Abstract
Background
Patterning and differentiation of developing musculatures require elaborate networks of transcriptional regulation. In Drosophila, significant progress has been made into identifying the regulators of muscle development and defining their interactive networks. One major family of transcription factors involved in these processes consists of homeodomain proteins. In flies, several members of this family serve as muscle identity genes to specify the fates of individual muscles, or groups thereof, during embryonic and/or adult muscle development. Herein, we report on the expression and function of a new Drosophila homeobox gene during both embryonic and adult muscle development.
Methodology/Principal Findings
The newly described homeobox gene, termed lateral muscles scarcer (lms), which has yet uncharacterized orthologs in other invertebrates and primitive chordates but not in vertebrates, is expressed exclusively in subsets of developing muscle tissues. In embryos, lms is expressed specifically in the four lateral transverse (LT) muscles and their founder cells in each hemisegment, whereas in larval wing imaginal discs, it is expressed in myoblasts that develop into direct flight muscles (DFMs), which are important for proper wing positioning. We have analyzed the regulatory inputs of various other muscle identity genes with overlapping or complementary expression patterns towards the cell type specific regulation of lms expression. Further we demonstrate that lms null mutants exhibit reduced numbers of embryonic LT muscles, and null mutant adults feature held-out-wing phenotypes. We provide a detailed description of the pattern and morphology of the direct flight muscles in the wild type and lms mutant flies by using the recently-developed ultramicroscopy and show that, in the mutants, all DFMs are present and present normal morphologies.
Conclusions/Significance
We have identified the homeobox gene lms as a new muscle identity gene and show that it interacts with various previously-characterized muscle identity genes to regulate normal formation of embryonic lateral transverse muscles. In addition, the direct flight muscles in the adults require lms for reliably exerting their functions in controlling wing postures.
Introduction
The musculatures in both vertebrate and invertebrate animals are composed of a large variety of different muscles that are distinguished according to their specific size, morphology, and physiological properties. Whereas much progress has been made in defining the regulatory processes of myogenesis as such, the understanding of the developmental mechanisms underpinning muscle diversity is much less complete. To date, the fruit fly Drosophila has been one of the most profitable models for dissecting the mechanisms regulating muscle diversity. In this system, a number of mechanisms that provide specific identities to individual muscles have been described, particularly for the development of larval muscles during embryogenesis. By contrast, our knowledge about the diversification of adult muscles, which takes place in a second round of muscle development during metamorphosis, is still much more limited.
Larval muscle development in Drosophila leads to the formation of ∼30 distinct muscle fibers arranged in a stereotyped pattern within each embryonic trunk hemisegment (reviewed in [1]). A large body of evidence has revealed that each of these muscles is seeded by a single myoblast, termed muscle founder cell, which already retains a defined identity that predetermines the characteristics of the particular muscle it will form. Upon myoblast fusion between founder myoblasts and fusion-competent myoblasts, which largely lack distinct identities, the identity of the founder cell is then imposed on the growing muscle syncytium and shapes its development. In the current view, the identity of individual founder cells is conferred through the expression and function of particular combinations of muscle identity genes (reviewed in [2], [3]). The muscle identity genes that have been characterized functionally to date all encode various types of transcription factors. The best-characterized muscle identity factors belong to the families of the homeodomain proteins (including Apterous/Ap, Slouch/S59, Ladybird/Lb, Even-skipped/Eve), Zinc-finger factors (Krüppel/Kr), basic helix-loop-helix factors (Nautilus/Nau), and the COE (Col/Olf1/EBF) transcription factors (Collier/Col) [4], [5], [6], [7], [8], [9], [10], [11]. In addition, the activities of these identity factors are modulated by transcription factors that are expressed within distinct broad domains in the somatic mesoderm, such as the homeodomain proteins Tinman/Tin, Muscle segment homeodomain/Msh, Six4, and Pox meso [12], [13], [14], [15]. Another notable example of this latter class of regulators are the Hox factors, which are expressed in broad domains along the anterior-posterior axis within the somatic mesoderm and are known to modulate the activities of muscle identity factors in a region-specific manner along this axis [16], [17], [18]. As has been shown in some of these cases, different muscle identity genes and regional regulators are part of hierarchical and cross-regulatory networks during the development of a particular muscle. As a result, some of the identity factors and regional factors are expressed only transiently whereas the expression of others continues until a mature fiber is formed. Ultimately, the functions of these transcription factors in muscle fate determination must be mediated by their transcriptional target genes, but our information about these targets and their roles in making muscles distinct is currently rather limited [19], [20].
During metamorphosis, the majority of the adult muscles are generated anew from the descendants of undifferentiated myoblasts, termed adult muscle precursors, that are set aside during embryogenesis and start proliferating during larval stages [21] (reviewed in [22], [23]). Currently, there is no clear evidence for muscle identity genes acting at the level of individual muscle founder cells during adult muscle development. However, like in embryos, Hox genes are known to play an important role during the regional diversification of muscle patterns along the anterior-posterior axis [24], [25], [26]. Another example for genes involved in adult muscle diversification is ladybird, which is widely expressed in leg disc-associated myoblasts and required for normal leg muscle development [27], [28]. Hence, this embryonic muscle identity gene is redeployed during metamorphosis to participate in the control of the development of large subset of myoblasts, namely those forming the leg muscles. An instructive example of myoblast diversification during metamorphosis has also been described in the wing disc. The wing disc-associated myoblasts generate two fundamentally different types of muscles, which on the one hand include the indirect flight muscles that power the flight, and on the other hand the direct flight muscles that control wing positioning during steering and flight stabilization. It has been demonstrated that the myoblasts giving rise to the indirect flight muscles (IFMs), which form the majority of the wing disc-associated myoblasts and are located in proximal areas of the wing disc of the presumptive notum, are marked by the expression of the homeobox gene vestigial/vg. Conversely, the myoblasts forming the direct flight muscles (DFMs), which are located in adjacent areas near the future wing hinge, are marked by high-level expression of the homeobox gene cut. In this latter population of myoblasts, high levels of Cut repress vestigial, whereas in the IFM-forming population of myoblasts Vg down-regulates cut to low expression levels. In addition, Vg represses apterous (ap), which can therefore only be activated in the high-cut myoblasts. ap then helps specifying these myoblasts as DFM myoblasts [29], [30], [31], [32]. Altogether, these regulatory interactions and the functions ascribed to vg and cut/ap in IFM versus DFM development point to some mechanistic analogies of muscle diversification during larval and adult muscle development.
As the currently-known collection of muscle identity genes is still not sufficient to explain the entire muscle pattern during embryogenesis, and even less so during formation of adult muscle diversity, our laboratories have been aiming to identify additional genes of this type. In this report, we describe a new homeobox gene, which we call lateral muscles scarcer (lms), that fulfils the criteria for a muscle identity gene. During embryogenesis, lms is expressed specifically in the founders and syncytial fibers of the lateral muscles LT1-LT4 as part of a regulatory network that includes ap, which exhibits a closely related expression pattern, as well as lb, Kr, and msh. We show that null mutations for lms, which are homozygous viable, cause defects in LT muscle development that consist of a reduction in the number of muscles and morphological aberrations. These defects occur with a relatively low expressivity, similar to those reported for ap, and double mutants for lms and ap show additive effects. During adult muscle development, lms is expressed in wing disc-associated myoblasts within a small area that overlaps with the presumptive DFM myoblasts marked by high-cut expression. The held-out wing phenotype of lms null mutant flies is compatible with a requirement of lms for normal DFM differentiation. Because detailed analysis of the DFMs in lms mutant flies showed that the DFMs are present and lack any overt morphological alterations, it appears that lms is needed for the acquisition of the requisite functional properties of these muscles rather than their formation and morphogenesis.
