Abstract
Chromosomal polymorphisms, such as inversions, are presumably involved in the rapid adaptation of populations to local environmental conditions. Reduced recombination between alternative arrangements in heterozygotes may protect sets of locally adapted genes, promoting ecological divergence and potentially leading to reproductive isolation and speciation. Through a comparative analysis of chromosomal inversions and microsatellite marker polymorphisms, we hereby present biological evidence that strengthens this view in the mosquito Anopheles funestus s.s, one of the most important and widespread malaria vectors in Africa. Specimens were collected across a wide range of geographical, ecological, and climatic conditions in Cameroon. We observed a sharp contrast between population structure measured at neutral microsatellite markers and at chromosomal inversions. Microsatellite data detected only a weak signal for population structuring among geographical zones (FST < 0.013, P < 0.01). By contrast, strong differentiation among ecological zones was revealed by chromosomal inversions (FST > 0.190, P < 0.01). Using standardized estimates of FST, we show that inversions behave at odds with neutral expectations strongly suggesting a role of environmental selection in shaping their distribution. We further demonstrate through canonical correspondence analysis that heterogeneity in eco-geographical variables measured at specimen sampling sites explained 89% of chromosomal variance in A. funestus. These results are in agreement with a role of chromosomal inversions in ecotypic adaptation in this species. We argue that this widespread mosquito represents an interesting model system for the study of chromosomal speciation mechanisms and should provide ample opportunity for comparative studies on the evolution of reproductive isolation and speciation in major human malaria vectors.
Keywords: chromosome inversions, Anopheles funestus, natural selection, local adaptation, malaria, Cameroon
Introduction
Chromosomal polymorphism (such as inversions, robertsonian fusions and fissions, and translocations) has been recognized as a major driving force in local adaptation, speciation processes, and evolution of sex chromosomes (King 1993; Noor et al. 2001; Rieseberg 2001; Hoffmann et al. 2004; van Doorn and Kirkpatrick 2007). The main evolutionary significance of inversions may come from the fact that they reduce recombination in heterozygotes, protecting genomic regions from introgression (Stump et al. 2007; Kulathinal et al. 2009). Chromosomal inversions can favor local adaptation by capturing sets of locally adapted genes, therefore promoting ecological divergence and fostering reproductive isolation between incipient species (Ortiz-Barrientos et al. 2002; Navarro and Barton 2003; Butlin and Roper 2005; Kirkpatrick and Barton 2006). In this sense, chromosomal inversions have been repeatedly regarded as genetic markers for ecotypic adaptation (Coluzzi et al. 1985; Hoffmann and Willi 2008). Strong and significant correlations between chromosomal inversion frequencies and environmental parameters have been reported in a number of plant and animal species (Coluzzi et al. 1979; Rodriguez-Trelles et al. 1996; Hoffmann et al. 2004). The adaptive significance of chromosomal inversions is further testified by the presence of parallel geographical clines of inversions on different continents, the rapid reestablishment of inversion frequencies after a new habitat is colonized, and documented seasonal shifts in inversion frequencies associated with environmental conditions in fruitflies and anopheline mosquitoes (Coluzzi et al. 1979; Balanya et al. 2003).
Afrotropical anopheline mosquitoes are emerging as suitable biological models for the study of the role of chromosomal inversions in ecological adaptation and speciation as exemplified by the rapidly growing literature available on this topic in the malaria mosquito A. gambiae (Coluzzi et al. 2002; della Torre et al. 2002). A conceptual model for ecological speciation within the A. gambiae complex was first proposed by Coluzzi (1982) more than 25 years ago. This “ecotypification” model explored the role of paracentric polymorphic chromosomal inversions in promoting and maintaining ecological divergence between populations in the context of rapidly fluctuating populations subject to recurrent niche expansions and contractions. In this model, sets of locally adapted alleles are selected in peripheral populations as the species expands into marginal habitats during phases of high population growth as typically observed in these mosquito species all through the rainy season. During phases of population contraction, which occur in the dry season, the peripheral populations become geographically and ecologically distinct from the species core. Then, by chance alone, sets of locally adapted alleles may be captured by a chromosomal inversion in peripheral populations, thereby preventing recombination during the next demographic expansion. Once an inversion is established, new locally adapted alleles are expected to accumulate within the inversion, strengthening ecological divergence and promoting assortative mating between ecotypes eventually leading to speciation. Empirical support for the ecotypification model of chromosomal speciation in A. gambiae was provided recently (Manoukis et al. 2008; Costantini et al. 2009; Simard et al. 2009).
Beside pure academic interest, a clear understanding of the mechanisms underlying the dynamics of ecological adaptation, ecological niche divergence, and speciation in major disease vectors has relevance for the epidemiology of the diseases being transmitted and has direct implications for vector control (Besansky et al. 2003; White et al. 2007). Together with A. gambiae, A. funestus is considered a major vector of the deadly malaria parasite Plasmodium falciparum in Africa (Gillies and de Meillon 1968). This highly anthropophilic mosquito, widely spread throughout sub-Saharan Africa, populates a wide range of habitats and presents a high level of chromosomal polymorphism (Coetzee and Fontenille 2004). Nonrandom distribution of chromosomal inversions, strong and significant heterozygote deficits, and linkage disequilibrium (LD) between inversions within natural populations of A. funestus have been described and interpreted as indicative of environmental adaptation and/or incipient speciation, a situation that is reminiscent of A. gambiae (Costantini et al. 1999; Cohuet et al. 2005; Guelbeogo et al. 2005; Michel, Guelbeogo, et al. 2005).
Here, we focus on the vector species A. funestus and present original data that strengthen the view that chromosomal inversions are strongly exposed to natural selection in this species. We collected samples of A. funestus from different ecological zones in Cameroon, a country in Central Africa with highly heterogeneous landscapes. We first compared the spatial population genetic structure inferred from presumably neutral nuclear microsatellite markers with that inferred from chromosomal inversions. In doing so, we show that the distribution of chromosomal polymorphism is at odds with neutral expectations. Then, using canonical correspondence analysis (CCA), we relate inversion frequencies to environmental data and elucidate the contribution of a set of eco-geographical variables (EGVs) to the distribution of chromosomal polymorphisms in natural populations of A. funestus. Our results strengthen the view that selection due to environmental factors probably is a major architect of the distribution of chromosomal inversion polymorphism in A. funestus. However, the shallow population structure revealed by microsatellite markers localized within and outside of the inversions suggests that recombination between chromosomal rearrangements still occurs and allows significant gene flow at least in genomic areas not targeted by natural selection as previously observed in A. gambiae (Stump et al. 2007; White et al. 2007, 2009). We highlight the relevance of our findings for comparative genomic studies and discuss their implications for malaria transmission and control.
Materials and Methods
Mosquitoes and Sampling Sites
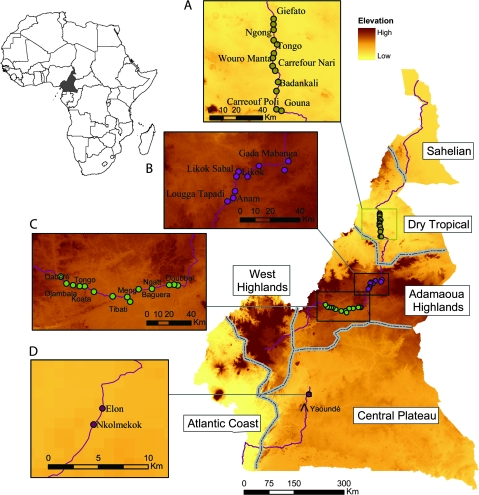
Mosquitoes were sampled in four different ecological zones distributed along a latitudinal transect across Cameroon from August to December 2005, which included a total of 36 villages (fig. 1 and table 1). In each village, mosquito specimens were collected by daytime indoor pyrethroid spraying (Service 1993). Among the 663 A. funestus females collected, 313 were at the half-gravid stage. Their ovaries were dissected in the field and placed individually in labeled 1.5 ml tubes containing Carnoy’s solution (three parts of 100% ethanol: one part of glacial acetic acid by volume) and stored at−20 °C for cytological analysis (below). Mosquitoes’ carcasses were stored individually at room temperature in labeled tubes with desiccant.
FIG. 1.
