties because isoform A would continue to fulfill the original function of the gene.
Duplication History of the FoxP Gene Subfamily
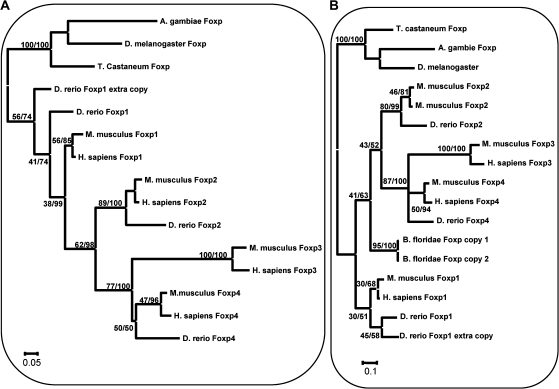
Using ML and Bayesian methods on sequences corresponding to isoform A of the forkhead domain and zinc finger domain, we could estimate the phylogenetic relationship between FoxP genes, which supports DmFoxP to be closer to FoxP1 than to any other FoxP vertebrate gene (fig. 5A).
FIG. 5.
Evolution of FoxP gene subfamily. Rooted ML phylogenetic trees generated with PAUP program with 100 replicates using the sequences from zinc finger and forkhead domains. (A) Vertebrates and arthropods. (B) Vertebrates, arthropods, and cephalochordates. In both phylogenies, FoxP from arthropods and from amphioxus is more closely related to FoxP1 from vertebrates than to any other FoxP. The value before the slash is the bootstrap and after is the posterior probability determined with the Bayesian method, with n = 4 in 2 × 106 generations. (A) The distance matrix was determined with the symmetrical model assuming heterogeneity in nucleotide mutation rates with a = 0.2584. (B) The distance matrix was determined with the generalized time reversible model assuming heterogeneity in nucleotide mutation rates with a = 0.758 and I = 0.3647.
In short, FoxP1 is more related to the ancestral state, as represented by the DmFoxP gene, both at the gene structure and at the sequence levels.
The topology of the phylogenetic tree in figure 5A is consistent with a serial FoxP duplication, suggesting that three duplication events occurred after the split of vertebrates from invertebrates. In this light, the first duplication generated FoxP1 and the ancestral form of all the other paralogues; a second duplication gave rise to FoxP2 and the ancestral forms of FoxP3 and FoxP4; and finally, a third event gave rise to FoxP3 and FoxP4. This duplication pattern is not well supported by bootstrap but corresponds to the tree with the highest likelihood.
We constructed another phylogeny with the two FoxP sequences (fig. 5B). The tree generated has lower bootstrap values and posterior probabilities but confirms the duplication pattern obtained in the previous analysis. The two amphioxus genes constitute a monophyletic group, which indicates that the duplication in this organism occurred after the split between vertebrates and cephalochordates. Both amphioxus FoxP copies have the same alternative splicing pattern as DmFoxP and M. musculus FoxP1. These two genes have a basal position on the tree relative to FoxP2/3/4, and regarding gene structure, both of them are closer to FoxP1 than to any other gene of the FoxP subfamily. These observations suggest that this duplication in amphioxus is recent and lineage specific.
Finally, the fifth FoxP copy present in D. rerio clusters with DrFoxP1 (fig. 5B), indicating that the duplication event that originated, it occurred after the split between teleosts and other vertebrates. Interestingly, based on its genomic sequence, we cannot predict a FoxP1-specific alternative splicing, suggesting that as for other duplication events, it retained isoform A and lost isoform B.
Predicted DNA Interactions of FoxP/FoxP1 Isoforms Show Bonding Differences
The conservation of alternative splicing in the FoxP/FoxP1 forkhead domain across Bilateria reveals a structural diversification that strongly suggests functional diversification between splicing isoforms. At this stage, we cannot experimentally determine the putatively different functions of the two isoforms of FoxP1. Nevertheless, we modeled their structures and evaluate their DNA-binding interactions in order to generate stronger hypotheses on their functional evolution.