Results
CG13424/lms orthologs are present in dipteran, chordate, and cnidarian lineages
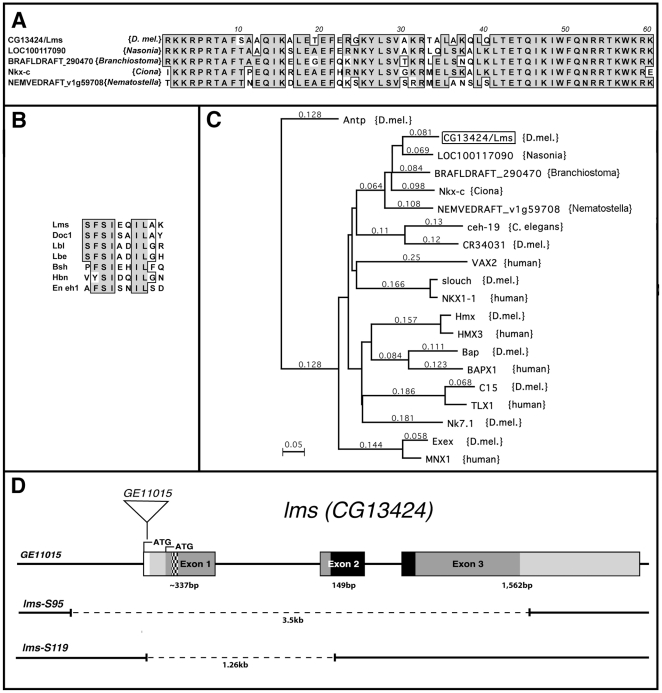
The Drosophila homeobox gene CG13424, subsequently named lateral muscles scarcer (lms), was reported to be expressed in specific somatic muscle founders (Berkeley Drosophila Genome Database, [33]), which suggested it as a candidate for a new regulator of muscle identity or differentiation. Database searches showed that orthologs of lms are present in the parasitoid wasp Nasonia, honey bee, the Cnidarian Nematostella, as well as in the chordates Branchiostoma and Ciona (Fig. 1A and data not shown), thus indicating an ancient origin of this gene. By contrast, in vertebrates and in C. elegans orthologs of lms are not present, suggesting that the corresponding genes have been lost in these lineages. Lms also contains a putative eh1 repression domain (in NK homeodomain proteins also known as TN domain) near its N-terminus (Fig. 1B) [34]. A phylogenetic tree for lms and its closest homologs in the above species as well as humans is shown in Fig. 1C, which indicates a sequence affinity of the Lms homeodomain with the NK homeodomain family. The Drosophila lms gene locus, which maps to 57A on the second chromosome, is shown in Fig. 1D.
Figure 1. Sequence conservation of Lms and imprecise P-excisions at the lms locus.
(A) Sequence alignments of the homeodomains from Lms (D. melanogaster) with its predicted orthologs from the sequenced genomes of Nasonia vitripennis (parasitoid wasp), Branchiostoma floridae (amphioxus, shown is one of two paralogs also known as Nedxa and Nedxb [60]), Ciona intestinalis (vase tunicate), and Nematostella vectensis (Cnidaria). (B) Sequence alignments of the most closely related eh1 (TN) domains from Drosophila transcription factors with that from Lms. (C) Phylogenetic tree using the homeodomain sequences from Lms (D. melanogaster) and the most closely related homeodomains from Branchiostoma, Nasonia, Ciona, Nematostella, D. melanogaster, and humans, showing that Drosophila and humans lack any paralogs and orthologs, respectively, of lms. (D) lms gene locus with P-insertion GE11015 and deletions generated via imprecise excision of this P-element underneath. Predicted exons are boxed, sequences covered by the longest known EST RE33150 are shaded. The open reading frame starting from the first ATG of RE33150 is shown in dark grey, the eh1 (TN) domain checkered, and the homeodomain in black. The open reading frame extends towards the 5′ in the genomic sequence to another ATG that is embedded in a less favorable translation initiation sequence and may not be present in the transcripts.
Expression pattern of lms during embryonic development
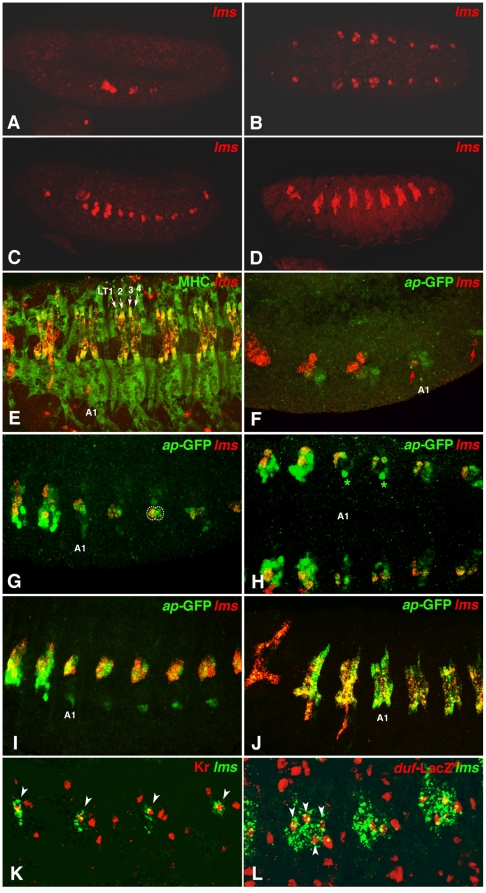
The embryonic expression pattern of lms was analyzed in greater detail by whole mount in situ hybridizations of wild type embryos and by comparing it to additional markers for the somatic mesoderm (Fig. 2). The gene lacks any maternal contribution and its expression occurs exclusively in the somatic mesoderm. During stage 11, lms mRNA is first detected in small clusters of mesodermal cells in each of the three thoracic hemisegments (Fig. 2A). During early stage 12, lms transcripts are also detected in abdominal segments, initially in a single mesodermal cell in each abdominal hemisegment (Fig. 2B). At late stage 12 and into early stage 13, the abdominal lms expression consists of clusters of three to four cells in each hemisegment and the size of the lms-positive thoracic cell clusters is increased (Fig. 2C). As shown in Fig. 2D in a stage 15 embryo, these lms-positive cell clusters develop into lateral somatic muscle fibers. Double stainings with a myosin heavy chain (MHC) antibody, which marks all somatic muscles, identifies the four lms positive syncytia in each abdominal segment as the Lateral Transverse muscles (LT1 - LT4, also known as muscles 21–24; Fig. 2E, [1]). LT1–LT4 are also positive for lms in the thoracic segments T2 and T3, whereas in T1, where LT muscles are absent [1], lms is expressed in muscle VT1 (see below, Fig. 2J).
Figure 2. lms mRNA expression during embryonic lateral transverse muscle (LTM) development.