Sampling sites. Topographic Map of Cameroon showing the study area divided into four ecological zones (A–D) and villages sampled in each zone (dots). Dotted lines delimit biogeographic domains as defined in Olivry (1986). Elevation Source: Shuttle Radar Topography Mission, http://www2.jpl.nasa.gov/srtm
Table 1.
Sampling Details.
| Number of Villages | Number of Specimens | Length (km) | Width (km) | Bioclimatic Domain | Elevation (m) | Temp. (°C) | Rainfall (mm) | WVP (%) | |
| Zone A | 13 | 286 | 70 | 6 | Dry tropical | 300 | 27 | 1,100 | 19.9 |
| Zone B | 10 | 117 | 25 | 37 | Highlands | 1,100 | 22 | 1,500 | 17.0 |
| Zone C | 12 | 213 | 25 | 106 | Highlands | 900 | 23 | 1,700 | 20.9 |
| Zone D | 2 | 47 | 2 | 2 | Rainforest | 500 | 25 | 1,600 | 24.3 |
NOTE.—Bioclimatic domain classification from Olivry (1986); elevation: mean elevation (in meters); rainfall: mean yearly rainfall (in mm); temp.: mean yearly temperature (in °C); WVP: mean yearly water vapor pressure values (in %).
Cytogenetics
Ovaries of half-gravid females were squashed and stained according to standard protocols to reveal the polytene chromosomes (Hunt 1973).The preparations were examined under a phase contrast microscope, and paracentric inversions were scored according to the A. funestus chromosome map (Sharakhov et al. 2004). The banding patterns of polytene chromosomes provide an excellent visualization of general chromatin structure, such as inversions or deletions, allowing determination of the three karyotypic states: standard homokaryotypes (encoded as +/+), inverted homokaryotypes (−/−) and heterozygotes for the inversion (+/−). For population genetics analyses, alternative chromosomal arrangements were considered as different alleles at one locus as previously described (Cohuet et al. 2005). Goodness of fit to Hardy–Weinberg expectations, observed (Ho) and expected (He) heterozygosity (Nei 1978, 1987) and LD between inversions were computed using FSTAT 2.9.3.2 (Goudet 2001).
DNA Extraction and Polymerase Chain Reaction Identification
Genomic DNA was extracted from the body of mosquitoes, which had been successfully karyotyped on all chromosomal arms, using the protocol described in Morlais et al. (2004). DNA was then resuspended in sterile water in individual tubes. Morphological identification of A. funestus s.s. was confirmed by molecular methods (Koekemoer et al. 2002; Cohuet et al. 2003). Two A. leesoni specimens were identified and excluded from the analysis. Finally, a total of 269 A. funestus s.s were used in this study.
Microsatellite Genotyping and Analysis
Sixteen physically mapped microsatellite loci were selected from published data (Sinkins et al. 2000; Cohuet et al. 2002; Sharakhov et al. 2004; Michel, Guelbeogo, et al. 2005). Microsatellite markers were distributed on all chromosome arms inside and outside common polymorphic chromosomal inversions (supplementary fig. S1, Supplementary Material online). Polymerase chain reaction (PCR) amplifications were performed for each locus individually in a 25 μl reaction volume containing 5× PCR buffer including 7.5 mM MgCl2 at pH 8.5 (Promega, France), 200 μM of each dNTP (Eurogentec, France), 0.5 units of Taq Polymerase (Promega, France), 10 pmol of each primer, and 5–10 ng of template DNA. The forward primer was 5′-end labeled with PET, VIC, 6FAM, or NED fluorescent dye (Eurogentec, Belgium) to allow for multiplexing prior to electrophoresis. PCR amplifications were carried out in a GeneAmp 9700 thermocycler (Applied Biosystems, France). Cycling conditions were 94 °C for 2 min followed by 36 cycles at 94 °C for 30 s, 54 °C for 30 s, and 72 °C for 30 s and a final extension step at 72 °C for 10 min. PCR products were diluted 1:75—1:100 depending on the fluorescent dye and loaded onto a AB3130sequencer (Applied Biosystems, France) as three multiplex sets. Alleles were scored at each locus using the software GeneMapper v4.0 (Applied Biosystems, France).
Because of low sample sizes of karyotyped females at the village scale (from 2 to 22 specimens per village), mosquitoes were pooled according to their ecological zone of origin to conduct population genetics analyses. Pooling was justified based on 1) the results of a former study of A. funestus in Cameroon, which revealed only a weak population structure among villages across the country (Cohuet et al. 2005) and 2) a preliminary population genetics analysis of our data based on a subset of villages with sample sizes above ten specimens that provided no evidence for significant population structuring within each ecological zone. We tested for the presence of null alleles using MICRO-CHECKER software (Van Oosterhout et al. 2004) for all loci. For each microsatellite locus and overall, we estimated Ho, He, and allelic richness (Rs) (el Mousadik and Petit 1996) using FSTAT 2.9.3.2 (Goudet 2001). Conformity to Hardy–Weinberg equilibrium (HWE) and LD between pairs of loci was tested using FSTAT 2.9.3.2 (Goudet 2001).
Genetic differentiation between ecological zones was examined using exact tests and F-statistics (Wright 1951) calculated according to Weir and Cockerham (1984) in FSTAT 2.9.3.2 (Goudet 2001). In order to compare levels of differentiation for chromosomal versus microsatellite markers, it is essential to take into account the fundamental differences in the nature and properties of each maker class. Chromosomal inversions are codominant biallelic markers, whereas microsatellite loci are codominant multiallelic markers. They have very different levels of heterozygosity and evolve following distinct mutational models. We used a procedure proposed by Hedrick (2005) and implemented in the RecodeData program (Meirmans 2006) to standardize levels of differentiation according to heterozygosity. This software recodes genetic data by maximizing heterozygosity in the total population, given the observed heterozygosity within subpopulations and derives standardized FST estimators, therefore allowing for direct comparisons between estimates obtained from markers with different mutation rates (Meirmans 2006). To compare genetic variation in chromosomal arrangements to that measured with microsatellite markers, we also tested LD between inversions and microsatellite markers using FSTAT 2.9.3.2 (Goudet 2001).
Homogeneity Across Loci and the Role of Selection
Conformity to expectation under neutral evolution was tested individually for each of the 16 microsatellite loci used in this study through a model-based test directed toward the detection of outlier loci. We used the software package FDIST2 (http://www.rubic.rdg.ac.uk/∼mab/software.html) to identify microsatellite loci showing unusually high FST values, taking into account their heterozygosity (Beaumont and Nichols 1996). We ran 500,000 coalescent simulations to characterize the joint distribution of FST and heterozygosity, using a 50-demes island model (with 4 sampled demes corresponding to the 4 sampled zones) with an average differentiation equal to that observed across loci (FST = 0.005). Although the true demographic history of A. funestus populations is likely to depart from the island model assumptions, the distribution of FST estimates so obtained should be robust to the vagaries of demographic history (Beaumont 2005). We used a stepwise mutation model as implemented in FDIST2. We obtained a close approximation to the expected joint distribution of the (FST, He) estimates following (Vitalis et al. 2001), using a 2D histogram of 100 × 100 square cells (see the Appendix in Vitalis et al. 2001). We used the averaged shifted histogram algorithm (Scott 1992) to smooth the simulated data and provide a more continuous region. For each observation (FST, He) in the sample data set, we computed its associated probability in the simulated data set (i.e., the relative frequency of this observation over the full set of simulated data under the null hypothesis of selective neutrality). Then, we derived the empirical P value for this observation as one minus the sum of all cell probabilities within the simulated data set that are more probable than the observed data.
Then, we aimed at testing whether chromosomal inversions showed outstanding FST values when compared with the microsatellite markers considered here as presumably neutral. Because homoplasy may not be properly accounted for by the above procedure proposed by Hedrick (2005) and Meirmans (2006), we approximated the expected distribution of FST conditional upon heterozygosity for chromosomal inversions, given the reference level of differentiation provided by the microsatellite loci. To that end, we modified the software package DFDIST (http://www.rubic.rdg.ac.uk/cgi-bin/MarkBeaumont/dirlist1.cgi; Beaumont and Balding 2004), which was specifically designed for the analysis of dominant markers (Bonin et al. 2006), in order to simulate codominant biallelic data (code available upon request). We therefore ran 500,000 coalescent simulations of biallelic markers to characterize the joint distribution of FST and heterozygosity, using a 50-demes island model (with 4 sampled demes corresponding to the 4 sampled zones) with an average differentiation equal to that observed across microsatellite loci (FST = 0.005). Here, we used the overall heterozygosity of the pooled sample, which makes the conditional density behave better, particularly for biallelic loci. We used the same algorithms as described above to characterize the joint distribution of FST and He and to calculate the empirical P values associated with each studied polymorphism.
