Abstract
Retroposition, a leading mechanism for gene duplication, is an important process shaping the evolution of genomes. Retrogenes are also involved in the gene structure evolution as a major player in the process of intron deletion. Here, we demonstrate the role of retrogenes in intron gain in mammals. We identified one case of “intronization,” the transformation of exonic sequences into an intron, in the primate specific retrogene RNF113B and two independent “intronization” events in the retrogene DCAF12L2, one in the common ancestor of primates and rodents and another one in the rodent lineage. Intron gain resulted from the origin of new splice variants, and both genes have two transcript forms, one with retained intron and one with the intron spliced out. Evolution of these genes, especially RNF113B, has been very dynamic and has been accompanied by several additional events including parental gene loss, secondary retroposition, and exaptation of transposable elements.
Keywords: intron gain, gene structure evolution, splice variant, RNF113, DCAF12
The majority of protein-coding genes in eukaryotes are interrupted by introns that are removed from the pre-mRNA by a RNA–protein complex called the spliceosome (Cavalier-Smith 1985; Crick 1979). Introns and the splicing machinery have been found in all eukaryotic species with fully sequenced genomes (Chow et al. 1977; Roy and Gilbert 2006). Comparative genomic studies have revealed striking conservation of intron positions in distant eukaryotes such as animals and plants (Fedorov et al. 2002; Rogozin et al. 2003; Carmel et al. 2007). On the other hand, many genome-wide comparisons of eukaryotic species demonstrated multiple intron losses and intron gains (Roy et al. 2003; Cho et al. 2004; Qiu et al. 2004; Coulombe-Huntington and Majewski 2007b; Li et al. 2009). However, it was found that intron gain is a very rare event in vertebrate evolution (Loh et al. 2007) and no intron gains into intact conserved coding regions of mammalian genes are known (Roy et al. 2003; Coulombe-Huntington and Majewski 2007a).
Comparative gene structure studies have not revealed any intron gain into existing exons in mammals. The only reported new introns were acquired, by and large, by either a fusion of retrogene with host genes or de novo from the genomic environment as a result of new exon capture (O'Neill et al. 1998; Vinckenbosch et al. 2006; Sela et al. 2007; Baertsch et al. 2008; Fablet et al. 2009). Here, we report two retrogenes, RNF113B and DCAF12, where the exon sequence was split by creation of a new intron as the result of mutations and emergence of new splice sites. The introns discovered by us represent cases of intron creation via recruitment of exonic sequence (intronization) proposed by Irimia et al. (2008) and Lahn and Page (1999).
Evolution of Introns
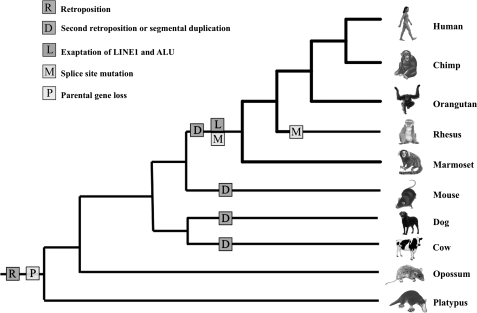
RNF113A is a retrogene encoding a ring finger protein of unknown function and is present in the genomes of all vertebrates. Interestingly, in mammalian genomes, only intronless copy exist, whereas in all other vertebrates, a ten-exon parental gene is present and no retrogenes were detected. Genomic sequence analysis showed that there are two copies of RNF113 in primates, rodents, carnivores, and even-toed ungulates and only one in the genomes of the other mammals we studied. The first copy of RNF113 was retroposed into the intronic region of NDUFA1 gene in the genome of the mammalian ancestor. Following the retroposition, the parental gene was lost. This likely took place before the divergence of Prototheria (Monotremes) and Theria (Marsupials and Placentals) because in the genomes of all species representing these lineages, the multiexon form of RNF113 is absent. After the mammalian radiation the RNF113A retrogene was duplicated, by retropositions or segmental duplications, in several lineages. Analysis of genomic locations of these copies suggests that the duplication events were independent in each lineage. For example, in rodents, the RNF113 copy (RNF113A2) was inserted into an intron of the 2900006K08Rik gene, whereas the primate specific gene, RNF113B, was copied into an intron of the FARP1 gene. The primate specific duplication happened before Old World Monkeys and New World Monkeys diverged (fig. 1).