(A) Stage 11 embryo (lateral view) showing earliest lms mRNA expression in thoracic clusters of somatic mesodermal cells. (B) Early stage 12 embryo (ventral view) showing lms mRNA expression in thoracic and abdominal progenitors of lateral transverse muscles (LTMs). Sites of procephalic expression may correspond to progenitors of yet unidentified head muscles (see also C, D). (C) Late stage 12 embryo (lateral view) showing lms mRNA expression in founder cells that begin to fuse to form LTMs. (D) Stage 15 embryo (lateral view) showing lms mRNA expression in LTM fibers. (E) High magnification view of stage 16 embryo counter-stained with MHC antibodies (green), which identifies the lms expressing muscles (red) of the thoracic segments T2 and T3 and the abdominal segments as LTM 1–4 (muscles 21–24) (A1: first abdominal segment). (F) Stage 11 apME680-GFP embryo showing lateral high magnification view of segments T1 to A2 stained for lms mRNA (red) and GFP protein (anti-GFP; green). In T2 and T3, lms expression partially overlaps with the ap-GFP expressing clusters, and in abdominal segments, lms expression occurs in a single cell within the ap-GFP clusters at this stage (arrow). (G) Early stage 12 apME680-GFP embryo (T2–A5) showing coexpression of lms mRNA (red) and ap-GFP (green) in two muscle progenitors in each abdominal segment (circled), with the anterior cell displaying higher lms mRNA levels. (H) Mid stage 12 apME680-GFP embryo (T2 –A4, ventral view) showing co-expressed lms mRNA (red) and ap-GFP (green) in 2–3 LTM muscle progenitors and/or founder cells per abdominal hemisegment. An additional, lms-negative ap-GFP-labeled cell appears ventrally adjacent that will form a ventral acute muscle (asterisks; see also I). (I) Stage 14 apME680-GFP embryo (T2 – A6, laterally) showing co-expression of lms mRNA (red) and ap-GFP (green) in LTM muscle precursors. (J) Stage 15 apME680-GFP embryo (T1 – A4, laterally) showing coincidence of lms mRNA (red) and ap-GFP (green) expression in abdominal LTM1-4 muscles, partial overlap in LT1-4 of T2 and T3, and exclusive lms expression in muscle VT1 of T1. (K) Stage 12 lateral high magnification view of embryo stained for lms mRNA (green) and Kr protein (red). The presumed founder cell of LT2 expresses both lms and Kr (arrow heads). (L) Late stage 12 lateral high magnification view or rP298-lacZ embryo stained for lms mRNA (green) and LacZ. Each lms cell cluster contains four founder nuclei for the muscles LT1 – 4 (arrow heads).
The muscle identity gene apterous (ap), encoding a LIM homeodomain containing transcription factor, is also expressed within the four LT muscles, as well as in two Ventral Acute muscles (VA2 and VA3) [35]. To compare the temporal and spatial expression patterns of lms and ap in these muscles and their progenitors, embryos carrying the apME680-GFP reporter were used for GFP/lms mRNA double stainings. The apME680 enhancer element recapitulates mesodermal ap expression in LT1–LT4 and in a single Ventral Acute muscle (VA) and their corresponding progenitors [35]. ap-GFP expression can be observed during stage 11 in small cell clusters within abdominal segments before lms expression is detectable (data not shown). During early stage 12, the single lms-positive cell observed in each abdominal hemisegment corresponds to one of the ap-GFP-positive cells within each abdominal cell cluster, whereas in T2 and T3 most of the lms-expressing cells are located dorsally adjacent to the ap-GFP-expressing cells (Fig. 2F). In T1, only lms expression but no ap-GFP can be detected (Fig. 2F). During mid stage 12, a second cell becomes positive for lms in each abdominal hemisegment and ap-GFP becomes restricted to the same two cells (Fig. 2G; the original, anterior cell has accumulated higher levels of lms mRNA). In T1 and T2, ap-GFP expression expands into the lms-positive cells (Fig. 2G). Double stainings for lms and the muscle identity factor Krüppel (Kr) at stage 12, which is expressed in the muscles LT2 & LT4 and their founders [6], show that the initial lms-positive cell corresponds to one of the Kr-positive cells (likely the LT2 founder or LT1/LT2 progenitor; Fig. 2K).
During late stage 12, lms and ap-GFP are co-expressed in up to four cells per abdominal hemisegment, which likely correspond to the four founder cells of the LT1 - 4 muscles (Fig. 2H). This assignment agrees with the presence of four founder nuclei labeled by duf (rP298)-LacZ within each segmental cluster of lms expression during late stage 12 (Fig. 2L, arrow heads). Upon myoblast fusion during stage 14, lms and ap-GFP are co-expressed in the thoracic and abdominal LT muscle precursors, although in the thorax ap-GFP still extends further ventrally (Fig. 2I; ap-GFP is also seen in lms-negative abdominal VA muscle precursors). Finally, in the fully-formed muscle fibers of the four Lateral Transverse muscles (LT1–LT4) from T2, T3, and each abdominal hemisegment, lms transcripts and apterous-GFP remain co-expressed, whereas in T1 only lms expression can be detected (Fig. 2J).
Taken together, these observations strongly indicate that lms and ap are co-expressed in the four founder cells of the Lateral Transverse Muscles LT1–LT4 and in the muscle fibers formed from them, although the spatial and temporal dynamics leading to this co-expression differs for the two genes.
Regulatory interactions among different muscle identity genes during LT muscle development
Apart from lms and ap, the muscle identity gene muscle segment homeobox (msh) is active in the founder cells of muscles LT1 - 4, and Krüppel (Kr) is expressed in LT2 & LT4 and their founders (among founders of additional muscles) [6], [13]. Conversely, the ladybird (lb) genes are expressed in the progenitors and founders of the segment boundary muscles (SBM) and lateral adult muscle precursors, which arise at positions directly adjacent to those of LT1 - 4 (Fig. 3A) [6], [7], [13]. To determine whether msh, ap, and lb exert any positive or negative regulatory inputs towards lms, we examined the effects of their loss or gain of function on lms expression.
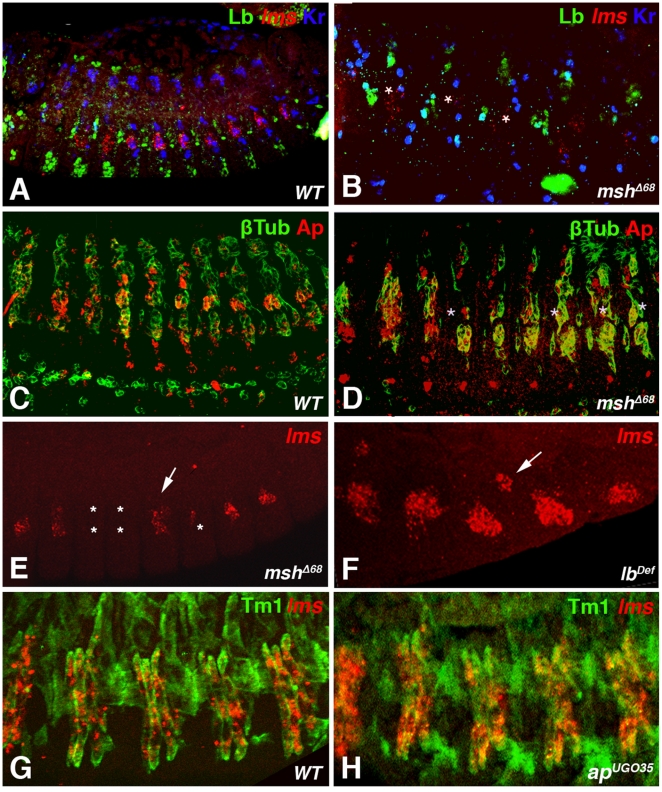
Figure 3. Effects of mutations of the muscle identity genes msh, lb, and ap on lms expression.