Genetic Cluster Analyses and Isolation by Distance
To infer population structure and to assign specimens to clusters without a priori knowledge of population units, a Bayesian model-based clustering algorithm was used as implemented in STRUCTURE v2.2 (Pritchard et al. 2000). Individual multilocus genotypic and karyotypic data were used to cluster individuals into K groups, minimizing HWE and gametic phase disequilibrium between loci within groups (Falush et al. 2003). In STRUCTURE v2.2, the number of distinct genetic clusters in the data set (K) was estimated from 1 to 12 by the posterior probability of data under each K, P(K|X) (Pritchard et al. 2000). Each run carried out 500,000 iterations after a burn-in period of 40,000, using the admixture model and correlated allele frequencies. Five replicates for each value of K were performed to check for the convergence of the Markov Chain Monte Carlo simulations (Fontaine et al. 2007).
Geographic distances can determine patterns of genetic differentiation across populations (Wright 1973). The correlation between genetic differentiation and the logarithm of geographic distances between mosquito collection sites (estimated through Geographical Positioning System, GPS) was tested using the Mantel test available in GENEPOP v4 (Rousset 1997). We used the statistics “êr” (Watts et al. 2007), that estimates an analogous of the parameter FST/(1 - FST) between pairs of individuals, to compute the regression. Bootstrapped confidence intervals for the slope of the regression (Leblois et al. 2003) were estimated across loci for microsatellites and inversions systems. The inverse of the regression slope provides an estimate of the “neighborhood size,” Dσ2, that is, the product of the effective population density (D) by the average squared parent–offspring distance (σ2) (Rousset 1997).
Eco-Geographic Determinants of Chromosomal Inversion Distributions
CCA was carried out to explore the relationships between chromosomal inversions and EGVs using the software CANOCO 4.0 (Ter Braak 1987). CCA shows the main pattern of variation in the distribution of chromosomal inversions as accounted for by the environmental variables and the distribution of the chromosomal inversions along each environmental variable. Thus, this technique allows a quick appraisal of how the distribution of chromosomal inversions varies with the environment. Six different EGVs were considered for each village: latitude (coordinates Universal Transverse Mercator [UTM]), longitude (coordinates UTM), altitude (m), rainfall (mm, yearly means averaged over the last 30 years), water vapor pressure (relative humidity in %, yearly means averaged over the last 30 years), and temperature (°C, yearly means averaged over the last 30 years). The first three EGVs were obtained directly from the field using portable GPS. The other three EGVs were extracted from the LocClim database developed by the Food and Agriculture Organization (http://www.fao.org/sd/2002/EN1203a_en.htm). Cytogenetic data were encoded as presence (1) or absence (0) of each chromosomal state (standard or inverted) for each inversion. Statistical significance of the canonical axes and environmental variables was tested with a Monte Carlo permutation test using 5,000 permutations, which allowed estimation of the 95% confidence interval around each estimate.
Results
Genetic Diversity
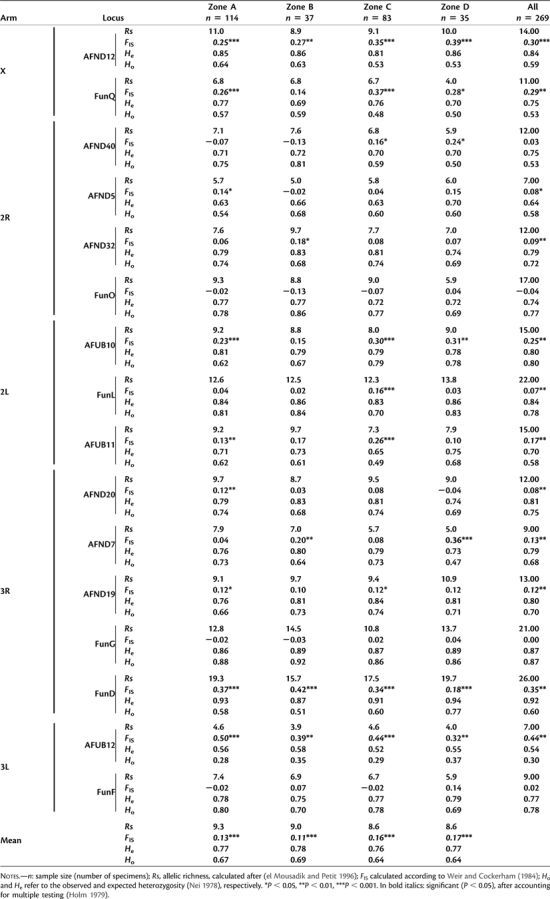
The genotypes of 269 A. funestus females collected in four distinct ecological zones in Cameroon (fig. 1) were determined at 16 microsatellite loci spread throughout the genome (supplementary fig. S1, Supplementary Material online). Mean allelic richness (Rs) was 9.2 alleles per microsatellite locus (table 2) ranging from 4.3 (locus AFUB12) to 18.1 (locus FUND). Mean allelic richness was slightly higher in Zone A (Rs = 9.3) as compared with other zones although no significant difference was observed among ecological zones (Wilcoxon signed-rank test, P = 0.72). Observed heterozygosity (Ho) ranged from 0.28 (AFUB12) to 0.92 (FUNG) and expected heterozygosity (He) from 0.52 (AFUB12) to 0.94 (FUND). There was significant difference among ecological zones neither in Ho (Wilcoxon signed-rank test, P = 0.71) nor in He (Wilcoxon signed-rank test, P = 0.98). No significant LD was found among pairs of microsatellite loci across the country and within zones. Significant heterozygote deficits were detected at some loci in each ecological zone (table 2). Locus FUND consistently showed significant heterozygote deficits across all ecological zones. Only AFND12, FUND, and AFUB12 showed some evidence for the presence of null alleles. However, subsequent analyses did not provide different results with these loci excluded and therefore, the results below are given for the entire set of microsatellite markers (Chapuis and Estoup 2007).
Table 2.
Polymorphism at 16 Microsatellite Loci in Anopheles funestus Collected in Four Ecological Zones of Cameroon.
 |
Full karyotypes along the chromosomal complement were scored for all 269 specimens. Seven paracentric inversions out of the 15 polymorphic chromosomal rearrangements known for A. funestus (Sharakhov et al. 2004) were found in our samples (table 3). Chromosomal inversions were recorded in all chromosomal arms except in the left arm of chromosome 2 and chromosome X. Inversions frequencies varied from 0.06 (2Rt) to 0.83 (3La). When considering all data together, significant deviation from HWE was detected for all inversion systems (exact test, P < 0.001). LD between all pairs of inversion systems was highly significant (G-test, P < 0.001) except between the 2Rh and 2Rt systems. LD between inversions have been reported in a large number of evolutionary contexts (Hoffmann and Rieseberg 2008). Epistatic effects between different inversions, protecting different coadapted genes, may explain LD pattern among inversions.
Table 3.
Polymorphism at Seven Chromosomal Inversion in Anopheles funestus Collected in Four Ecological Zones in Cameroon.
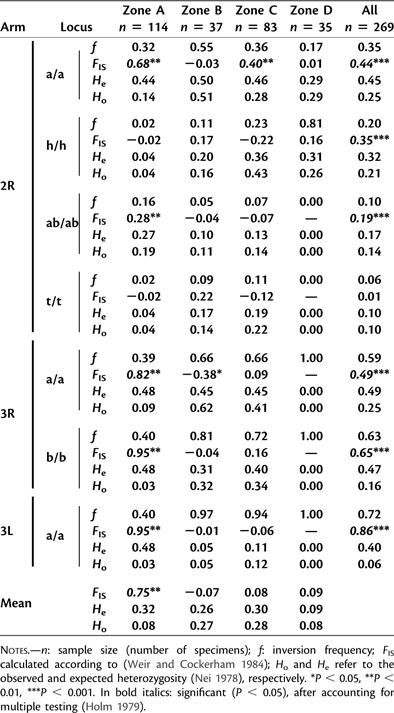 |
Although some microsatellite loci mapped within the DNA region encompassed by an inversion (supplementary fig. S1, Supplementary Material online), no significant LD was detected between microsatellite loci and inversion systems within zones and at the country scale (G-test, P > 0.05, after Bonferroni correction), which suggests that microsatellites alleles and chromosomal inversions assort independently from each other.