FIG. 1.
Schematic tree representing major events during evolution of RNF113 gene in mammalian lineage. Color version of the figure can be found in supplementary Data (Supplementary Material online).
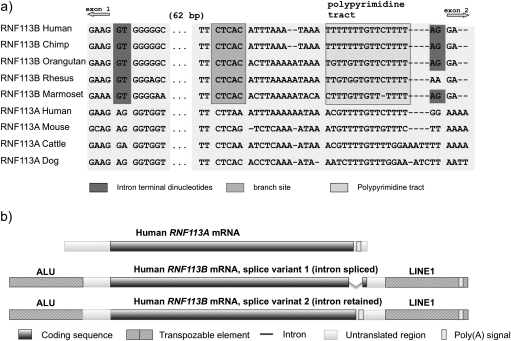
After the retroposition/duplication, the primate specific RNF113B gene underwent rapid evolution including intron gain. The presence of the intron is surprising, however, it is supported by several GenBank mRNA sequences (accession numbers: AF539427, BC025388, and BC017585). To confirm the existence of the intron and learn about its origin, we compared RNF113B sequences from available primate genomes (human, marmoset, macaque, orangutan, and chimpanzee) with sequences of other mammalian RNF113A genes. Sequence alignment revealed that the intron of RNF113B is not a de novo insertion but rather originated from the exonic sequence (fig. 2a). A double point mutation, AG → GT, generated the donor site (fig. 2a). The origin of acceptor site is not so clear. One possibility is that a point mutation, GG → AG, created acceptor site. Another option is that the acceptor site was brought during the exonization of L1 element, merged at the 3’ end of RNF113B (fig. 2b). The newly generated splice sites together with the branch site and the polypyrimidine tract likely enabled recognition of the new intron by the U2 spliceosome (fig. 2a). The 105 bp intron contains 59 nucleotides of previously coding sequence and 46 nucleotides from the 3’ UTR.
FIG. 2.
(a) Alignment of mammalian RNF113A and primate RNF113B genomic sequences at the acceptor and donor sites. (b) Structure of human RNF113A mRNA and two splice variants of RNF113B. Color version of the figure can be found in supplementary Data (Supplementary Material online).
Generation of splice sites most probably occurred in the primate specific RNF113B copy since neither human RNF113A gene, which gave a rise to primate RNF113B, nor RNF113A genes from other mammals have AG or GT at the donor and acceptor positions. Splicing signals were formed before the Old World Monkeys and New Monkeys split. Interestingly, loss of the splicing boundaries subsequently converted the intron into a “retained intron” in some primates. In rhesus, for example, acceptor was lost due to a point mutation (AG → AA change) (fig. 1b).
The creation of splicing signals was accompanied not only by exaptation of an L1 element but also by exonization of an Alu element. The L1 element inserted within the 3′ end of the gene could have contributed the acceptor site and provided a new polyA signal used for the new splice variant (fig. 2a). The complete AluSx element transposed upstream the gene was exapted at the 5′ end and most probably delivered some regulatory elements.
Sequencing of the human RNF113B cDNA using primers flanking the intronic sequence revealed that RNF113B produces two variant transcripts. One variant has two exons, as described above, and the other one is a single exon transcript similar to RNF113A. Consequently, most primates have three transcripts of RNF113: one from the RNF113A retrogene and two from the RNF113B (fig. 2b). Rodents, cow, and dog have two transcripts, each coming from different copy of RNF113, and all other mammals have only one RNF113 transcript. The presence of the splice variants in the retrogene is very surprising and has only been reported once before (Lahn and Page 1999).
