Abstract
The c-myc proto-oncogene regulates the expression of 15% to 20% of all genes, depending on the cell type, and the regulation is usually modest (1.5- to 2.0-fold). The authors discovered that in addition to regulating mRNA abundance, c-Myc regulates the formation of the 7-methylguanosine cap on many mRNAs, including transcriptional target genes and others not transcriptionally activated. Because the 7-methylguanosine cap is required for effective translation, enhanced methyl cap formation leads to increased protein production from Myc-responsive genes that exceeds the transcriptional induction. Increased cap methylation is linked to Myc-dependent enhanced activity of 2 critical kinases, TFIIH and p-TEFb, which phosphorylate the RNA polymerase II carboxy-terminal domain (CTD). Phosphorylation of the CTD recruits RNGTT and RNMT, the enzymes involved in mRNA capping, to the nascent transcript. Evidence is accumulating that enhanced cap methylation makes a significant contribution to Myc-dependent gene regulation and protein production.
Keywords: Myc, gene expression, translation, cap methylation, cell proliferation
Introduction
Since the discovery that deregulated expression of Myc promotes tumor formation and progression, the question of how Myc proteins function has been a subject of intense investigation. The Myc proteins were initially identified as transcription factors that activate and repress specific protein-encoding genes transcribed by RNA polymerase II.1,2 More recently, Myc proteins were found to regulate miRNA expression and upregulate the activity of RNA polymerase I and III, whose gene products include tRNAs and rRNA.3 Because Myc regulates protein-encoding genes, the miRNAs that control their expression, and the RNAs required to translate mRNA into proteins, it is not surprising that a fairly universal response to Myc expression is that of an increase in protein synthesis. However, the coordinated expression of different RNAs is not sufficient for protein synthesis, because RNA polymerase II transcripts require modification and processing to become competent for translation. Myc proteins have recently been discovered to regulate mRNA cap methylation, a modification that regulates the processing steps required to produce mature transcripts.4,5 This review explores the mechanism of Myc-induced mRNA cap methylation, the direct biochemical function of this modification, and its biological consequences.
mRNA Cap Methylation Is Essential for Gene Expression
RNA polymerase II transcripts are modified and regulated in a series of steps that prepare the RNA for initiation and sustained translation.5,6 Exons are spliced together to create an open reading frame; the 3′ end of the transcript is cleaved and polyadenylated; and RNA is exported from the nucleus into the cytoplasm, where translation factors bind to the RNA to direct recruitment of the ribosomal subunits and translation. While being translated, transcripts must be protected from premature degradation by exonucleases. These processes are governed, to a greater or lesser extent, by the methyl cap found at the 5′ end of mRNA. Furthermore, the methyl cap has the potential to promote transcription elongation (see below).
The methyl cap is an inverted 7-methylguanosine group joined to the first transcribed nucleotide by a triphosphate bridge (Figure 1). The methyl cap is not encoded by DNA but is rather formed by a series of enzymatic reactions that are conserved from yeast to human (Figure 2).5,7 The enzymes that catalyze this RNA modification are essential for cell viability.8–10 Nascent RNA is transcribed with a triphosphate on the first transcribed nucleotide. A triphosphatase removes the terminal phosphate, and a guanylyl transferase adds an inverted guanosine monophosphate group to create the unique 5′-5′ triphosphate linkage, G5′ppp5′N, where N is the first transcribed nucleotide. In mammals, these 2 enzymatic activities are present on 1 polypeptide called RNGTT (RNA guanylyltransferase and 5′ triphosphatase or capping enzyme). The cap is subsequently methylated by a distinct enzyme, RNMT (RNA guanine-7 methyltransferase). RNMT is effective at methylating guanosine only when it is in the cap structure and when the cap structure is attached to an oligonucleotide. Therefore, RNA must be capped before the cap structure can be methylated.
Figure 1.
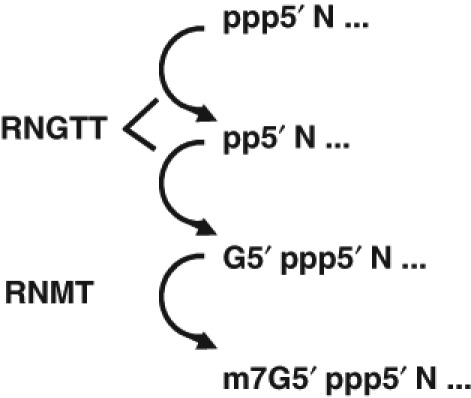
Enzymatic steps of mRNA capping. The primary mRNA transcript is synthesized with a 5′ triphosphate (pppN). The enzyme RNGTT (CE, or capping enzyme) acts as a phosphatase to remove the terminal phosphate (ppN). The RNGTT enzyme then uses GTP to create a G5′ppp5′N cap. This cap structure is subsequently recognized by a second enzyme RNMT to methylate the terminal guanosine.