Population Structure
Genetic differentiation measured across the 16 microsatellite loci was very low among the four ecological zones, with a mean FST of 0.005 [95% CI: 0.001–0.010]. Although very low, population differentiation was statistically significant for all population pairs (P < 0.05) except between Zone B and Zone C (table 4). Consistent with the weak genetic differentiation observed, the STRUCTURE analysis failed to cluster the multilocus genotypes in more than a single group [P(K = 1 | X) = 1.00; see the supplementary fig. S2A, Supplementary Material online]. Nonetheless, we found a significant correlation between the genetic distances measured between pairs of individuals (êr) and the logarithm of Euclidian geographic distances between them (one-tailed Mantel test; P = 0.009). From the slope of the regression (b = 0.002 [95% CI: 0.001–0.004]), we calculated an overall estimate of Dσ2 based on microsatellite data of 46.35 individuals.
Table 4.
Estimate of Genetic Differentiation (FST).
| Zone A | Zone B | Zone C | Zone 4 | |
| n = 114 | n = 37 | n = 83 | n = 35 | |
| Microsatellites | ||||
| Zone A | 0.0067** (0.0296) | 0.0038** (0.0163) | 0.0047** (0.0206) | |
| Zone B | 0.2253** (0.3219) | 0.0025* (0.0108) | 0.0129** (0.0572) | |
| Zone C | 0.1907** (0.2772) | 0.0209* (0.0292) | 0.0075** (0.0322) | |
| Zone D | 0.4826** (0.6447) | 0.3944** (0.4769) | 0.2539** (0.3292) | |
| Inversions |
Derived from microsatellite (above diagonal) and chromosomal inversions (below diagonal) frequency data among A. funestus populations from Cameroon. n: sample size (number of specimens); in brackets, He-corrected FST (see text).*P < 0.05, **P < 0.01. In bold italics: significant (P < 0.05) after accounting for multiple testing (Holm 1979).
In contrast to microsatellite markers, chromosomal inversions exhibited strong and significant genetic differentiation between ecological zones except between Zone B and Zone C (table 4). Pairwise FST estimates were much larger for inversion systems than for microsatellites, ranging from 0.021 to 0.483. Using standardized estimates of population differentiation (Hedrick 2005; Meirmans 2006), we found significant differences in FST values between chromosomal inversions and microsatellite markers (Wilcoxon signed-rank test, P < 0.05). The highest estimate of genetic differentiation was observed between Zone A and Zone D, which are the most geographically distant (fig. 1) and display highly different habitat conditions. From the STRUCTURE clustering analysis, we found the highest posterior probability of the data for K = 4 putative clusters ([P(K = 4 | X) = 0.83], supplementary fig. S2B, Supplementary Material online). Two of these clusters segregated the standard and inverted homokaryotypes for all inversions systems, respectively (e.g., yellow and green clusters on supplementary fig. S2C, Supplementary Material online). The other two clusters (in blue and red on supplementary fig. S2C, Supplementary Material online) consisted in a mixture of standard, inverted, and heterozygous karyotypes for all inversions systems. Although standard homokaryotypes were more prevalent in the northern populations and inverted homokarotypes in southern ones, no obvious geographical organization of the genetic clusters was revealed. However, we found evidence for isolation by distance across the entire study area with a highly significant correlation between pairwise estimates of genetic distances (êr) and the logarithm of Euclidian geographic distances between individuals (one-tailed Mantel test, P < 0.001). From the slope of the regression (b = 0.224 [95% CI: 0.107–0.522]) obtained from chromosomal inversions, we estimated a Dσ2 at 0.35 individuals. This estimate is two orders of magnitude lower than that obtained from microsatellite markers. Because the effective density D must be the same for chromosomal inversions and microsatellite markers, this suggests that the effective dispersal of chromosomal inversions is extremely limited.
Homogeneity Across Loci and the Role of Selection
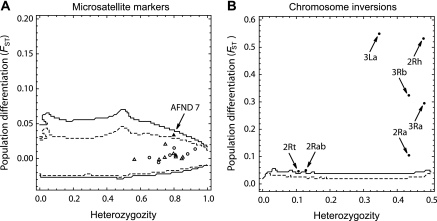
Single-locus FST estimates obtained from microsatellite loci ranged from 0 to 0.038. We tested for homogeneity in differentiation level, as measured by FST, across microsatellite loci among the four sampled areas using the software package FDIST2 (Beaumont and Nichols 1996). When plotted against their respective He value, FST estimates obtained from all loci (either within or outside polymorphic chromosomal inversions) mapped within the 99% confidence envelope of FST estimates expected under neutrality (fig. 2A). Only locus AFND7 revealed a marginally significant departure from neutral expectations (P = 0.018), with an FST estimate slightly higher than expected, lying outside the 95% confidence envelope. Discarding AFND7, the overall mean FST estimate remained unchanged at 0.005 [CI 95%: 0.001–0.006] across the 15 remaining loci. There was no apparent difference across locus-specific FST estimates lying within and outside polymorphic chromosomal inversions (fig. 2A).
FIG. 2.
Neutrality tests. single-locus FST estimates among geographic populations of A. funestus in Cameroon, plotted against expected heterozygosity (Nei 1987). (A) Data from 16 microsatellites markers located inside (circles) and outside (triangles) common polymorphic chromosomal inversions. (B) Data form seven chromosomal inversion systems. In both panels, outlier loci are pointed by arrows and referred to by their names. Dashed lines and solid lines represent the 95% and 99% confidence limits under the hypothesis of neutrality of the loci, respectively.
We generated the neutral distribution of FST estimates for biallelic codominant markers, given the level of differentiation measured at presumably neutral marker loci. To do so, we conducted coalescent simulations using a modified version of the software package DFDIST (Beaumont and Balding 2004). We used the multilocus FST estimate obtained from the 15 presumably neutral microsatellite loci (FST = 0.005, see above) to perform the coalescent simulations. As shown in figure 2B, all chromosomal inversion systems departed significantly from neutral expectations based on microsatellite data (P < 2.10−6, except for 2Rab and 2Rt for which P = 0.003 and P = 0.005, respectively). The two inversions 2Rab and 2Rt occur at very low frequency in our data set (table 3), which may reduce statistical power to detect departure from neutrality in this case. The significantly higher differentiation of chromosomal inversions as compared with that expected at migration–drift equilibrium given the microsatellite data, strongly suggests that gene flow is extremely limited for chromosomal inversions systems, and/or that chromosomal inversions are exposed to strong natural selection and local adaptation.
Inversions and Environment
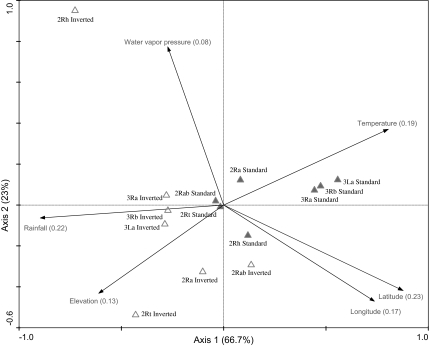
To further test whether chromosomal inversions are exposed to environmentally driven selection, we investigated how their distribution varies with environmental variables (i.e., latitude, longitude, altitude, rainfall, temperature, and water vapor pressure) using CCA. A graphical representation of the contribution of each EGV to the global chromosomal polymorphism distribution is given in figure 3. The first four axes of the CCA were statistically significant (Monte Carlo permutation test, F = 2.71, P = 0.0176), with the first two axes accounting for 89.7% of the total variance in the data set. The most informative EGV was latitude, explaining 23% of the system variance followed by rainfall and temperature (22% and 19%, respectively). All EGVs, except water vapor pressure, showed a highly significant contribution to the spatial pattern of chromosomal inversion distribution (P < 0.05).
FIG. 3.