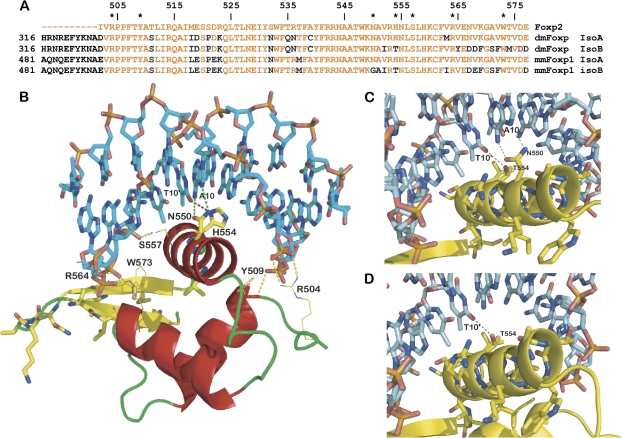
The structures of human FOXP2 (Stroud et al. 2006) and that of the mouse HNF-3 (Clark et al. 1993) in complex with DNA provide insights into the protein–DNA interactions of this domain family. DmFoxP and MmFoxP1 forkhead domains are sufficiently close to the FoxP2 protein (85.5% [Dm] and 88.1% [Mm] identity over 76 aa) as to base our modeling on this structure (fig. 6A). FoxP2 is shown to bind to DNA as a monomer as well as a dimer formed through domain swapping. Domain swapping of FoxP2 has been suggested to be of functional importance and possibly allowing heterodimerization between different vertebrate FoxP members (Stroud et al. 2006). The two alternative DmFoxP proteins differ in 11 residues located toward the C terminus of the HTH domains (fig. 6A and B). Residues participating in critical protein–DNA interactions in FoxP2 (N550, H554, R553, S557, W573; FoxP2 numbering) are all conserved in the DmFoxP-A isoform. Among those residues, only His554 changes to Thr in DmFoxP-B. This is an important change as a His residue in this position is conserved among all vertebrate FoxP proteins and forms critical contacts with DNA at T10' of the FoxP2-binding site. A Thr residue at this position is at less favorable position to form a hydrogen bond with T10′, thus we conclude that DmFoxP-B might differ from DmFoxP-A in binding to target DNA sequence at the position indicated with small letters: AAACAaATTTC (fig. 6C).
FIG. 6.
DNA interactions of FoxP1 isoforms. (A) Alignment of the FoxP2 forkhead domain with dmFoxP-A, dmFoxP-B and mmFoxP1A, and mmFoxP1B. Residues identical to FoxP2 are colored in red. Residues participating in protein–DNA interactions are marked with *. (B) Model of dmFoxP-A; residues that differ between AS forms are shown in stick representation, and residues participating in protein–DNA interactions are labeled. (C) Model of the recognition helix of DmFoxP-B. (D) Model of MmFoxP1-B in which only a weak single sequence–specific hydrogen bond can be formed with the cognate FoxP2 DNA sequence. For consistency, amino acid numbering in (B), (C), and (D) follows numbering of the FoxP2 protein.
The unique difference between the two isoforms affecting the hydrophobic core of the protein is at the equivalent position of Val552 in FoxP2, which changes to Ile in isoform B; this change is a common variation between FoxP proteins and is expected to have no effect in the stability of the domain. The remaining nine differences between the two isoforms are located in strands S1 and S2 and their connecting loop. These positions demonstrate variability between vertebrate FoxP proteins, and none of these alterations is predicted to affect DNA binding or protein stability. Nevertheless, it is conceivable that these differences between isoforms alter protein–protein interactions of the forkhead domain to the rest of the FoxP protein or its interactions with other proteins with which it may form heterodimers, such as Hox proteins (see below). As mentioned, the determination of FoxP2 structure showed that FoxP proteins can form a dimer through domain swapping. Domain swapping requires the presence of an Ala at position 539 (Stroud et al. 2006). It has been suggested that if a Pro residue occupies this position, like in other Fox family transcription factors, domain swapping is prevented. In DmFoxP, this position is occupied by Cys in both isoforms and is consistent with the formation of a swapped domain arrangement. In any case, both DmFoxP isoforms share identical residues in all positions previously judged as important for domain swapping and are not expected to differ in this respect.
In short, our analysis suggests that the forkhead domains of the two DmFoxP isoforms are properly folded and that their domains are functional. The main difference is the substitution of the critical DNA-binding residue His554 by Thr altering the interactions of this domain with the proposed recognition sequence 5′-CAAATT-3′ at position 3.