(A) to (D) show lateral and (E, F) dorso-lateral views of stage 13 embryos stained to reveal the effects of msh and lb loss of function mutations on expression of lms or ap. Triple-stained embryos for anti-Lb anti-Kr and lms transcripts are shown in (A and B) and double-stained for β3-tubulin (βTub) and ap transcripts in (C and D). (A) and (C) Wild type embryos. (B) Strongly reduced lms expression in homozygous mshΔ68 embryos is associated with the loss of Kr staining in LT2 and LT4 muscles and with expanded Lb expression domain (SBM muscle). (D) In segments with reduced ap expression in the absence of msh, β3-tubulin-labeled muscle precursors are still detected. (E) and (F) show lms expression patterns in homozygous mshΔ68 and in ladybird-deficient embryos (homozygous Df(3R)3/1). msh and lb mutations have opposite effects on lms expression. In msh null mutants (E) lms transcripts are either absent in all or in a subset of LT muscle precursors (asterisks) or the lms expression levels are reduced (arrow). Loss of lb (F) leads to an expanded expression of lms in lateral domains. Occasionally, ectopic expression of lms in more dorsal clusters of muscle cells can be detected (arrow). (G, H) Lateral views of stage 15 embryos (G: wild type, H: homozygous apUGO35; four abdominal hemisegments are shown with a focus on LT muscles) stained for lms transcripts and with anti-TM1 to reveal the muscle pattern, showing that in ap mutant embryos lms expression is largely unaffected.
As shown in Fig. 3B (see also Table 1), deletion of msh results in a severe reduction of lms and a loss of Kr expression in the founders of the LT muscles. In addition, the expression of ap in the founders and precursors the LT muscles is strongly reduced (Fig. 3D, asterisks, compare with Fig. 3C). At later stages, when myoblast fusion has initiated, lms expression does appear in the precursors of the LT muscles in msh mutants, but the expression is still reduced or absent in many of the segments (Fig. 3E). These effects are in line with the reported loss of many LT muscles in msh mutants [13] and suggest that this loss may in part be due to the failure in the proper activation of these three muscle identity genes in their muscle founders.
Table 1. Quantification of genetic effects on embryonic lms expression.
| lms expression/genetic context | Unchanged*n/% | Enlarged**n/% | Reduced**n/% | Ectopicn/% | Lossn/% | Total hemisegments |
| lb def | 17/36% | 20/42% | 0 | 10/22% | 0 | 47 |
| 24B>lbe | 12/29% | 0 | 30/71% | 0 | 0 | 42 |
| msh Δ68 | 9/19% | 0 | 28/60% | 0 | 10/21% | 47 |
*less than 30% difference of expression area between wt and a genetic context measured on projections from confocal stacks with detection of lms transcripts. Age matched stage 13 embryos oriented laterally were imaged and counted (Olympus, Fluoview Analysis tool was used for calculating expression area).
**more than 30% difference of expression area (mm2) between wt and a genetic context measured on projections from confocal stacks with detection of lms transcripts. Age matched stage 13 embryos oriented laterally were imaged and counted (Olympus, Fluoview Analysis tool was used for calculating expression area).
In embryos carrying a deficiency for the two lb genes, lbe and lbl, lms expression in lateral clusters appears slightly expanded (Fig. 3F; Table 1). Occasionally, lms expression is also seen in a few ectopic mesodermal cells that lie close to the segmental borders, possibly at the positions of SBM myoblasts that have lost their identity in the lbdef context.
As shown in Fig. 3H for late stage apUGO35 null mutants, loss of ap function does not grossly affect lms expression in LT muscles (and their founders, data not shown). As has been reported, mutation of ap causes occasional losses of individual LT muscles [5] (see also below). As we observe that any changes of lms expression in ap mutants closely parallel these effects on the numbers of LT muscles, these changes could be an indirect consequence of mis-specification of individual muscle fates rather than direct effects of ap on lms expression. The mild effects of ap mutation on lms expression could either be due to the absence of regulatory inputs or to functional redundancy of ap with other regulators.
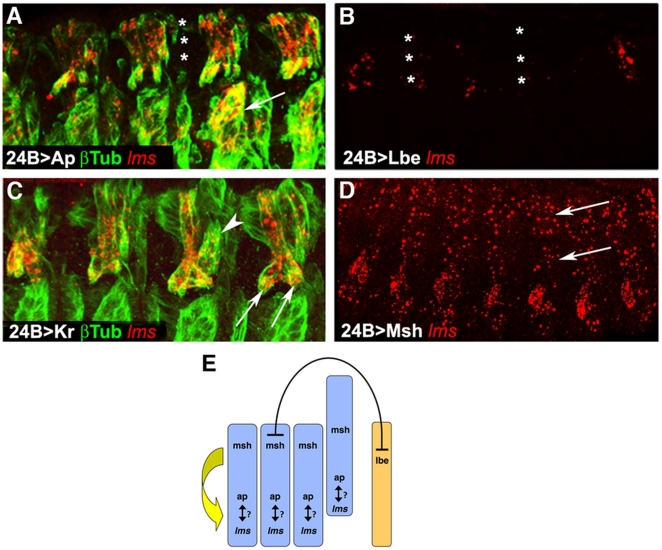
To complement the loss of function studies with gain of function experiments, we forced the expression of ap, lbe, Kr, and msh pan-mesodermally by using the 24B-Gal4 driver in combination with the corresponding UAS-constructs [36]. Panmesodermal expression of ap leads to malformation of the Lateral Transverse muscles (LT1 - 4, Fig. 4A) [17]. Additionally, a loss of the SBM (♯8) muscle can be detected (Fig. 4A, asterisks). Notably, ap misexpression results in lms expression in the three Ventral Acute muscles (VA1 - VA3, Fig. 4A, arrow). In the wildtype situation, no lms expression can be observed in these muscles.
Figure 4. Effects of ectopic mesodermal expression of the muscle identity genes ap, lbe, Kr, and msh, on lms expression.
(A) Lateral view of stage 15 24B-GAL4;UAS-ap embryo stained with anti-β3-tubulin (βTub) and for lms transcripts. Four abdominal hemisegments are shown revealing that panmesodermal Ap expression leads to an ectopic activation of lms in ventral muscles (arrow). Asterisks indicate position of segment border muscle (SBM) that is barely detected in the Ap gain of function context. (B) Lateral view of stage 14 24B-GAL4; UAS-lbe embryo stained for lms transcripts and showing four abdominal segments. In the lbe gain of function context lms expression is lost or reduced (asterisks). (C) Lateral view of stage 15 24B-GAL4;UAS-Kr embryo stained with anti-β3-tubulin (βTub) and for lms transcripts, showing four abdominal segments. Kr gain of function leads to muscle pattern defects such as LT muscle bending (arrows) or LT muscle shortening (arrowheads) without affecting lms expression. (D) Lateral view of stage 14 24B-GAL4; UAS-msh embryo stained for lms transcripts. lms expression is ectopically induced in mesodermal cells at dorso-lateral and dorsal locations (arrows). (E) Scheme showing regulatory interactions between msh, lbe, ap and lms identity genes in lateral muscles. LT1-LT4 are in blue whereas SBM is in yellow (all interactions are meant to be cell-autonomous and shown as they occur during the muscular precluster, progenitor, and founder cell stages). It is currently not known whether the interactions shown are direct or whether they are indirect, e.g., being a result of cell fate transformations.
Ectopic expression of lbe via 24B-Gal4 line results in enlarged and duplicated SBMs, increased numbers of lateral adult muscle precursors, and other muscle defects [7]. With regard to lms, we observe a severe reduction, and in some segments complete loss, of mRNA expression within the areas of the presumptive LT muscles (Fig. 4B; Table 1). It has been shown that increased SBM formation in ladybird mis-expressing embryos occurs at the expense of the LT musculature, which is replaced by unfused mono-nucleated myoblasts [7]. Based on these data and on the observed effects of ladybird loss and gain of function on lms, we conclude that ladybird may normally repress LT muscle founder genes, including lms, in SBM founders, whereas ectopic expression of ladybird is able to repress lms (and additional identity genes) in neighboring LT founders, leading to their failure to form LT muscles.