Eco-geographical chromosome inversion distributions. CCA diagram showing the ordination of chromosomal inversions for each homozygote (standard or inverted) along the first two canonical axes (explaining 89.7% of variance). Open triangles represent STANDARD homokaryotes. Full triangles represent INVERTED homokaryotes. EGVs are passively plotted on the graph. Alt, altitude; temp, temperature; rain, rainfall; lat, latitude; long, longitude; WVP: water vapor pressure.
The first axis, which explains 66.7% of the total variance in chromosomal polymorphism, can be ascribed to an increasing aridity gradient (from left to right): We observe elevated water vapor pressure and rainfall, which are characteristic of the humid conditions in the rainforest biome on the left-hand side of the axis, whereas high temperatures, which characterize more arid conditions, are found on the right-hand side. The ordination diagram (fig. 3) shows a clear separation among alternative arrangements (i.e., standard vs. inverted) for each inversion system. Standard arrangements are positively associated with warmer and drier environments (except 2Rab +/+ and 2Rt +/+, which show almost no variance) and map accordingly in the right-hand part of the diagram shown in figure 3. Inverted arrangements are associated with more humid and cooler climates and map in the left-hand part of the diagram.
The second axis of the CCA, which explains 23% of total variance, is more difficult to interpret; it represents an environmental gradient mostly affected by the “highlands” bioclimatic domain correlated with water vapor pressure, altitude, and temperature. Thus, we interpret it as a gradient separating cooler highlands from warmer and humid lowlands and valleys. Inversions 2Rt and 2Rh show the highest level of divergence along this axis: Inversion 2Rt is associated with higher elevation, whereas inversion 2Rh is associated with humid lowlands (fig. 3).
Discussion
The comparative analysis of chromosomal inversions and microsatellite markers polymorphisms in A. funestus populations from Cameroon revealed a contrasting pattern of population genetic subdivision. Presumably, neutral microsatellite markers depicted a weak and shallow population structure, with very low levels of genetic differentiation among populations, whereas chromosomal inversions revealed a very strong population structure. Strong deviation from the expected migration–drift equilibrium based on microsatellite markers suggested that gene flow is limited for chromosomal inversions, and/or that chromosomal inversions are targets for local adaptation. The latter hypothesis was further confirmed by the nonrandom distribution of alternative chromosomal arrangements across different environmental conditions.
Previous genetic structure analysis based on microsatellite markers revealed important gene flow among A. funestus populations with a weak, if any, role of geographic distance in shaping natural population structure (Kamau et al. 2003; Cohuet et al. 2005; Michel, Guelbeogo, et al. 2005; Michel, Ingrasci, et al. 2005), as we observed in the present study. As reported in the widely codistributed A. gambiae, retention of ancestral polymorphism together with evidence for homoplasy at microsatellite loci and, more importantly, molecular signatures of recent population expansion and gene flow result in a shallow population structure (Donnelly et al. 2001; Krzywinski and Besansky 2003; Lehmann et al. 2003; Michel, Ingrasci, et al. 2005). However, significant levels of genetic differentiation have been reported between sympatric populations characterized by different levels of chromosomal polymorphism (i.e., the “Folonzo” and “Kiribina” chromosomal forms described by Costantini et al. 1999 and Guelbeogo et al. 2005) in Burkina Faso (Michel, Guelbeogo, et al. 2005) or between allopatric populations of A. funestus across the inhospitable Great Rift Valley complex (Braginets et al. 2003) with the same or a subset of the microsatellite markers used in this study. Our results indeed suggest that the amount of gene flow between geographical populations of A. funestus in Cameroon is high enough to prevent population splitting and demographic independence at this geographical scale. However, the unique experimental design of our study, in which every specimen was both karyotyped on all arms of their chromosomal complement and genotyped at microsatellite loci, allows us to investigate the role of chromosomal inversions in local adaptation and speciation in A. funestus. Interestingly, no significant LD was detected between microsatellite markers and inversions. Although chromosomal inversions reduce recombination, particularly in the vicinity of breakpoints, that is not completely stopped, and the interaction between gene flux and selection is expected to produce a mosaic of more- and less- differentiated genomic regions within the inversion and not necessarily close to the breakpoints (Navarro et al. 1997; White et al. 2007, 2010). Several factors, such as the number of genes under selection or the age of the inversion, can affect the recombination rates within a rearrangement. Recent studies in A. gambiae have confirmed that variation in LD patterns within the inversion is not homogeneous (White et al. 2007, 2009). They estimated that, without selection and according to the estimated level of recombination that occurs within the 2La inversion of A. gambiae (see above), LD should drop to undetectable levels within less than 4,600 generation (i.e., 400 years) (White et al. 2007, 2009). These general trends are likely to apply to A. funestus as well, although no estimate of inversion age or recombination rates in this malaria vector is available yet. However, taking into account the large and standing distribution of these inversions throughout the species range in Africa, we may consider that the age of inversions is an important factor that would explain low LD between inversions and microsatellite markers.
In contrast to microsatellite markers, chromosomal inversions polymorphism depicted a situation whereby alternative chromosomal arrangements (i.e., standard vs. inverted) segregated nonrandomly in different environments across diverse ecological zones in Cameroon. Again, this situation is reminiscent of what has been observed in A. gambiae (Coluzzi et al. 1979, 1985; Powell et al. 1999; Toure et al. 1998) and provides strong support for a role of chromosomal inversions or the genes they contain in local adaptation and ecological specialization in both vector species. As such, inversions might contribute to local adaptation by maintaining tight linkage between locally advantageous alleles in the face of gene flow from other populations carrying different sets of alleles. We have shown that estimates of population differentiation based on chromosomal inversions were at odds with the expected migration–drift equilibrium based on presumably neutral markers (fig. 2). Furthermore, alternative chromosomal arrangements segregated differently in the ordination space defined by a number of EGVs (fig. 3). This suggests that the population structure depicted by these chromosomal arrangements is likely the result of local adaptation, which could lead to propitious conditions for incipient speciation, increasing genetic drift between geographically isolated populations (Manoukis et al. 2008). Strong LD between inversions in the face of linkage equilibrium between microsatellite loci and inversions further strengthens this view, suggesting that epistasis between locally adapted alleles captured by different inversions and/or parallel selection–migration balance resulting from local adaptation in multiple traits generated the observed pattern (Li and Nei 1974; Kirkpatrick and Barton 2006; White et al. 2007). In this sense, two recent studies have revealed that both molecular forms in A. gambiae shared the same chromosomal arrangements in areas where they were found in sympatry (Costantini et al. 2009; Simard et al. 2009). The most likely explanation for the observed pattern of chromosomal polymorphism is the action of directional and/or balancing selection acting within the M and S forms on alternative arrangements according to the prevailing eco-geographical conditions. Thus, inversions almost certainly predated the speciation process in A. gambiae.
Chromosomal inversions are typically regarded as a genetic mechanism protecting sets of (locally adapted) genes by reducing recombination in heterozygotes (Hoffmann and Rieseberg 2008). The very low genetic differentiation measured at microsatellite markers between populations characterized by alternative arrangements (i.e., standard vs. inverted) was consistent across loci, independently of their physical location within or outside polymorphic chromosomal inversions. This suggests that recombination suppression is not complete within the inversion and that residual gene flow (through double crossing-over or gene conversion) between alternative arrangements is sufficient to homogenize genetic variation in genomic regions not directly targeted by selection. Recent genomic investigations in A. gambiae indeed provided evidence that, although recombination is drastically reduced within inversions, the amount of residual gene flow between alternative arrangements is sufficient to allow rapid homogenization (Stump et al. 2007; White et al. 2007, 2009). These empirical findings have been recently supported by theoretical approaches (Feder and Nosil 2009). In A. funestus, genetic independence between karyotypes might be prevented by hybridization (see table 3) and migration across the country (estimate of Dσ2 based on microsatellite data of 46.35 individuals), homogenizing variation at neutral microsallite loci, as it has been previously shown (Cohuet et al. 2005). On the other hand, inversions exposed to environmental selection (fig. 3) and possible epistatic effects between inversions (high LD between inversions) might lead to a strong population structure, as observed across heterogeneous landscapes in Cameroon.