We then asked whether the alternative spliced forms of MmFoxP1 differ in an analogous manner. The mouse and Drosophila alternative splice forms share high homology, and modeling of the mouse proteins fits with the observations described above for DmFoxP. However, in addition to the H554/T alteration in the DNA-binding cleft, a second critical amino acid equivalent to FoxP2 N550 is changed to a Gly in the mouse isoform B. Asn550 in FoxP2 forms bidentate hydrogen bonds to adenine 10, bonds which are lost in the mouse FoxP1B isoform (fig. 6C and D). This alteration along with the H554/T is predicted to profoundly affect the sequence specificity of the domain as those are the only two residues forming direct contacts with bases in the FoxP2 complex.
As the nonspecific interactions with the DNA backbone are virtually identical between the two alternative forms and FoxP2, their affinity for DNA should be largely retained. Nevertheless, both in Drosophila and mouse, the alternative splicing isoforms are expected to have relaxed specificity with the mouse isoform being more strongly affected.
Discussion
Both gene duplication and alternative splicing are processes that significantly contribute to proteome diversity for they affect the evolution of protein sequence, structure, and function. Our analysis of the FoxP gene subfamily evolution revealed its pattern of gene duplication and alternative splicing acquisition and loss. By establishing a clear historical pattern for the evolution of the FoxP gene subfamily, our data generate clear hypotheses for the functional evolution of gene duplicates in the vertebrate gene complex. Specifically, we describe a novel alternative spliced form of the bilaterian FoxP gene that can produce two proteins with predicted differences in DNA binding and, presumably, transcriptional regulation. Such differential expression is utilized in hemocytes, opening the possibility that some functional aspect of this cell type biology may be regulated through alternative splicing. Moreover, our analysis unveils new levels of regulation for vertebrate FoxP1 action, a central gene in vertebrate development, which should be amenable to experimental dissection. Particularly, it will be interesting to determine if, in motor neuron specification and connectivity, this extra layer of FoxP1 regulation is used on top of the combinatorial mechanism involving Hox genes and FoxP1 expression levels (Dasen et al. 2008; Rousso et al. 2008).
Also, in the Drosophila model system, in which the same logic of Hox gene combinatorial coding has been established in the patterning of the CNS (Hirth et al. 1998), the description of this gene provided by our work suggests further interesting parallels with the vertebrate system that are amenable to experimental testing. Furthermore, another aspect in which the comparison between vertebrate and invertebrate systems can be extremely informative with respect to the intertwined roles of Hox and Fox regards the role of apoptosis in sculpting the CNS (Economides et al. 2003; Rogulja-Ortmann et al. 2008). Indeed, the dissection of the developmental genetics of the interplay between Hox and Fox in a comparative framework can potentially yield fundamental insights into the evolutionary origin and fine tuning of CNS architecture across Bilateria.
Gene duplication and alternative splicing can function in a concerted fashion during gene family evolution, but only in a small proportion of cases, a correspondence between isoforms and gene duplicates across different species has been verified (Pacheco et al. 2004; Talavera et al. 2007). In most cases, alternative splicing generates putative deleterious protein changes arguing against an adaptive equivalence between this mechanism and gene duplication (Ohta 1991; Talavera et al. 2007; Tress et al. 2007). In this respect, our study reveals an exceptional case in which the evolution of alternative splicing in the ancestral gene and maintained in the FoxP1 orthologue produces two isoforms with putatively functional differences through the deployment of different forkhead domains with distinct DNA interaction potential.
Moreover, analysis of human and mouse genomes has revealed that the number of isoforms is inversely correlated with the size of the gene families and that single-copy genes typically have a higher level of alternative splicing (Kopelman et al. 2005; Su et al. 2006). Su and colleagues take their analysis one step further and propose that when a gene with alternative splicing is duplicated, each of the new duplicates retains a part of the functional diversity performed previously by different isoforms. After this subfunctionalization, the acquisition of new functional isoforms can contribute to neofunctionalization, a scenario consistent with the model of He and Zhang (2005). Despite the lack of a complete functional data set, our data are in agreement with this hypothesis since the ancestral gene exhibits an alternative splicing pattern absent in its duplicates. Also, in the mouse, each of the gene duplicates has secondarily acquired new and specific alternative splicing patterns. How orthologues and paralogues depart from their shared origin and functional redundancy and adopt unique targets that shape different cell transcriptomes into higher-level differentiation is at the core of the evolutionary process (Ohno 1970; Ohta 1991; Otto and Yong 2002; Kafri et al. 2006). This description of the evolutionary path and gene structure diversification undertaken by the FoxP gene family provides a coherent framework to further dissect the roles each copy has evolved into, from cognition and immunity to CNS and heart development.