Pan-mesodermal Kr expression leads to mis-arrangements and patterning defects of the somatic musculature, including reduced numbers and altered shapes of the LT muscles (Fig. 4C). The remaining LT muscles are often bent at their ventral endings and others appear thicker and shortened (Fig. 4C, arrows and arrow head). In these embryos, lms mRNA is still expressed within the residual LT muscles. The absence of any ectopic expression of lms suggests that Kr is either not sufficient to activate lms on its own or it lacks any inputs altogether towards lms regulation.
Ectopic expression of msh via 24B-Gal4 leads to patterning defects in the dorsal musculature as well as disorganization of the ventral muscles [13]. Mis-expression of msh has also an effect on the expression of lms. Beside the regular expression of lms within the LT muscles, ectopic lms expression in lateral and in particular in dorsal areas of the somatic mesoderm is observed (Fig. 4D, arrows).
Taken together, misexpression of msh and to a lesser extent of ap causes ectopic expression of lms, suggesting that in the normal situation these two muscle-identity genes contribute to the transcriptional activation and maintenance of lms expression in the founders and precursors of the developing LT muscles. Conversely, our data suggest that ladybird normally has repressive inputs on lms expression, analogous to its reported effects on the muscle identity gene slouch, in order to prevent the inappropriate activation of these genes in the progenitors and founders of SBM muscles and lateral adult muscle precursors by yet undefined upstream regulators (Fig. 4E).
Generation of lms null alleles and consequences of loss of lms for LT muscle development
For the analysis of lms function during muscle development we generated null alleles by using the GE11015 P-element insertion in the lms locus (Fig. 1D). GE11015 is inserted 11 ntd. downstream of the computationally predicted start of the open reading frame, although there is currently no experimental evidence that this part of the gene locus is transcribed and that the ATG upstream of the insertion is being used as a translation start codon. The facts that the 5′ of the longest available EST begins ∼100 bp downstream of the insertion site, the second ATG of the predicted open reading frame has a better match to Drosophila consensus sequences for translation start sites, and the GE11015 strain is fully viable with normal embryonic expression of lms (data not shown) would suggest that GE11015 is inserted just upstream of the transcription start site of lms. From an imprecise excision screen with GE11015 we recovered several semilethal lines that we characterized by genomic PCR and sequencing of the deletion breakpoints. One allele, lmsS95, carries a deletion of both sides flanking the insertion site and removes the complete coding sequence of lms without affecting the coding sequences of the two adjacent genes located on either side. The deletion from a second allele, lmsS119, extends from the insertion site into the beginning of exon 2 (Fig. 1D). Based upon these molecular defects, both alleles are predicted to be functional nulls. Due to their weakness, adult homozygous flies for neither of the two alleles can be maintained as homozygous lines.
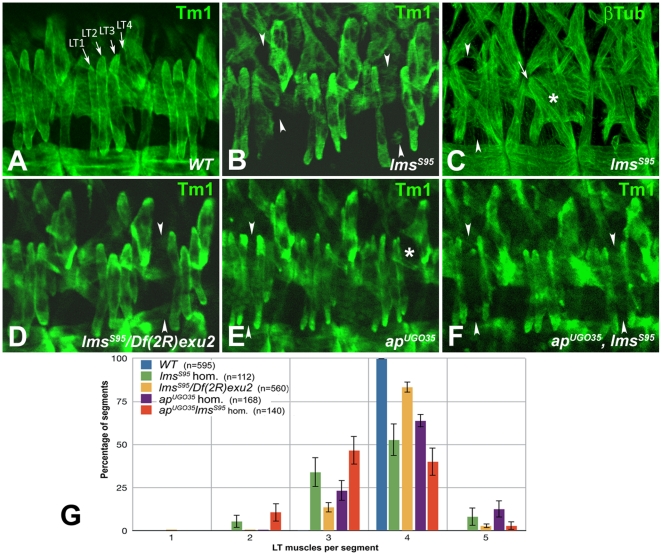
The embryonic muscle phenotypes of lms mutants were examined with general muscle markers and by crossing in apME680-GFP as a specific marker for the LT muscle pattern. In stage 16 embryos homozygous for lmsS95, we frequently observe the loss of one of the LT muscles in individual abdominal segments (Fig. 5B, compare with A). Generally, the LT muscles that are still present exhibit normal morphologies and attachments. However, in rarer cases, especially when two LT muscle fibers are missing in a hemisegment, the remaining LT muscles can be attached to segmental instead of intrasegmental attachment sites (Fig. 5C). The same range of phenotypes is also observed in embryos homozygous for lmsS119 (data not shown) and in embryos with the excision alleles in trans to the larger deficiency, Df(2R)exu2 (Fig. 5D). Overall, the observed muscle phenotypes are very reminiscent of the LT muscle phenotypes reported for ap mutant embryos [5] (Fig. 5E). Because lms and ap are co-expressed in developing LT muscles and mutations in both genes cause relatively mild LT muscle phenotypes, we generated ap, lms double mutant embryos to test whether the two genes exhibit partial functional redundancy in regulating LT muscle formation. However, in ap, lms double mutant embryos most of the LT muscles are also being formed (Fig. 5F). For a more detailed analysis of possible genetic interactions between lms and ap during LT muscle development, we undertook a quantitative analysis of the LT muscle phenotype of double mutant embryos and compared it to those of the single mutants. As shown in Fig. 5G, the expressivity of the LT muscle phenotype in lms single mutant embryos (green and yellow columns) is similar to that of ap single mutants (purple columns). In ap, lms double mutant embryos (red columns), the number of hemisegments with only three or two LT muscles present is significantly higher than for each of the single mutant genotypes. However, the effects appear additive rather than interactive, which indicates that the two genes function in parallel to ensure the formation of normal numbers of LT muscles.
Figure 5. Embryonic LT muscle phenotypes in lms mutants and ap, lms double mutants.
(A) to (F) show representative examples of phenotypes in three abdominal segments from stage 16 embryos with the denoted genotypes that were stained with anti tropomyosin (Tm1) or anti β3-tubulin (βTub) as indicated to visualize muscle patterns. (A) Wild type embryo with four LT muscles (arrows) in every segment. (B) Homozygous lmsS95 embryo with a LT4 and a LT3 muscle missing in the segments depicted on the left hand and right hand side, respectively (arrow heads). (Note that due to the slightly more dorsal orientation of the embryo as compared to embryo in (A), the VL muscles are not included in the Z-scan.) (C) lmsS95 with stronger than average disruptions of LT muscles. In left hand segment, one LT muscle is missing (arrow heads), and in segment in center, LT1 is connected to an abnormal attachment site (arrow) whereas the remaining LT muscles are replaced by aberrant syncytia with undefined identities (asterisk). (D) lmsS95/Df(2R)exu2 embryo with an LT1 muscles missing (arrow heads). (E) Homozygous apUGO 35 embryo with mis-arranged LT muscles, causing a gap (arrow heads), and possible transformation of LT4 (asterisk) into a second LT3 muscle. (F) Homozygous apUGO 35, lmsS95 double mutant embryo with only three LT muscles in each of the left and right hand segments (arrow heads; perhaps with the regular LT3 missing and LT4 transformed into LT3). (G) Numerical evaluation of LT muscle phenotypes. Genotypes are color-coded as shown, with “n” denoting the assessed number of abdominal segments for each genotype. Confidence intervals were calculated at 95% confidence levels. Note that sporadic occurrences of a fifth LT muscle have also been reported for controls and may be genetic background-dependent [7].