Although neither reduced recombination nor selection alone is expected to maintain inversion polymorphism and divergence, migration–selection balance between multiple loci within an inversion and/or selection against deleterious recessives are the most likely to maintain inversion polymorphism in natural populations and to generate high FST between ecotypes (Navarro et al. 1997; Kirkpatrick and Barton 2006; White et al. 2007). This was experimentally demonstrated in Drosophila (Schaeffer and Anderson 2005) and our data suggest that this general principle may also apply to chromosomal polymorphism and divergence in A. funestus. Furthermore, selection against heterozygotes (e.g., underdominance) has been pointed out as a mechanism to establish inversions (King 1993). This is a controversial mechanism because an underdominant inversion is established only by drift in a population and it requires special conditions such as small population size, weak selection against heterokaryotypes, or meiotic drive (Kirkpatrick and Barton 2006). However, if underdominance is genic (interactions between alleles within an inversion, resulting in maladaptive heterozygotes) and not structural (problems during meiosis) as it seems occur for paracentric inversions in dipteran species (Hoffmann and Rieseberg 2008), local adaptation theory could explain how underdominant inversions are established (Kirkpatrick and Barton 2006). Although several inversions do not show any deficit in heterozygotes across the country, others, such as 3Ra, are dependent upon the eco-geographical zone, suggesting a fitness cost at the heterozygous state. Finally, other mechanisms involving prezygotic barriers and assortative mating could participate to the inversion distribution patterns as it has been described in Rhagoletis (Feder et al. 2003). Unfortunately, conventional formal genetics approaches based on experimental crossings are yet impossible to conduct in A. funestus because of the scarcity of laboratory colonies available. Furthermore, high-resolution genomics approaches are still awaiting complete genome sequencing. Once available, these tools will provide considerable opportunities for comparative genomics between A. gambiae and A. funestus and will help elucidate the exact role of common polymorphic chromosomal inversions in ecological adaptation of such devastating pests and pinpoint the genes involved in these processes. Further theoretical and empirical studies aiming at measuring migration rates, fitness, or selection on inversions and on the genes they contain should further help deciphering the biological and ecological mechanisms underlying such adaptive potential.
The ability of both A. gambiae and A. funestus to colonize a wide range of habitats and to cope with dramatic seasonal fluctuations in environmental conditions have direct consequences on malaria transmission dynamics: It contributes to increase the intensity of parasites transmission locally during the most favorable season and to extend malaria transmission in both space and time (Fontenille and Simard 2004). In these two major African malaria vectors, ecological plasticity is mirrored at the genetic level by high amounts of chromosomal polymorphisms of proven adaptive value (Coluzzi et al. 2002; Pombi et al. 2008). Moreover, behavioral variations with direct relevance for vector efficiency, such as the ability to bite humans and rest indoors, have been shown to be associated with chromosomal polymorphisms in A. gambiae (Bryan et al. 1982). Inversions polymorphism within vector populations have also been invoked to explain vector control failures in West Africa (Coluzzi 1982; Molineaux and Gramiccia 1980). Dissecting the molecular make up of chromosomal inversions and unraveling the genetic processes by which they are produced, raise in frequency and are maintained within natural vector populations, is of paramount importance for vector control strategies to be efficient and sustainable. This is particularly true because of the considerable reservoir of standing and possibly cryptic variation embodied in chromosomal inversions (Pombi et al. 2008). A more detailed understanding of the population genetic structure of major African malaria vectors and, more importantly, of the interplay between genetic polymorphisms and ecological or behavioral variations is required for a number of reasons. First, it will provide a better assessment of the feasibility of innovative vector control strategies based on the use of genetically modified organisms with altered vector competence. Second, it will contribute to devise implementation guidelines and protocols and to insure comprehensive monitoring of the intervention. Finally, such studies are also warranted to assess the impact of anthropogenic modifications of the environment (e.g., urbanization, deforestation, and agriculture) and global climate change on vector distributions and fitness and their consequences on the evolution of malaria transmission risks in Africa (Tanser et al. 2003; Guerra et al. 2006; Pinto et al. 2007; Boete 2009).
Supplementary Material
Supplementary figures S1 and S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
We thank the Laboratoire de Recherche sur le Paludisme at OCEAC, Yaoundé for their excellent field assistance. F.S. and D.A. gratefully acknowledge Joe Bajo and Kass Telbiehr for continuously prompting useful discussions throughout the study. Authors’ contribution: D.A., A.C., D.F., and F.S. conceived and designed the experiments. D.A. and A.C. performed the experiments. D.A., M.C.F., A.C., R.V., and F.S. analyzed the data. D.A., D.F., and F.S. coordinated the fieldwork. D.A., R.V., and F.S. wrote the manuscript with editorial suggestions from the other coauthors. Financial support was provided by the Institut de Recherche pour le Développement. Fieldwork was supported in part by the National Institutes of Health (R01-AI063508 to Nora J. Besansky). D.A. was supported by a student fellowship grant from Fundación CAJA MADRID (Madrid, Spain).
References
- Balanya J, Serra L, Gilchrist GW, Huey RB, Pascual M, Mestres F, Sole E. Evolutionary pace of chromosomal polymorphism in colonizing populations of Drosophila subobscura: an evolutionary time series. Evolution. 2003;57:1837–1845. doi: 10.1111/j.0014-3820.2003.tb00591.x. [DOI] [PubMed] [Google Scholar]
- Beaumont MA. Adaptation and speciation: what can F-st tell us? Trends Ecol Evol. 2005;20:435–440. doi: 10.1016/j.tree.2005.05.017. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Balding DJ. Identifying adaptive genetic divergence among populations from genome scans. Mol Ecol. 2004;13:969–980. doi: 10.1111/j.1365-294x.2004.02125.x. [DOI] [PubMed] [Google Scholar]
- Beaumont MA, Nichols R. Evaluating loci for use in the genetic analysis of population structure. Proc R Soc Lond Ser B Biol Sci. 1996;263:1619–1626. [Google Scholar]
- Besansky NJ, Krzywinski J, Lehmann T, Simard F, Kern M, Mukabayire O, Fontenille D, Toure Y, Sagnon NF. Semipermeable species boundaries between Anopheles gambiae and Anopheles arabiensis: evidence from multilocus DNA sequence variation. Proc Natl Acad Sci U S A. 2003;100:10818–10823. doi: 10.1073/pnas.1434337100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boete C. Anopheles mosquitoes: not just flying malaria vectors especially in the field. Trends Parasitol. 2009;25:53–55. doi: 10.1016/j.pt.2008.10.005. [DOI] [PubMed] [Google Scholar]
- Bonin A, Taberlet P, Miaud C, Pompanon F. Explorative genome scan to detect candidate loci for adaptation along a gradient of altitude in the common frog (Rana temporaria) Mol Biol Evol. 2006;23:773–783. doi: 10.1093/molbev/msj087. [DOI] [PubMed] [Google Scholar]
- Braginets OP, Minakawa N, Mbogo CM, Yan G. Population genetic structure of the African malaria mosquito Anopheles funestus in Kenya. Am J Trop Med Hyg. 2003;69:303–308. [PubMed] [Google Scholar]
- Bryan JH, Deco MA, Petrarca V, Coluzzi M. Inversion polymorphism and incipient speciation in Anopheles gambiae s.str. in The Gambia, West Africa. Genetica. 1982;59:167–176. [Google Scholar]
- Butlin R, Roper C. Evolutionary genetics: microarrays and species origins. Nature. 2005;437:199–201. doi: 10.1038/437199a. [DOI] [PubMed] [Google Scholar]