Figure 2.
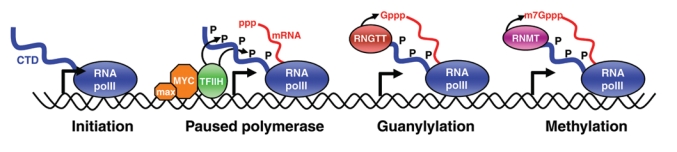
mRNA cap formation is coupled to phosphorylation of the RNA polymerase II carboxy-terminal domain (CTD). RNA polymerase II forms a preinitiation complex at the promoter with an unphosphorylated CTD (Initiation). The TFIIH basal transcription factor is recruited to phosphorylate the CTD on serine-5 (paused polymerase). Myc can enhance the level and recruitment of TFIIH to promoters. The phosphorylated CTD recruits the first capping enzyme (RNGTT) to add the reverse guanosine to the 5′ end of the mRNA (guanylylation). The cap methylation enzyme (RNMT) is also recruited by the phospho-CTD to methylate the GpppN-cap to yield m7G-pppN (methylation). Only the methylated cap is recruited by eIF4E in the cytoplasm to load efficiently onto ribosomes for translation.
The methyl cap is formed predominantly, if not entirely, shortly after the initiation of transcription. The capping enzymes are recruited to the 5′ end of RNA by binding to the carboxy-terminal domain (CTD) of RNA polymerase II (Figure 2). The CTD contains a series of repeats based on a 7–amino acid sequence that receives posttranslational modifications during the transcription cycle.11 TFIIH phosphorylates the CTD at the initiation of transcription, which promotes recruitment of the capping enzymes, RNGTT and RNMT, to the nascent transcript as it emerges from the polymerase machinery.12 The capping enzymes have been observed in the gene body13 and have been associated with transcription elongation, although this mechanism does not appear to require the enzymatic activity of the guanylyltransferase or cap methyltransferase.14,15 In vitro, the mammalian capping enzyme, RNGTT, relieves transcriptional repression imposed by NELF (negative elongation factor), thus favoring promoter clearance.15 In yeast, the capping enzymes have been demonstrated to bind to p-TEFb (positive transcription elongation factor), although the role of this interaction in mammalian systems is unclear at present.16,17
As described above, the methyl cap has the potential to promote several posttranscriptional processing events.5 The methyl cap binds to the cap binding complex (CBC), a heterodimer of CBC80 and CBC20. The interaction of CBC with the methyl cap promotes correct splicing of the first intron, polyadenylation, and nuclear export of RNA. The methyl cap also binds to the eIF4F (elongation initiation factor 4F) complex via eIF4E, which promotes recruitment of the 40S ribosomal subunit and the initiation of translation. The unique 5′-5′ triphosphate linkage in the cap also protects RNA against attack by exonucleases. The studies that revealed the potential of the methyl cap in mammalian systems were carried out in vitro using synthetic RNA, so we have little idea about how genes are regulated by the methyl cap in vivo. What is clear is that the methyl cap is essential for mammalian mRNA translation and cell viability.8,18
mRNA Cap Methylation Is a Significant Component of the Myc Response
It is important to consider the relative contribution of transcriptional and cap-mediated responses to Myc. Although its transcriptional effects are broad, the majority of Myc-responsive genes are only weakly modulated in terms of mRNA abundance, usually 1.5- to 2.0-fold. In contrast to this weak transcriptional induction, there is frequently a 3- to 6-fold increase in cap-methylated mRNA, which is more than double the transcriptional response (Figure 3).19,20 This increased cap methylation manifests itself in greater loading of mRNAs onto polysomes and significant increases in protein levels that exceed the modest transcriptional induction of Myc target genes.20 Hence, increased cap methylation may contribute as much to the induction of Myc-responsive protein expression as transcriptional induction. Although transcription and cap methylation can be mechanistically linked, Myc appears to regulate both processes independently. Myc-induced cap methylation does not depend on increased transcription, because Myc mutants that are defective for transcription remain competent to promote cap methylation.20 Interestingly, some targets of Myc-induced cap methylation are not transcriptional targets and can exhibit increased translation and protein levels without any change in mRNA levels.20 There is no evidence that Myc-induced transcription depends on cap methylation.20,21 The gene features that promote responsiveness to cap methylation remain undefined.
Figure 3.