Supplementary Material
Supplementary table S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank José Pereira-Leal, Jocelyne Demengeot, Thiago Carvalho, and Patrícia Beldade for the critical reading of the manuscript; two anonymous reviewers for constructive criticism; Leonor Sarmento and Andreia Lino for help with the mouse tissue RNA extractions; and Barbara Vreede and Alexis Hazbun for technical support. This work was supported by Fundação para a Ciência e a Tecnologia, Portugal (POCTI/BIA-BDE/60950/2004 and PPCDT/BIA-BDE/60950/2004).
References
- Adell T, Muller WE. Isolation and characterization of five Fox (Forkhead) genes from the sponge Suberites domuncula. Gene. 2004;334:35–46. doi: 10.1016/j.gene.2004.02.036. [DOI] [PubMed] [Google Scholar]
- Amores A, Force A, Yan YL, et al. (13 co-authors) Zebrafish hox clusters and vertebrate genome evolution. Science. 1998;282:1711–1714. doi: 10.1126/science.282.5394.1711. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Banerjee-Basu S, Baxevanis AD. Structural analysis of disease-causing mutations in the P-subfamily of forkhead transcription factors. Proteins. 2004;54:639–647. doi: 10.1002/prot.10621. [DOI] [PubMed] [Google Scholar]
- Brunkow ME, Jeffery EW, Hjerrild KA, Paeper B, Clark LB, Yasayko SA, Wilkinson JE, Galas D, Ziegler SF, Ramsdell F. Disruption of a new forkhead/winged-helix protein, scurfin, results in the fatal lymphoproliferative disorder of the scurfy mouse. Nat Genet. 2001;27:68–73. doi: 10.1038/83784. [DOI] [PubMed] [Google Scholar]
- Chothia C, Gough J, Vogel C, Teichmann SA. Evolution of the protein repertoire. Science. 2003;300:1701–1703. doi: 10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- Dasen JS, De Camilli A, Wang B, Tucker PW, Jessell TM. Hox repertoires for motor neuron diversity and connectivity gated by a single accessory factor, FoxP1. Cell. 2008;134:304–316. doi: 10.1016/j.cell.2008.06.019. [DOI] [PubMed] [Google Scholar]
- de Rosa R, Grenier JK, Andreeva T, Cook CE, Adoutte A, Akam M, Carroll SB, Balavoine G. Hox genes in brachiopods and priapulids and protostome evolution. Nature. 1999;399:772–776. doi: 10.1038/21631. [DOI] [PubMed] [Google Scholar]
- Economides KD, Zeltser L, Capecchi MR. Hoxb13 mutations cause overgrowth of caudal spinal cord and tail vertebrae. Dev Biol. 2003;256:317–330. doi: 10.1016/s0012-1606(02)00137-9. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Ferland RJ, Cherry TJ, Preware PO, Morrisey EE, Walsh CA. Characterization of Foxp2 and Foxp1 mRNA and protein in the developing and mature brain. J Comp Neurol. 2003;460:266–279. doi: 10.1002/cne.10654. [DOI] [PubMed] [Google Scholar]
- Haesler S, Wada K, Nshdejan A, Morrisey EE, Lints T, Jarvis ED, Scharff C. FoxP2 expression in avian vocal learners and non-learners. J Neurosci. 2004;24:3164–3175. doi: 10.1523/JNEUROSCI.4369-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Zhang J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics. 2005;169:1157–1164. doi: 10.1534/genetics.104.037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirth F, Hartmann B, Reichert H. Homeotic gene action in embryonic brain development of Drosophila. Development. 1998;125:1579–1589. doi: 10.1242/dev.125.9.1579. [DOI] [PubMed] [Google Scholar]
- Hu H, Wang B, Borde M, Nardone J, Maika S, Allred L, Tucker PW, Rao A. Foxp1 is an essential transcriptional regulator of B cell development. Nat Immunol. 2006;7:819–826. doi: 10.1038/ni1358. [DOI] [PubMed] [Google Scholar]