- Chapuis MP, Estoup A. Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol. 2007;24:621–631. doi: 10.1093/molbev/msl191. [DOI] [PubMed] [Google Scholar]
- Coetzee M, Fontenille D. Advances in the study of Anopheles funestus, a major vector of malaria in Africa. Insect Biochem Mol Biol. 2004;34:599–605. doi: 10.1016/j.ibmb.2004.03.012. [DOI] [PubMed] [Google Scholar]
- Cohuet A, Dia I, Simard F, Raymond M, Rousset F, Antonio-Nkondjio C, Awono-Ambene PH, Wondji CS, Fontenille D. Gene flow between chromosomal forms of the malaria vector Anopheles funestus in Cameroon, Central Africa, and its relevance in malaria fighting. Genetics. 2005;169:301–311. doi: 10.1534/genetics.103.025031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohuet A, Simard F, Berthomieu A, Raymond M, Fontenille D, Weill M. Isolation and characterization of microsatellite DNA markers in the malaria vector Anopheles funestus. Mol Ecol Notes. 2002;2:498–500. [Google Scholar]
- Cohuet A, Simard F, Toto JC, Kengne P, Coetzee M, Fontenille D. Species identification within the Anopheles funestus group of malaria vectors in Cameroon and evidence for a new species. Am J Trop Med Hyg. 2003;69:200–205. [PubMed] [Google Scholar]
- Coluzzi M. Spatial distribuion of chromosomal inversions and speciation in Anopheline mosquitoes. In: Barigozzi C, editor. Mechanisms of speciation. New York: Alan R Liss; 1982. pp. 113–115. [Google Scholar]
- Coluzzi M, Petrarca V, Di Deco MA. Chromosomal inversion intergradation and incipient speciation in Anopheles gambiae. Boll Zool. 1985;52:45–63. [Google Scholar]
- Coluzzi M, Sabatini A, della Torre A, Di Deco MA, Petrarca V. A polytene chromosome analysis of the Anopheles gambiae species complex. Science. 2002;298:1415. doi: 10.1126/science.1077769. [DOI] [PubMed] [Google Scholar]
- Coluzzi M, Sabatini A, Petrarca V, Dideco MA. Chromosomal differentiation and adaptation to human environments in the Anopheles gambiae complex. Trans R Soc Trop Med Hyg. 1979;73:483–497. doi: 10.1016/0035-9203(79)90036-1. [DOI] [PubMed] [Google Scholar]
- Costantini C, Ayala D, Guelbeogo W, et al. (12 co-authors) Living at the edge: biogeographic patterns of habitat segregation conform to speciation by niche expansion in Anopheles gambiae. BMC Ecol. 2009;9:16. doi: 10.1186/1472-6785-9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costantini C, Sagnon N, Ilboudo-Sanogo E, Coluzzi M, Boccolini D. Chromosomal and bionomic heterogeneities suggest incipient speciation in Anopheles funestus from Burkina Faso. Parassitologia. 1999;41:595–611. [PubMed] [Google Scholar]
- della Torre A, Costantini C, Besansky NJ, Caccone A, Petrarca V, Powell JR, Coluzzi M. Speciation within Anopheles gambiae—the glass is half full. Science. 2002;298:115–117. doi: 10.1126/science.1078170. [DOI] [PubMed] [Google Scholar]
- Donnelly MJ, Licht MC, Lehmann T. Evidence for recent population expansion in the evolutionary history of the malaria vectors Anopheles arabiensis and Anopheles gambiae. Mol Biol Evol. 2001;18:1353–1364. doi: 10.1093/oxfordjournals.molbev.a003919. [DOI] [PubMed] [Google Scholar]
- el Mousadik A, Petit RJ. Chloroplast DNA phylogeography of the argan tree of Morocco. Mol Ecol. 1996;5:547–555. doi: 10.1111/j.1365-294x.1996.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feder JL, Nosil P. Chromosomal inversions and species differences: when are genes affecting adaptive divergence and reproductive isolation expected to reside within inversions? Evolution. 2009;63:3061–3075. doi: 10.1111/j.1558-5646.2009.00786.x. [DOI] [PubMed] [Google Scholar]
- Feder JL, Roethele FB, Filchak K, Niedbalski J, Romero-Severson J. Evidence for inversion polymorphism related to sympatric host race formation in the apple maggot fly, Rhagoletis pomonella. Genetics. 2003;163:939. doi: 10.1093/genetics/163.3.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontaine MC, Baird SJE, Piry S, et al. (19 co-authors) Rise of oceanographic barriers in continuous populations of a cetacean: the genetic structure of harbour porpoises in Old World waters. BMC Biol. 2007;5 doi: 10.1186/1741-7007-5-30. 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontenille D, Simard F. Unravelling complexities in human malaria transmission dynamics in Africa through a comprehensive knowledge of vector populations. Comp Immunol Microbiol Infect Dis. 2004;27:357–375. doi: 10.1016/j.cimid.2004.03.005. [DOI] [PubMed] [Google Scholar]
- Gillies MT, de Meillon B. The anophelinae of Africa, south of the Sahara. Johannesburg (South Afirca): The South African Institute for Medical Research; 1968. [Google Scholar]
- Goudet J. FSTAT: a program to estimate and test gene diversities and fixation indices. Version 2.9.3. 2001. Available from http://www.unil.ch./izea/softwares/fstat.html. [Google Scholar]
- Guelbeogo WM, Grushko O, Boccolini D, Ouedraogo PA, Besansky NJ, Sagnon NF, Costantini C. Chromosomal evidence of incipient speciation in the Afrotropical malaria mosquito Anopheles funestus. Med Vet Entomol. 2005;19:458–469. doi: 10.1111/j.1365-2915.2005.00595.x. [DOI] [PubMed] [Google Scholar]
- Guerra CA, Snow RW, Hay SI. A global assessment of closed forests, deforestation and malaria risk. Ann Trop Med Parasitol. 2006;100:189–204. doi: 10.1179/136485906X91512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedrick PW. A standardized genetic differentiation measure. Evolution. 2005;59:1633–1638. [PubMed] [Google Scholar]
- Hoffmann AA, Rieseberg LH. Revisiting the impact of inversions in evolution: from population genetic markers to drivers of adaptive shifts and speciation? Annu Rev Ecol Evol Syst. 2008;39:21–42. doi: 10.1146/annurev.ecolsys.39.110707.173532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann AA, Sgro CM, Weeks AR. Chromosomal inversion polymorphisms and adaptation. Trends Ecol Evol. 2004;19:482–488. doi: 10.1016/j.tree.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Hoffmann AA, Willi Y. Detecting genetic responses to environmental change. Nat Rev Genet. 2008;9:421–432. doi: 10.1038/nrg2339. [DOI] [PubMed] [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- Hunt RH. A cytological technique for the study of Anopheles gambiae complex. Parassitologia. 1973;15:137–139. [PubMed] [Google Scholar]
- Kamau L, Munyekenye GO, Koekemoer LL, Hunt RH, Coetzee M. A survey of the Anopheles funestus (Diptera: Culicidae) group of mosquitoes from 10 sites in Kenya with special emphasis on population genetic structure based on chromosomal inversion karyotypes. J Med Entomol. 2003;40:664–671. doi: 10.1603/0022-2585-40.5.664. [DOI] [PubMed] [Google Scholar]
- King M. Species evolution: the role of chromosome change. Cambridge (United Kingdom): Cambridge University Press; 1993. [Google Scholar]
- Kirkpatrick M, Barton N. Chromosome inversions, local adaptation and speciation. Genetics. 2006;173:419. doi: 10.1534/genetics.105.047985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koekemoer LL, Kamau L, Hunt RH, Coetzee M. A cocktail polymerase chain reaction assay to identify members of the Anopheles funestus (Diptera: Culicidae) group. Am J Trop Med Hyg. 2002;66:804–811. doi: 10.4269/ajtmh.2002.66.804. [DOI] [PubMed] [Google Scholar]
- Krzywinski J, Besansky NJ. Molecular systematics of Anopheles: from subgenera to subpopulations. Annu Rev Entomol. 2003;48:111–139. doi: 10.1146/annurev.ento.48.091801.112647. [DOI] [PubMed] [Google Scholar]
- Kulathinal RJ, Stevison LS, Noor MAF. The genomics of speciation in Drosophila: diversity, divergence, and introgression estimated using low-coverage genome sequencing. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000550. e1000550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblois R, Estoup A, Rousset F. Influence of mutational and sampling factors on the estimation of demographic parameters in a “continuous” population under isolation by distance. Mol Biol Evol. 2003;20:491–502. doi: 10.1093/molbev/msg034. [DOI] [PubMed] [Google Scholar]
- Lehmann T, Licht M, Elissa N, Maega BT, Chimumbwa JM, Watsenga FT, Wondji CS, Simard F, Hawley WA. Population structure of Anopheles gambiae in Africa. J Hered. 2003;94:133–147. doi: 10.1093/jhered/esg024. [DOI] [PubMed] [Google Scholar]