lms function during adult muscle development in Drosophila
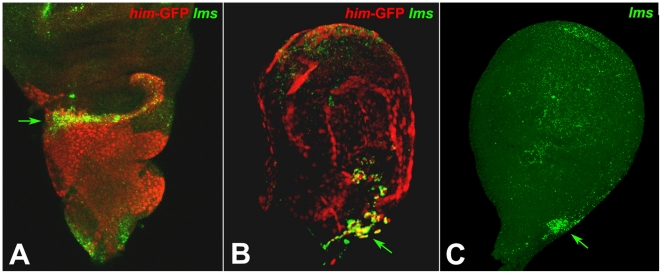
To examine whether lms also fulfils a role during the development of the adult musculature, we stained imaginal discs for lms mRNA expression. We used discs from a line expressing GFP driven by regulatory regions of the Him gene, which is expressed in all adepithelial cells of the wing and leg discs that will form the adult musculatures of the thorax and its appendices [30], [37], [38], [39], [40]. As shown in Fig. 6A, during late third instar the Him-GFP-labeled adepithelial cells cover the entire notum and hinge area of the wing disc. By contrast, lms is only expressed in the adepithelial cells within a narrow zone along the distal border of this area, which is located near the hinge region of the disc. The adepithelial cells of this zone are known to give rise to the direct flight muscles (DFMs) and, as seen in Fig. 6A, lms expression occurs predominantly within the anterior ∼1/2 of this zone. Likewise, lms is also restricted to a small area of adepithelial cells in third instar leg discs (Fig. 6B). The lms expressing cells are located in the stalk region of the disc outside of the leg proper area. Because clonal analysis had suggested that direct flight muscle 51 is derived from the leg disc, it is conceivable that some of this expression corresponds to myoblasts of presumptive DFM 51 in mesothoracic leg discs [38]. However, the observed expression in similar domains in all leg discs (including those of T1 and T3) would suggest that most of these cells give rise to muscles other than the DFMs, possibly including some that extend from the thorax into the coxae.
Figure 6. Expression of lms in adepithelial cells of wing and leg discs.
Shown are 3rd instar wing and leg discs. (A, B) show GFP-expressing adepithelial cells associated with wing (A) and leg (B) imaginal discs dissected from the transgenic Him-GFP larvae and stained with anti-GFP (red) and for lms transcripts (green). (A) lms transcripts accumulate specifically in a subset of adepithelial cells located in most distal positions of the thoracic part of the wing disc (arrow). (B) In the late 3rd instar leg discs from Him-GFP larvae lms is expressed in a restricted subpopulation of adepithelial cells (arrow) located outside of the leg disc proper within the stalk region. (C) Highly restricted lms expression in leg discs at the same position as in (B) (arrow) is already detected in early 3rd instar larvae.
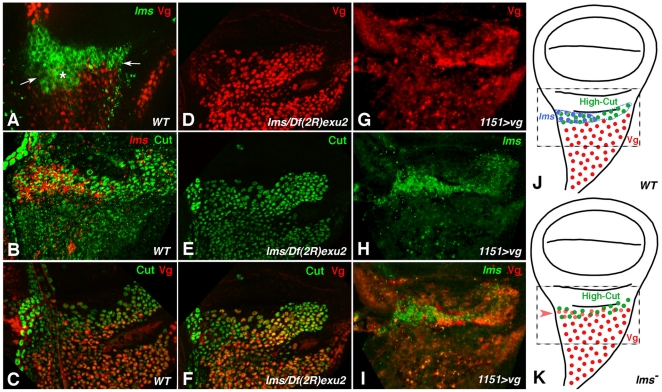
The restricted expression pattern of lms in wing discs is interesting in light of previous reports that the distal adepithelial myoblasts giving rise to direct flight muscles are marked by high levels of expression of the homeobox gene cut, whereas the remaining myoblasts giving rise to the indirect flight muscles are marked by the expression of vg and low levels of cut during late third instar [30]. Therefore we performed additional experiments to compare the spatial expression and clarify the regulatory relationships among cut, vg, and lms. In wild type wing discs, most of the lms expression occurs in distal adepithelial cells that are adjacent to the Vg-expressing cells (Fig. 7A, arrow), except for a small area of overlap (Fig. 7A, asterisk). Accordingly, lms shows extensive co-expression with high-level Cut in the anterior ∼1/2 of the distal domain, albeit with a slight extension into the low-Cut domain (Fig. 7B). Together with the published data, these observations confirm that lms expression is largely restricted to a subset of the adepithelial myoblasts giving rise to direct flight muscles (Fig. 7J).
Figure 7. Regulatory interactions between lms and vg during patterning of adepithelial cells in wing discs.
Shown are high magnification views centering on the wing hinge areas of 3rd instar wing discs (distal is up, proximal is down; anterior to the left; areas shown correspond to dashed rectangles in J, K). A-C: wild type; D-F: lmsS95/Df(2R)exu2; G-I: 1151-GAL4>vg). (A) lms mRNA expression (green) occurs in areas distally adjacent to the areas of Vg expression (red) in the adepithelial cell layer (arrows indicate border between the two domains), although there is also a small region of overlap (asterisk). (B) lms expression within the area displaying high levels of Cut protein (“high Cut domain”), which forms direct flight muscles. (C) Normal expression of Vg in presumptive indirect flight muscle myoblasts and high-level Cut expression in adjacent direct flight muscle myoblasts, respectively. (D, E, F) In lms mutant wing discs, Vg expression is expanded into the Cut domain. (G, H, I) lms mRNA expression in a largely normal pattern in wing disc with ectopic vg expression in all adepithelial cells. (J, K) Schematic drawings of Vg and Cut expression in wildtype and lms mutant disc, respectively, illustrating the expansion of Cut expression into anterior portions of the Vg domain upon loss of lms activity (area shown in panels A – I is indicated by dashed rectangle; blue dots in J represents high-level lms expression and region outlined with blue dotted line low-level lms expression area).
To test whether the expression of cut and vg is regulated by lms, we stained wing discs from lms mutant third instar larvae for Cut and Vg proteins. As shown in Fig. 7D-F, Vg expression is strongly expanded into the high-Cut domain in these discs such that almost all nuclei with high-Cut also contain Vg. Hence, lms contributes to the negative regulation of vg in the myoblasts of the presumptive direct flight muscles and high-Cut, which is unaltered in lms mutants, does not appear to be sufficient to repress vg (Fig. 7K). Next, we tested whether vg has any regulatory effects on lms expression. For this purpose, we forced expression of vg in all adepithelial cells with the 1151-Gal4 driver [25], [41]. Although we do observe a wing posture phenotype under these conditions (see also Fig. 8D), we do not detect any obvious repression of lms expression with ectopic vg (Fig. 7G-I). Thus it appears that vg is not sufficient to exert repressive effects on lms that could have fully explained the largely complementary expression of the two genes in the wing discs.