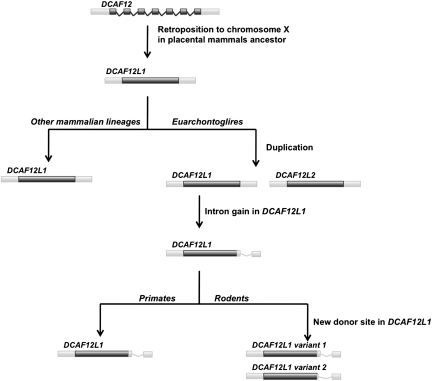
A second case involves DCAF12 (DDB1 and CUL4 associated factor 12), which encodes a WD repeat-containing protein that interacts with the COP9 signalosome (Jin et al. 2006). Although the gene is present in vertebrate and insect genomes, only placental mammals have retrocopies of this gene. One copy, DCAF12L2, has the same location in all placental mammals and therefore most likely was retroposed in the placental mammals ancestor. Another copy, DCAF12L1, is present only in Euarchontoglires (a clade which includes rodents and primates). It likely emerged as a result of tandem duplication of DCAF12L2 as it is located next to the DCAG12L1 gene. There were two events that changed the splicing pattern in DCAF12L2. First, an intronization event occurred in the common ancestor of primates and rodents. Second, an alternative donor site emerged in rodents only (fig. 3). The limited available data and sequence divergence make any conclusions in regard to the exact pattern of splice site evolution infeasible. However, there is convincing experimental evidence confirming both splicing events (fig. 3): splicing at the shared rodent–primate intron, boundaries are confirmed by two expressed sequence tags (ESTs) (AK034343 and AK047360), and usage of the rodent alternative donor site is confirmed by four ESTs (AK038557, BC068319, AK034472, and AK039767).
FIG. 3.
Pattern of DCAF12 duplication and “intronization” events in mammalian genomes. Color version of the figure can be found in supplementary Data (Supplementary Material online).
Retrogene Expression
Numerous studies revealed a tendency of retrogenes to be expressed exclusively in testis. It was suggested that the hypertranscription present in the meiotic and postmeiotic spermatogenic cells makes possible transcription of DNA that is usually not transcribed. This may facilitate transcription of retrocopies in the testis during their early evolution (reviewed in (Kaessmann et al. 2009). Another hypothesis explains the high expression of retrogenes in testis by the fact that these are, in most cases, retrocopies of spermatogenesis-related genes located on the X chromosome. Because the X chromosome is inactivated during meiosis, retroposition to autosomes enables escape from inactivation and expression during spermatogenesis (Turner 2007).
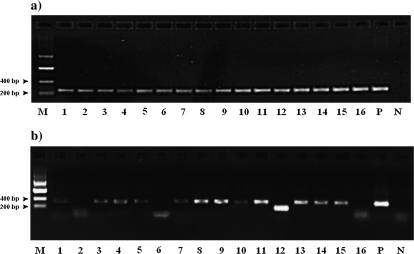
The retroposition of both genes studied here, RNF113 and DCAF12, was in the opposite direction, from autosomes to chromosome X. In the case of RNF113, the parental gene is detectable by sequence similarity as an apparent pseudogene on chromosome 9. The parental multiexon DCAF12 gene is coincidentally also located on chromosome 9. RNF113A and both DCAF12 retrogenes are on chromosome X. We surveyed the expression pattern of all human RNF113 transcripts (one from RNF113A and two from RNF113B) in 16 human tissues (fig. 4) (for methods, see Supplementary Material online). RNF113A was expressed in all studied tissues, including testes. Interestingly, RNF113B exhibited tissue-specific splicing; while the unspliced form of RNF113B was expressed in all tissues but testis, the spliced variant was expressed in testis, prostate, thymus, and lung. Both RNF113B splice variants were present in thymus, prostate, and lung, but in all of these tissues, the form with the intron spliced out had much lower expression level than the single exon primary form. Relatively high expression of the new form of RNF113B, form with the intron spliced out, was observed only in testis.
FIG. 4.
Expression pattern of RNF113A and two forms of RNF113B (195 bp product with intron spliced; 295 bp product-form with intron retained) in 16 human tissues: 1: heart, 2: brain, 3: placenta, 4: lung, 5: liver, 6: skeletal muscle, 7: kidney, 8: pancreas, 9: spleen, 10: thymus, 11: prostate, 12: testis, 13: ovary, 14: small intestine w/o mucosal lining, 15: colon, 16: peripheral leukocytes, P: genomic DNA, and N: water.
According to the EST data, the human DCAF12 gene is widely expressed. EST sequences present in the dbEST database represent almost 40 libraries and show the highest expression in testis and trachea. The retrogene DCAF12L1 is expressed only in kidney and testis and a second human retrogene, DCAF12L2, is expressed in eye and testis. Therefore, both retrogenes show very different expression patterns than their parental genes, with very limited and low expression level and notable expression in testis.