- Kafri R, Levy M, Pilpel Y. The regulatory utilization of genetic redundancy through responsive backup circuits. Proc Natl Acad Sci U S A. 2006;103:11653–11658. doi: 10.1073/pnas.0604883103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- Kiya T, Itoh Y, Kubo T. Expression analysis of the FoxP homologue in the brain of the honeybee, Apis mellifera. Insect Mol Biol. 2008;17:53–60. doi: 10.1111/j.1365-2583.2008.00775.x. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA, Koonin EV. Origin of alternative splicing by tandem exon duplication. Hum Mol Genet. 2001;10:2661–2669. doi: 10.1093/hmg/10.23.2661. [DOI] [PubMed] [Google Scholar]
- Kopelman NM, Lancet D, Yanai I. Alternative splicing and gene duplication are inversely correlated evolutionary mechanisms. Nat Genet. 2005;37:588–589. doi: 10.1038/ng1575. [DOI] [PubMed] [Google Scholar]
- Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- Lai E, Prezioso VR, Smith E, Litvin O, Costa RH, Darnell JE., Jr. HNF-3A, a hepatocyte-enriched transcription factor of novel structure is regulated transcriptionally. Genes Dev. 1990;4:1427–1436. doi: 10.1101/gad.4.8.1427. [DOI] [PubMed] [Google Scholar]
- Lee HH, Frasch M. Survey of forkhead domain encoding genes in the Drosophila genome: classification and embryonic expression patterns. Dev Dyn. 2004;229:357–366. doi: 10.1002/dvdy.10443. [DOI] [PubMed] [Google Scholar]
- Li S, Weidenfeld J, Morrisey EE. Transcriptional and DNA binding activity of the Foxp1/2/4 family is modulated by heterotypic and homotypic protein interactions. Mol Cell Biol. 2004;24:809–822. doi: 10.1128/MCB.24.2.809-822.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MM, Li S, Yang H, Morrisey EE. Foxp4: a novel member of the Foxp subfamily of winged-helix genes co-expressed with Foxp1 and Foxp2 in pulmonary and gut tissues. Gene Expr Patterns. 2002;2:223–228. doi: 10.1016/s1567-133x(02)00058-3. [DOI] [PubMed] [Google Scholar]
- Mazet F, Yu JK, Liberles DA, Holland LZ, Shimeld SM. Phylogenetic relationships of the Fox (Forkhead) gene family in the Bilateria. Gene. 2003;316:79–89. doi: 10.1016/s0378-1119(03)00741-8. [DOI] [PubMed] [Google Scholar]
- O'Neill JW, Bier E. Double-label in situ hybridization using biotin and digoxigenin-tagged RNA probes. Biotechniques. 1994;17 870, 874–875. [PubMed] [Google Scholar]
- Ohno S. Evolution by gene duplication. New York: Springer-Verlag; 1970. [Google Scholar]
- Ohta T. Multigene families and the evolution of complexity. J Mol Evol. 1991;33:34–41. doi: 10.1007/BF02100193. [DOI] [PubMed] [Google Scholar]
- Otto SP, Yong P. The evolution of gene duplicates. Adv Genet. 2002;46:451–483. doi: 10.1016/s0065-2660(02)46017-8. [DOI] [PubMed] [Google Scholar]
- Pacheco TR, Gomes AQ, Barbosa-Morais NL, Benes V, Ansorge W, Wollerton M, Smith CW, Valcarcel J, Carmo-Fonseca M. Diversity of vertebrate splicing factor U2AF35: identification of alternatively spliced U2AF1 mRNAS. J Biol Chem. 2004;279:27039–27049. doi: 10.1074/jbc.M402136200. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Rogulja-Ortmann A, Renner S, Technau GM. Antagonistic roles for Ultrabithorax and Antennapedia in regulating segment-specific apoptosis of differentiated motoneurons in the Drosophila embryonic central nervous system. Development. 2008;135:3435–3445. doi: 10.1242/dev.023986. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Rousso DL, Gaber ZB, Wellik D, Morrisey EE, Novitch BG. Coordinated actions of the forkhead protein Foxp1 and Hox proteins in the columnar organization of spinal motor neurons. Neuron. 2008;59:226–240. doi: 10.1016/j.neuron.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schon C, Wochnik A, Rossner A, Donow C, Knochel W. The FoxP subclass in Xenopus laevis development. Dev Genes Evol. 2006;216:641–646. doi: 10.1007/s00427-006-0073-8. [DOI] [PubMed] [Google Scholar]