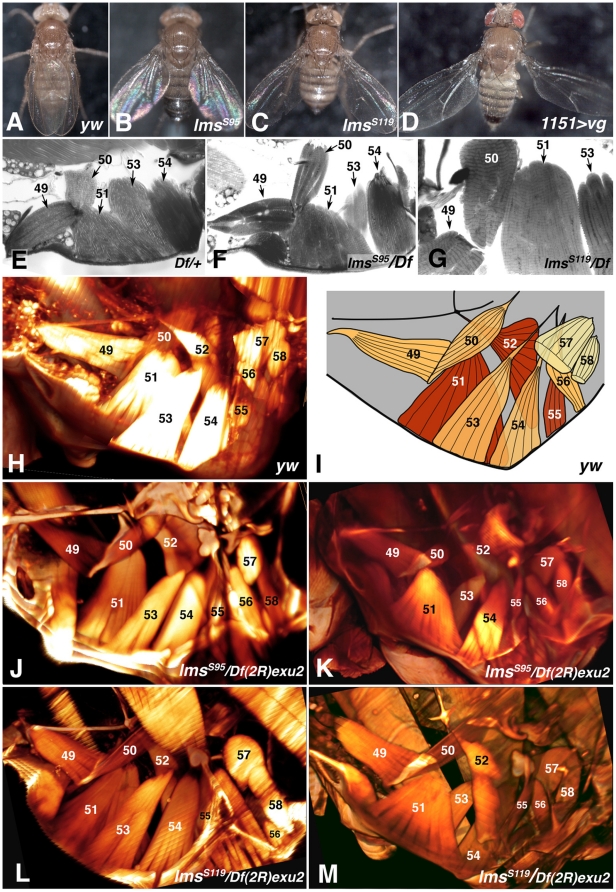
Figure 8. Wing posture phenotype and morphological analysis of direct flight musculature in adult lms mutant flies.
(A) Control fly (yw). (B) lmsS95 homozygous mutant fly. (C) Homozygous lmsS119 fly. (D) 1151-Gal4; UAS-vg fly with ectopic expression of vg in DFM myoblasts. (E) Plastic section showing DFMs from control fly (Df(2R)exu2/+). (F) Section showing DFMs from lmsS95/Df(2R)exu2 fly. (G) Section from lmsS119/Df(2R)exu2 fly at higher magnification showing the striated, non-fibrillar DFMs 49, 50, 51, 53 that appear normal. (H) and (J – M) show 3D-reconstructions obtained from stacks of images acquired by ultramicroscopy from whole mount flies. In some cases, certain areas from individual layers that covered important muscles underneath have been omitted for better clarity (see movies S2, S3, S4 with the complete reconstruction). (H) Control fly (yw) scanned from the inside. The muscles are numbered according to Miller, 1950 [46]. (I) Schematic drawing of direct flight muscle pattern as seen in (H) (maroon: outer muscle layer; orange: intermediate layer; yellow: inner layer). Black lines demark external cuticle and sclerites. (J) lmsS95/Df(2R)exu2 mutant fly scanned from the inside. (K) lmsS95/Df(2R)exu2 mutant fly scanned from the outside. (L) lmsS119/Df(2R)exu2 mutant fly scanned from the inside. (M) lmsS119/Df(2R)exu2 mutant (same fly as in G) scanned from the outside.
lms mutant flies frequently show a held-out wing phenotype, in which the wings are held at various angles (typically ∼45°) from the body axis instead of parallel to it (Fig. 8B, C, compare to A). The phenotype is seen both in flies that are homozygous for the lms null alleles and in flies that carry an lms mutation in trans to larger deficiencies at the locus, although the penetrance and expressivity can vary presumably due to genetic background effects (e.g., lmsS119/lmsS119 or lmsS119/Df(2R)BSC400 escaper flies raised at 18oC: ∼80% penetrance of wing posture defects; lmsS119/Df(2R)exu2: ∼40% penetrance). The same held-out phenotype is also seen upon RNAi knockdown of lms in adepithelial cells (1151-GAL4>lms-IR; data not shown). The flies with normal wing postures or with mildly held-out wings from the above-described genotypes are able to fly, but most individuals with more strongly held-out wings show poor flying capabilities or are unable to fly. In a flying assay with a 500 ml graduated cylinder according to Benzer [42], ∼45% of lmsS119/Df(2R)exu2 flies with held-out wings (n = 66) landed on the bottom, whereas for the lmsS119/Df(2R)exu2 flies with normal wing postures (n = 77) and yw control flies (n = 168) this number was only ∼8 and 18%, respectively (data not shown). Likewise, when lmsS119/lmsS119 and lmsS95/Df(2R)BSC400 escaper flies with held-out wings were dropped from a height of 100 cm, all of them landed within an area of ∼25 cm diameter. When these flies were kept in an open dish or on a tip, they walked and jumped when touched, but did not fly away (e.g., see Movie S1).
The observed wing posture phenotype is similar to that of flies with ectopic vg expression in adepithelial myoblasts, albeit somewhat milder (Fig. 8D). The effects of ectopic vg have been connected to disruptions of direct flight muscles [30]. Because of the similarity of these wing phenotypes, the known role of direct flight muscles in controlling the wing posture [43], and the observed expression of lms within the domain of the myoblasts that give rise to direct flight muscles, we surmised that lms has a specific function in regulating DFM development.
Initially, we examined the direct flight musculature in dissected flies and plastic sections, which did not reveal any obvious defects in the pattern and ultrastructure (Fig. 8E – G). Therefore, we decided to analyze the DFMs in fixed whole mount thoraces via ultramicroscopy and 3D reconstruction of the scanned images, which produces detailed views of the musculature and other internal structures of the fly [44], [45]. As shown in Fig. 8H, I in a view from the inside of the thorax towards the wing attachment (see Movies S2, S3, S4), this technique has allowed us to reconstruct the morphology of Drosophila DFMs with great detail comparable to scanning electron micrographs [46], [47], [48] while providing the additional advantage of 3D views. Next, we used this method to examine the direct flight musculatures in lms mutant flies that featured strong held-out-wing phenotypes. Figures 8J and L show 3D reconstructions of the direct flight muscles from lmsS95/Df(2R)exu2 and lmsS119/Df(2R)exu2 flies, respectively, viewed from the inside like the control in Fig. 8H, and panels K and J depict these muscles from the same flies as viewed from the outside. Importantly, all muscles are present with their characteristic shapes, arrangements, and connections to the proper attachment sites in these mutant flies. Although there are minor differences in muscle size (e.g., compare reduced thickness of muscles 53 from the mutants in Fig. 8J with the corresponding muscle 53 from control in Fig. 8H), given the low sample size of flies that can be processed with this method (see Materials and Methods) it is unclear whether these subtle differences are related to the absence of lms function.
Discussion
The homeobox gene lms is the first representative among its orthologs in insects and primitive chordates (Ciona and amphioxus) that has now been characterized in terms of its expression and function. Although some expression data are available for Nkx-C, its ortholog from Ciona intestinalis, the exact tissues of expression of this gene remain to be characterized (http://ghost.zool.kyoto-u.ac.jp/cgi-bin3/photoget2.cgi?citb089b03; [49]). Of note, in Drosophila the expression of lms is highly restricted and only found in specific domains of cells within the somatic mesoderm and the muscles derived from them. The lms gene is active within the somatic mesoderm during both larval and adult myogenesis, which suggested that it functions during both of these phases of muscle development.
In the embryo, lms is expressed like a typical muscle identity gene. Its expression in progenitors, founders and syncytia of the lateral muscles LT1 – LT4 is very similar to the mesodermal expression of the LIM homeobox gene apterous (ap), except that ap is activated slightly earlier in the corresponding myogenic preclusters. We have shown that ap exerts regulatory inputs towards lms, which become most apparent upon ectopic expression of ap. However in ap mutants, lms is still expressed largely normally and the same is true for the expression of ap in lms mutants. Thus, although regulatory interactions between the two genes do exist, their expression seems to be established largely independently from one another by related upstream activators. Two candidates for these include msh and lb. msh expression is significantly broader but overlaps with lms, and loss of msh function causes delayed and less robust lms expression. Conversely, lb is expressed in adjacent cells and appears to play a role in the spatial restriction of lms expression. Because single mutations for any of the tested candidate genes do not lead to a total disruption of lms expression, either these regulators act redundantly or there are additional yet unidentified regulators of lms expression that play more indispensable roles.