Conclusions
Retroposition, a major mechanism for gene duplication, is an important process shaping the evolution of genomes (Brosius 1991; Marques et al. 2005). Our study confirms the unusual role of retrogenes in shaping the genomes and underscores the importance of mobile elements in evolution. It also reveals that retrogenes may be responsible for a wealth of species-specific features including species-specfic introns and splice variants.
Previous analyses of introns in the vertebrate genomes did not uncover any intron gain in mammals (Roy et al. 2003). Our study clearly shows that creation of introns has occurred during mammalian evolution. The failure of previous studies to find intron gains can be explained by the fact that they were focused on different intron gain mechanisms and did not consider exon intronization. In addition, they looked at conserved among studied species genes, while we focused on young and in many cases lineage-specific retrogenes.
Interestingly, the retrogenes studied here exhibit testis-specific expression typically associated with genes escaping from the X chromosome despite their opposite history (retroposition from autosome to X). This biased expression pattern may not be exclusively related to meiotic genes, sex chromosome inactivation, and dosage compensation (Marques et al. 2005; Vinckenbosch et al. 2006; Potrzebowski et al. 2008). The same pattern of high expression level in testis is observed in young, primate-specific splice variant of retrogene RNF113B as well as in both retroposed copies of DCAF12 retroposed on the human X chromosome. The older, unspliced variant of RNF113B, as well as an earlier retrocopy RNF113A, displays more diverse expression patterns. Therefore, testis-specific expression could be a common feature of all newly evolved transcripts regardless of their chromosomal localization and may reflect a transcriptional noise due to “hypertranscription” in testis, facilitating the activation of new transcripts (Kleene et al. 1998).
The small number of observed intron gain in retrogenes may reflect that this is a rare event. Alternatively, the low number of observations could reflect the difficulties in identification of such events. One major complication lies in annotation problems and the common expectation that retrogenes do not have introns. Genome-wide comparative studies currently underway have already showed that intron gain in retrogenes could be more frequent than we expected but that annotations remain a major obstacle in uncovering this phenomenon.
Supplementary Material
Supplementary Data are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Jurgen Brosius and two reviewers for their comments and insightful suggestions. I.B.R. was supported by the Intramural Research Program of the National Library of Medicine at National Institutes of Health/U.S. Department of Health and Human Services.
References
- Baertsch R, Diekhans M, Kent WJ, Haussler D, Brosius J. Retrocopy contributions to the evolution of the human genome. BMC Genomics. 2008;9:466. doi: 10.1186/1471-2164-9-466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. Retroposons–seeds of evolution. Science. 1991;251:753. doi: 10.1126/science.1990437. [DOI] [PubMed] [Google Scholar]
- Carmel L, Wolf YI, Rogozin IB, Koonin EV. Three distinct models of intron dynamics in the evolution of eukaryotes. Genome Res. 2007;17:1034–1044. doi: 10.1101/gr.6438607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalier-Smith T. Selfish DNA and the origin of introns. Nature. 1985;315:283–284. doi: 10.1038/315283b0. [DOI] [PubMed] [Google Scholar]
- Cho S, Jin SW, Cohen A, Ellis RE. A phylogeny of caenorhabditis reveals frequent loss of introns during nematode evolution. Genome Res. 2004;14:1207–1220. doi: 10.1101/gr.2639304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ. An amazing sequence arrangement at the 5' ends of adenovirus 2 messenger RNA. Cell. 1977;12:1–8. doi: 10.1016/0092-8674(77)90180-5. [DOI] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Majewski J. Characterization of intron loss events in mammals. Genome Res. 2007b;17:23–32. doi: 10.1101/gr.5703406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulombe-Huntington J, Majewski J. Intron loss and gain in Drosophila. Mol Biol Evol. 2007a;24:2842–2850. doi: 10.1093/molbev/msm235. [DOI] [PubMed] [Google Scholar]