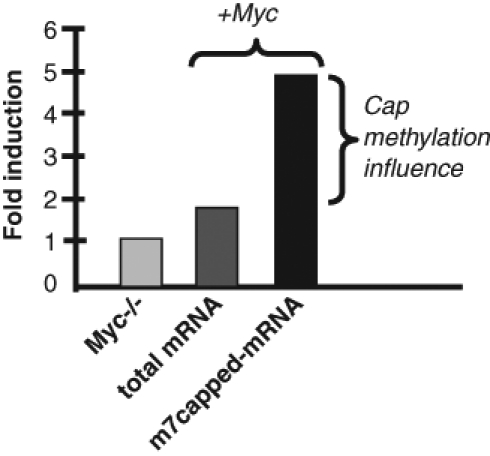
Enhanced mRNA cap methylation makes a significant contribution to the Myc response. Data are presented for an average of 8 Myc target genes from a previous study. Myc-null fibroblasts were reconstituted with exogenous Myc, and total mRNA levels were measured by RT-PCR. mRNAs were also selected with antibodies for the presence of a m7G cap. In comparison to the parental cells, Myc induced a 1.8-fold average increase in total mRNA abundance. However, the average increase in m7G-containing mRNAs was 4.9-fold, which reflects mRNA abundance and an increased fraction of mRNA with a methylated cap. Because m7G-containing mRNAs are translated much more efficiently than mRNAs with a nonmethylated cap, the net increase in protein levels are more reflective of the m7G-induction.
Mechanism of Myc-Induced Cap Methylation
Myc is a DNA-binding protein that predominantly localizes around transcription initiation sites.22,23 Myc binds to TFIIH and promotes its recruitment to transcription initiation sites.20 As described above, TFIIH phosphorylation of the RNA polymerase II CTD promotes increased recruitment of the enzymes that catalyze formation of the methyl cap (Figure 2).12 Myc also promotes recruitment of histone acetyltransferases to transcription initiation sites, and the resultant relaxation of chromatin may promote TFIIH recruitment.24 In correlation with increased TFIIH recruitment, Myc promotes increased RNA polymerase II phosphorylation on serine-5 of the CTD, which correlates with increased cap methylation20 (Figure 2).
Myc-induced TFIIH recruitment is not sufficient to promote cap methylation.21 Cellular methylation reactions utilize the methyl donor, S-adenosyl methionine (SAM), which is converted to S-adenosyl homocysteine (SAH) during the methylation reaction.5 SAH inhibits methylation reactions, and Myc-induced cap methylation cannot proceed without removal of SAH.21 Myc upregulates the enzyme S-adenosyl homocysteine hydrolase (SAHH), which hydrolyzes SAH, thus permitting cap methylation (Figure 3B). Myc upregulates SAHH by promoting its transcription and cap methylation.
Enhanced Cap Methylation Is Essential for Myc-Induced Protein Synthesis and Cell Proliferation
Myc promotes cap methylation on a significant subset of transcripts, including most Myc transcriptional targets and some nontranscriptional target genes.19,20 The significance of the upregulation of cap methylation for Myc biological function was addressed following the finding that SAHH upregulation is required for Myc-induced cap methylation.21 SAHH is not required for activated transcription; therefore, by inhibiting SAHH expression or activity, Myc-induced cap methylation could be selectively inhibited. Inhibition of cap methylation resulted in a reduction in Myc-induced protein synthesis, cell proliferation, and cell transformation, thus demonstrating that cap methylation is required for the core Myc functions.
Future Directions
The regulation of cap methylation makes a significant contribution to the induction of protein levels for Myc target genes, perhaps equaling transcriptional induction for most genes. Although cap methylation is coupled to transcription through CTD phosphorylation, the levels of methyl cap on a given mRNA can be regulated independent of mRNA level. Surprisingly, several genes that are critical for transcription and cell cycle regulation can be regulated at the level of cap methylation and concomitantly increased protein levels without any change in mRNA levels.20,25 However, better methods for separating methyl-capped from capped mRNAs will be required to characterize the full extent of this nontranscriptional Myc activity, as well as cap methylation in general. Even small changes in Myc levels (2-fold or less) can affect oncogenic transformation by Myc itself or for other pathways such as APC/WNT.26,27 Hence, inhibiting cap methylation represents a viable therapeutic target in cancer cells because it would block a key Myc activity likely required for oncogenic transformation. The broader question emerges: How many other transcription factors increase cap methylation along with their transcriptional effects? E2F1 transcription factor can enhance cap methylation on its target genes,19 but whether Myc and E2F1 are unique in this capacity remains to be explored. Differential cap methylation may represent a general mechanism for enhanced protein production that augments transcriptional induction across a broad spectrum of the genome.
Acknowledgments
This review was funded by a grant from the National Cancer Institute to M.D.C. and by an Medical Research Council Career Development Award to V.H.C.