- Shi C, Zhang X, Chen Z, Sulaiman K, Feinberg MW, Ballantyne CM, Jain MK, Simon DI. Integrin engagement regulates monocyte differentiation through the forkhead transcription factor Foxp1. J Clin Invest. 2004;114:408–418. doi: 10.1172/JCI21100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu W, Lu MM, Zhang Y, Tucker PW, Zhou D, Morrisey EE. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development. 2007;134:1991–2000. doi: 10.1242/dev.02846. [DOI] [PubMed] [Google Scholar]
- Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006;14:159–166. doi: 10.1016/j.str.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Su Z, Wang J, Yu J, Huang X, Gu X. Evolution of alternative splicing after gene duplication. Genome Res. 2006;16:182–189. doi: 10.1101/gr.4197006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. PAUP*. Phylogenetic Analysis Using Parsimony (*and other methods) Sunderland (MA): Sinauer Associates; 2001. [Google Scholar]
- Talavera D, Vogel C, Orozco M, Teichmann SA, de la Cruz X. The (in)dependence of alternative splicing and gene duplication. PLoS Comput Biol. 2007;3:e33. doi: 10.1371/journal.pcbi.0030033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura S, Morikawa Y, Iwanishi H, Hisaoka T, Senba E. Expression pattern of the winged-helix/forkhead transcription factor Foxp1 in the developing central nervous system. Gene Expr Patterns. 2003;3:193–197. doi: 10.1016/s1567-133x(03)00003-6. [DOI] [PubMed] [Google Scholar]
- Teufel A, Wong EA, Mukhopadhyay M, Malik N, Westphal H. FoxP4, a novel forkhead transcription factor. Biochim Biophys Acta. 2003;1627:147–152. doi: 10.1016/s0167-4781(03)00074-5. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirouvanziam R, Davidson CJ, Lipsick JS, Herzenberg LA. Fluorescence-activated cell sorting (FACS) of Drosophila hemocytes reveals important functional similarities to mammalian leukocytes. Proc Natl Acad Sci U S A. 2004;101:2912–2917. doi: 10.1073/pnas.0308734101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tress ML, Martelli PL, Frankish A, et al. (46 co-authors) The implications of alternative splicing in the ENCODE protein complement. Proc Natl Acad Sci U S A. 2007;104:5495–5500. doi: 10.1073/pnas.0700800104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Q, Brown CT, Davidson EH, Oliveri P. Sea urchin Forkhead gene family: phylogeny and embryonic expression. Dev Biol. 2006;300:49–62. doi: 10.1016/j.ydbio.2006.09.031. [DOI] [PubMed] [Google Scholar]
- Weigel D, Jurgens G, Kuttner F, Seifert E, Jackle H. The homeotic gene fork head encodes a nuclear protein and is expressed in the terminal regions of the Drosophila embryo. Cell. 1989;57:645–658. doi: 10.1016/0092-8674(89)90133-5. [DOI] [PubMed] [Google Scholar]
- Wijchers PJ, Burbach JP, Smidt MP. In control of biology: of mice, men and Foxes. Biochem J. 2006;397:233–246. doi: 10.1042/BJ20060387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yagi H, Nomura T, Nakamura K, et al. (11 co-authors) Crucial role of FOXP3 in the development and function of human CD25+CD4+ regulatory T cells. Int Immunol. 2004;16:1643–1656. doi: 10.1093/intimm/dxh165. [DOI] [PubMed] [Google Scholar]
- Yu JK, Mazet F, Chen YT, Huang SW, Jung KC, Shimeld SM. The Fox genes of Branchiostoma floridae. Dev Genes Evol. 2008;218:629–638. doi: 10.1007/s00427-008-0229-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.