- Crick F. Split genes and RNA splicing. Science. 1979;204:264–271. doi: 10.1126/science.373120. [DOI] [PubMed] [Google Scholar]
- Fablet M, Bueno M, Potrzebowski L, Kaessmann H. Evolutionary origin and functions of retrogene introns. Mol Biol Evol. 2009; 26:2147–2156. doi: 10.1093/molbev/msp125. [DOI] [PubMed] [Google Scholar]
- Fedorov A, Merican AF, Gilbert W. Large-scale comparison of intron positions among animal, plant, and fungal genes. Proc Natl Acad Sci U S A. 2002;99:16128–16133. doi: 10.1073/pnas.242624899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Rukov JL, Penny D, Vinther J, Garcia-Fernandez J, Roy SW. Origin of introns by 'intronization' of exonic sequences. Trends Genet. 2008;24:378–381. doi: 10.1016/j.tig.2008.05.007. [DOI] [PubMed] [Google Scholar]
- Jin J, Arias EE, Chen J, Harper JW, Walter JC. A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol Cell. 2006;23:709–721. doi: 10.1016/j.molcel.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Kaessmann H, Vinckenbosch N, Long M. RNA-based gene duplication: mechanistic and evolutionary insights. Nat Rev Genet. 2009;10:19–31. doi: 10.1038/nrg2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleene KC, Mulligan E, Steiger D, Donohue K, Mastrangelo MA. The mouse gene encoding the testis-specific isoform of Poly(A) binding protein (Pabp2) is an expressed retroposon: intimations that gene expression in spermatogenic cells facilitates the creation of new genes. J Mol Evol. 1998;47:275–281. doi: 10.1007/pl00006385. [DOI] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Retroposition of autosomal mRNA yielded testis specific gene family on human Y chromosome. Nat Genet. 1999;21:429–433. doi: 10.1038/7771. [DOI] [PubMed] [Google Scholar]
- Li W, Tucker AE, Sung W, Thomas WK, Lynch M. Extensive, recent intron gains in Daphnia populations. Science. 2009;326:1260–1262. doi: 10.1126/science.1179302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh Y-H, Brenner S, Venkatesh B. Investigation of loss and gain of introns in the compact genomes of pufferfishes (Fugu and Tetraodon) Mol Biol Evol. 2007;25:526–535. doi: 10.1093/molbev/msm278. [DOI] [PubMed] [Google Scholar]
- Marques AC, Dupanloup I, Vinckenbosch N, Reymond A, Kaessmann H. Emergence of young human genes after a burst of retroposition in primates. PLoS Biol. 2005;3:e357. doi: 10.1371/journal.pbio.0030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill RJ, Brennan FE, Delbridge ML, Crozier RH, Graves JA. De novo insertion of an intron into the mammalian sex determining gene, SRY. Proc Natl Acad Sci U S A. 1998;95:1653–1657. doi: 10.1073/pnas.95.4.1653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrzebowski L, Vinckenbosch N, Marques AC, Chalmel F, Jegou B, Kaessmann H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008;6:e80. doi: 10.1371/journal.pbio.0060080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu WG, Schisler N, Stoltzfus A. The evolutionary gain of spliceosomal introns: sequence and phase preferences. Mol Biol Evol. 2004;21:1252–1263. doi: 10.1093/molbev/msh120. [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Wolf YI, Sorokin AV, Mirkin BG, Koonin EV. Remarkable interkingdom conservation of intron positions and massive, lineage-specific intron loss and gain in eukaryotic evolution. Curr Biol. 2003;13:1512–1517. doi: 10.1016/s0960-9822(03)00558-x. [DOI] [PubMed] [Google Scholar]
- Roy SW, Fedorov A, Gilbert W. Large-scale comparison of intron positions in mammalian genes shows intron loss but no gain. Proc Natl Acad Sci U S A. 2003;100:7158–7162. doi: 10.1073/pnas.1232297100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy SW, Gilbert W. The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet. 2006;7:211–221. doi: 10.1038/nrg1807. [DOI] [PubMed] [Google Scholar]
- Sela N, Mersch B, Gal-Mark N, Lev-Maor G, Hotz-Wagenblatt A, Ast G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JM. Meiotic sex chromosome inactivation. Development. 2007;134:1823–1831. doi: 10.1242/dev.000018. [DOI] [PubMed] [Google Scholar]
- Vinckenbosch N, Dupanloup I, Kaessmann H. Evolutionary fate of retroposed gene copies in the human genome. Proc Natl Acad Sci U S A. 2006;103:3220–3225. doi: 10.1073/pnas.0511307103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.