- Li WH, Nei M. Stable linkage disequilibrium without epistasis in subdivided populations. Theor Popul Biol. 1974;6:173–183. doi: 10.1016/0040-5809(74)90022-7. [DOI] [PubMed] [Google Scholar]
- Manoukis NC, Powell JR, Toure MB, Sacko A, Edillo FE, Coulibaly MB, Traore SF, Taylor CE, Besansky NJ. A test of the chromosomal theory of ecotypic speciation in Anopheles gambiae. Proc Natl Acad Sci U S A. 2008;105:2940–2945. doi: 10.1073/pnas.0709806105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirmans PG. Using the AMOVA framework to estimate a standardized genetic differentiation measure. Evolution. 2006;60:2399–2402. [PubMed] [Google Scholar]
- Michel AP, Guelbeogo WM, Grushko O, Schemerhorn BJ, Kern M. Molecular differentiation between chromosomally defined incipient species of Anopheles funestus. Insect Mol Biol. 2005;14:375. doi: 10.1111/j.1365-2583.2005.00568.x. [DOI] [PubMed] [Google Scholar]
- Michel AP, Ingrasci MJ, Schemerhorn BJ, et al. (13 co-authors) Rangewide population genetic structure of the African malaria vector Anopheles funestus. Mol Ecol. 2005;14:4235–4248. doi: 10.1111/j.1365-294X.2005.02754.x. [DOI] [PubMed] [Google Scholar]
- Molineaux L, Gramiccia G. The Garki Project. Research on Epidemiology and Control of Malaria in the Sudan Savannah of West Africa. Bull World Health Organ. 1980 [Google Scholar]
- Morlais I, Poncon N, Simard F, Cohuet A, Fontenille D. Intraspecific nucleotide variation in Anopheles gambiae: new insights into the biology of malaria vectors. Am J Trop Med Hyg. 2004;71:795–802. [PubMed] [Google Scholar]
- Navarro A, Barton NH. Chromosomal speciation and molecular divergence—accelerated evolution in rearranged chromosomes. Science. 2003;300:321–324. doi: 10.1126/science.1080600. [DOI] [PubMed] [Google Scholar]
- Navarro A, Betran E, Barbadilla A, Ruiz A. Recombination and gene flux caused by gene conversion and crossing over in inversion heterokaryotypes. Genetics. 1997;146:695. doi: 10.1093/genetics/146.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics. 1978;89:583–590. doi: 10.1093/genetics/89.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. Molecular evolutionary genetics. New York: Columbia University Press; 1987. [Google Scholar]
- Noor MA, Cunningham AL, Larkin JC. Consequences of recombination rate variation on quantitative trait locus mapping studies. Simulations based on the Drosophila melanogaster genome. Genetics. 2001;159:581–588. doi: 10.1093/genetics/159.2.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivry JC. Fleuves et Rivières du Cameroun. Paris (France): ORSTOM; 1986. [Google Scholar]
- Ortiz-Barrientos D, Reiland J, Hey J, Noor MAF. Recombination and the divergence of hybridizing species. Genetica. 2002;116:167. [PubMed] [Google Scholar]
- Pinto J, Lynd A, Vicente JL, et al. (13 co-authors) Multiple origins of knockdown resistance mutations in the Afrotropical mosquito vector Anopheles gambiae. PLoS One. 2007;2:e1243. doi: 10.1371/journal.pone.0001243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombi M, Caputo B, Simard F, Di Deco MA, Coluzzi M, Della Torre A, Costantini C, Besansky NJ, Petrarca V. Chromosomal plasticity and evolutionary potential in the malaria vector Anopheles gambiae sensu stricto: insights from three decades of rare paracentric inversions. BMC Evol Biol. 2008;8:309. doi: 10.1186/1471-2148-8-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell JR, Petrarca V, della Torre A, Caccone A, Coluzzi M. Population structure, speciation, and introgression in the Anopheles gambiae complex. Parassitologia. 1999;41:101–113. [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieseberg LH. Chromosomal rearrangements and speciation. Trends Ecol Evol. 2001;16:351–358. doi: 10.1016/s0169-5347(01)02187-5. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Trelles F, Alvarez G, Zapata C. Time-series analysis of seasonal changes of the O inversion polymorphism of Drosophila subobscura. Genetics. 1996;142:179–187. doi: 10.1093/genetics/142.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219–1228. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer SW, Anderson WW. Mechanisms of genetic exchange within the chromosomal inversions of Drosophila pseudoobscura. Genetics. 2005;171:1729–1739. doi: 10.1534/genetics.105.041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott DW. Multivariate density estimation: theory, practice, and visualization. New York: John Wiley; 1992. [Google Scholar]
- Service M. Mosquito ecology field sampling methods. New York: Elsevier Applied Science; 1993. [Google Scholar]
- Sharakhov I, Braginets O, Grushko O, et al. (12 co-authors) A microsatellite map of the African human malaria vector Anopheles funestus. J Hered. 2004;95:29–34. doi: 10.1093/jhered/esh011. [DOI] [PubMed] [Google Scholar]
- Simard F, Ayala D, Kamdem G, et al. Ecological niche partitioning between Anopheles gambiae molecular forms in Cameroon: the ecological side of speciation. BMC Ecol. 2009;9:17. doi: 10.1186/1472-6785-9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinkins SP, Hackett BJ, Costantini C, Vulule J, Ling YY, Collins FH, Besansky NJ. Isolation of polymorphic microsatellite loci from the malaria vector Anopheles funestus. Mol Ecol. 2000;9:490–492. doi: 10.1046/j.1365-294x.2000.00871-2.x. [DOI] [PubMed] [Google Scholar]
- Stump AD, Pombi M, Goeddel L, Ribeiro JM, Wilder JA, della Torre A, Besansky NJ. Genetic exchange in 2La inversion heterokaryotypes of Anopheles gambiae. Insect Mol Biol. 2007;16:703–709. doi: 10.1111/j.1365-2583.2007.00764.x. [DOI] [PubMed] [Google Scholar]
- Tanser FC, Sharp B, le Sueur D. Potential effect of climate change on malaria transmission in Africa. Lancet. 2003;362:1792–1798. doi: 10.1016/S0140-6736(03)14898-2. [DOI] [PubMed] [Google Scholar]
- Ter Braak C. CANOCO—Fortran program for canconical community ordination. Ithaca (NY): Microcomputer Power; 1987. [Google Scholar]
- Toure YT, Petrarca V, Traore SF, Coulibaly A, Maiga HM, Sankare O, Sow M, Di Deco MA, Coluzzi M. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia. 1998;40:477–511. [PubMed] [Google Scholar]
- van Doorn GS, Kirkpatrick M. Turnover of sex chromosomes induced by sexual conflict. Nature. 2007;449:909. doi: 10.1038/nature06178. [DOI] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Vitalis R, Dawson K, Boursot P. Interpretation of variation across marker loci as evidence of selection. Genetics. 2001;158:1811–1823. doi: 10.1093/genetics/158.4.1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts PC, Rousset F, Saccheri IJ, Leblois R, Kemp SJ, Thompson DJ. Compatible genetic and ecological estimates of dispersal rates in insect (Coenagrion mercuriale: Odonata: Zygoptera) populations: analysis of ‘neighbourhood size’ using a more precise estimator. Mol Ecol. 2007;16:737–751. doi: 10.1111/j.1365-294X.2006.03184.x. [DOI] [PubMed] [Google Scholar]
- Weir B, Cockerham C. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–1370. doi: 10.1111/j.1558-5646.1984.tb05657.x. [DOI] [PubMed] [Google Scholar]
- White BJ, Cheng C, Sangare D, Lobo NF, Collins FH, Besansky NJ. The population genomics of trans-specific inversion polymorphisms in Anopheles gambiae. Genetics. 2009;183:275–288. doi: 10.1534/genetics.109.105817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Cheng C, Simard F, Costantini C, Besansky NJ. Genetic association of physically unlinked islands of genomic divergence in incipient species of Anopheles gambiae. Mol Ecol. 2010;19:925–939. doi: 10.1111/j.1365-294X.2010.04531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White BJ, Hahn MW, Pombi M, Cassone BJ, Lobo NF, Simard F, Besansky NJ. Localization of candidate regions maintaining a common polymorphic inversion (2La) in. Anopheles gambiae. PLoS Genet. 2007;3:e217. doi: 10.1371/journal.pgen.0030217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The genetical structure of populations. Ann Eugen. 1951;15:323–354. doi: 10.1111/j.1469-1809.1949.tb02451.x. [DOI] [PubMed] [Google Scholar]
- Wright S. The origin of the F-statistics for describing the genetic aspects of population structure. Honolulu (HI): University of Hawaii Press; 1973. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.