Footnotes
The authors declared no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- 1. Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol 2006;16:253-64 [DOI] [PubMed] [Google Scholar]
- 2. Meyer N, Penn LZ. Reflecting on 25 years with MYC. Nat Rev Cancer 2008;8:976-90 [DOI] [PubMed] [Google Scholar]
- 3. Kenneth NS, White RJ. Regulation by c-Myc of ncRNA expression. Curr Opin Genet Dev 2009;19:38-43 [DOI] [PubMed] [Google Scholar]
- 4. Furuichi Y, Shatkin AJ. Viral and cellular mRNA capping: past and prospects. Adv Virus Res 2000;55:135-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Cowling VH. Regulation of mRNA cap methylation. Biochem J 2010;425:295-302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Shatkin AJ, Manley JL. The ends of the affair: capping and polyadenylation. Nat Struct Biol 2000;7:838-42 [DOI] [PubMed] [Google Scholar]
- 7. Shatkin AJ. Capping of eucaryotic mRNAs. Cell 1976;9:645-53 [DOI] [PubMed] [Google Scholar]
- 8. Chu C, Shatkin AJ. Apoptosis and autophagy induction in mammalian cells by small interfering RNA knockdown of mRNA capping enzymes. Mol Cell Biol 2008;28:5829-36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mao X, Schwer B, Shuman S. Yeast mRNA cap methyltransferase is a 50-kilodalton protein encoded by an essential gene. Mol Cell Biol 1995;15:4167-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shibagaki Y, Itoh N, Yamada H, Nagata S, Mizumoto K. mRNA capping enzyme: isolation and characterization of the gene encoding mRNA guanylytransferase subunit from Saccharomyces cerevisiae. J Biol Chem 1992;267:9521-8 [PubMed] [Google Scholar]
- 11. Chapman RD, Heidemann M, Hintermair C, Eick D. Molecular evolution of the RNA polymerase II CTD. Trends Genet 2008;24:289-96 [DOI] [PubMed] [Google Scholar]
- 12. Bentley DL. Rules of engagement: co-transcriptional recruitment of pre-mRNA processing factors. Curr Opin Cell Biol 2005;17:251-6 [DOI] [PubMed] [Google Scholar]
- 13. Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat Struct Mol Biol 2008;15:71-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schroeder SC, Zorio DA, Schwer B, Shuman S, Bentley D. A function of yeast mRNA cap methyltransferase, Abd1, in transcription by RNA polymerase II. Mol Cell 2004;13:377-87 [DOI] [PubMed] [Google Scholar]
- 15. Mandal SS, Chu C, Wada T, Handa H, Shatkin AJ, Reinberg D. Functional interactions of RNA-capping enzyme with factors that positively and negatively regulate promoter escape by RNA polymerase II. Proc Natl Acad Sci U S A 2004;101:7572-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Guiguen A, Soutourina J, Dewez M, et al. Recruitment of P-TEFb (Cdk9-Pch1) to chromatin by the cap-methyl transferase Pcm1 in fission yeast. EMBO J 2007;26:1552-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Viladevall L, St Amour CV, Rosebrock A, et al. TFIIH and P-TEFb coordinate transcription with capping enzyme recruitment at specific genes in fission yeast. Mol Cell 2009;33:738-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Muthukrishnan S, Both GW, Furuichi Y, Shatkin AJ. 5′-Terminal 7-methylguanosine in eukaryotic mRNA is required for translation. Nature 1975;255:33-7 [DOI] [PubMed] [Google Scholar]
- 19. Cole MD, Cowling VH. Specific regulation of mRNA cap methylation by the c-Myc and E2F1 transcription factors. Oncogene 2009;28:1169-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Cowling VH, Cole MD. The Myc transactivation domain promotes global phosphorylation of the RNA polymerase II carboxy-terminal domain independently of direct DNA binding. Mol Cell Biol 2007;27:2059-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fernandez-Sanchez ME, Gonatopoulos-Pournatzis T, Preston G, Lawlor MA, Cowling VH. S-adenosyl homocysteine hydrolase is required for Myc-induced mRNA cap methylation, protein synthesis, and cell proliferation. Mol Cell Biol 2009;29:6182-91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Brown SJ, Cole MD, Erives AJ. Evolution of the holozoan ribosome biogenesis regulon. BMC Genomics 2008;9:442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hulf T, Bellosta P, Furrer M, et al. Whole-genome analysis reveals a strong positional bias of conserved dMyc-dependent E-boxes. Mol Cell Biol 2005;25:3401-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer 2004;4:562-8 [DOI] [PubMed] [Google Scholar]
- 25. Cowling VH. Enhanced mRNA cap methylation increases cyclin D1 expression and promotes cell transformation. Oncogene 2010;29:930-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shachaf CM, Gentles AJ, Elchuri S, et al. Genomic and proteomic analysis reveals a threshold level of MYC required for tumor maintenance. Cancer Res 2008;68:5132-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Yekkala K, Baudino TA. Inhibition of intestinal polyposis with reduced angiogenesis in ApcMin/+ mice due to decreases in c-Myc expression. Mol Cancer Res 2007;5:1296-303 [DOI] [PubMed] [Google Scholar]