Loss of lms can cause the absence of individual LT muscles or in some cases morphological changes, particularly insertions into inappropriate attachment sites. The absence of an LT muscle could be due to a transformation of its identity into another, although we have not observed any clear examples of that. Alternatively, loss of lms function could lead to a failure of a muscle founder to acquire any specific identity or to progress only partially towards acquiring a normal LT muscle identity. We favor this second interpretation, which is compatible with the occasional presence of a small, amorphous syncytium at the position of a missing fiber and the observation of mis-attached and mis-shapen LT muscle fibers. Interestingly, similar phenotypes with comparable low expressivity were also described for ap mutants. To explain the low expressivity, it was proposed that additional factors can partially compensate for the loss of ap function [5]. Because the expression of lms in LT muscles and their progenitors is very similar to that of ap, Lms was a very good candidate for such a factor. However, in ap, lms double mutants the majority of LT muscles are still present as well, thus ruling out that the two genes are required for LT muscle specification in a mutually-redundant fashion. Rather, the roughly additive effects on the expressivity in the double mutants indicate that the two genes act in parallel with each other and in combination with yet other, perhaps more critical genes during the specification of LT muscle identities. These likely include Kr and msh, functional loss of which leads to a loss of over 30% and 50% of the LT muscles, respectively [6], [13], as well as yet unknown genes.
Altogether, it appears that lms (and ap) is needed for securing the robustness of the program determining LT muscle identities. Our findings reinforce the view that there is a significant degree of redundancy built into the muscle specification program in Drosophila. It is increasingly clear that the expressivities of phenotypes upon loss of function of different muscle identity genes occupy a wide range. Whereas lms and ap fall into the low end of this range, the expressivity of msh and Kr phenotypes is low for some muscle lineages and intermediate for others [13], [19]. At the high end of this spectrum are mutations for slou, col, and eve, which affect essentially all muscle lineages in which these identity genes are expressed [8], [9], [11]. In addition, it must be considered that identity genes act within a hierarchically structured network of interactions and at different steps of muscle development. Some of them (e.g., slou, eve, lb) appear to have major roles during the initial diversification of founder cells, whereas others (e.g. ap, lms) may act mainly or purely in the execution of identity programs of specified muscle precursors and the acquisition of individual muscle features such as shape, attachment, and distinct functional properties.
The presence of a wing posture phenotype in almost all lms mutant flies, albeit with variable severity, argues for a rather strict requirement for lms during adult muscle differentiation. The major domain of expression during this phase occupies the area of wing disc-associated myoblasts marked by high-cut expression that give rise to the direct flight muscles (DFMs). Interestingly, ap is also activated in these cells, but unlike in embryos, in this case significantly later than lms, namely in pupal stages. Reduction of ap activity has been shown to severely disrupt the formation of DFMs [29]. By contrast, all DFMs are formed and are arranged normally in lms null mutants, which implies that lms is not required for DFM muscle specification and morphogenesis. Instead, we presume that lms is needed in these muscles to fulfill their proper functions, which include the adjustment of wing positions and steering during flight [50]. It is conceivable that ap acts together with lms in this pathway, even though ap has additional roles in regulating the formation or survival of DFMs. Only the lms mutant flies with mild or absent held-out wings phenotypes are able to fly, but we have not examined their steering behaviour, which still may be disrupted. As shown herein, loss of lms leads to ectopic expression of vg in the presumptive DFM myoblasts. This effect could in part explain the functional defects of the resulting DFMs as GAL4/UAS-driven vg is known to interfere with normal DFM development [30]. In our hands, 1151-GAL4-driven vg caused the reported held-out wing phenotype but, as with lms mutants, analysis of these flies via ultramicroscopy did not reveal any differences in the DFM muscle pattern (data not shown). Whereas most genes with held-out or held-up wing phenotypes encode various muscle proteins [9], [51], a few others such as Dichaete and mirror are expressed in proximal areas of the disc epithelium and, when mutated, cause disruptions of wing structures in the hinge region [52], [53]. By contrast, lms mutants with held-out wings show normal morphologies of proximal wing elements (data not shown), which together with the myoblast-specific expression pattern of lms reinforces the notion of a DFM-specific role of lms.
A similar held-out wing phenotype as for lms and ectopic vg was observed for Wnt2 mutant flies, which show a loss, mis-attachment, or ectopic location of usually several of the DFMs in each fly [47]. Presumably as a result of these DFM patterning or attachment phenotypes, Wnt2 mutant flies hold out their wings more strongly as compared to lms mutants and are also unable to fly. The late expression at the epidermal wing hinge of Wnt2 and its temporal requirement, which occurs only during pupariation, rules out a role of Wnt2 in inducing lms, which is already expressed during third instar. However, it remains to be examined whether Wnt2 is needed for the maintenance of lms expression (and/or induction of ap) in the developing DFM myoblasts, which would be analogous to the known role of Wingless in the maintenance of vg in the presumptive indirect flight muscle (IFM) myoblasts [30].
Materials and Methods
Drosophila stocks and genetics
The GE11010 P-element insertion line, which contains an insertion 11nt downstream of the predicted start codon of the lms open reading frame (or 5′ to it if the first ATG of the longest cDNA available, EST RE33150, is being used), was purchased from Genexel (Korea) through a CNRS license. Df(2R)exu2 and Df(2R)BSC400 containing deletions uncovering the lms locus was obtained from the Bloomington Stock Center, as were the lines used for the excision screen. Df(3R)3/1 (deleting bap, lbl, lbe and C15) was used as a ladybird deficiency [54]. apME680-GFP and apUGO35 were a gift form J. Botas (Baylor, Houston, TX [35]). 1151-Gal4 driving expression in the adepithelial cells during larval development was a gift from K. VijayRaghavan (National Centre for Biological Sciences, Bangalore) [25], [41]. Him-GFP was a gift from J. W. Posakony (Univ. California, San Diego) [37], [40]. UAS-vg was a gift from H. Skaer (Cambridge University, UK) [30]. UAS-msh, mshΔ68 mutants, msh-lacZ, and rP298-lacZ were obtained from A. Nose (Tokyo University of Pharmacy and Life Sciences, Japan) [13], [55]. UAS-ap was a gift from J. Thomas (Salk Institute, La Jolla, CA) [5]. UAS-Kr was kindly provided by the Jäckle lab (MPI f. biophys. Chemie, Göttingen) [6]. Pan-mesodermal expression of muscle-identity genes were achieved with the 24B-Gal4 driver line [36] and the embryos were left to develop at 28°C for 12 hours before fixation. lacZ- or GFP-marked balancers were used throughout for the identification of homozygous mutant embryos.
For the generation of lmsS95 apUGO35 double mutants, 50 female flies transheterozygous for the two mutations were crossed to Sco/SM6. To identify recombined chromosomes the lines derived from individual balanced offspring were tested for lethality in trans to apUGO35 and via PCR for the lms deletion.
Generation of lms deletions
For P-excision mutagenesis, males from the GE11015 homozygous strain were crossed with virgins from a strain containing the Δ2–3 transposase on a CyO chromosome. Males carrying the P-element insertion together with the transposase were crossed with w; Sco/CyO. White eyed male progeny (either carrying Sco or CyO) were crossed individually with w; Df(2R)exu2/SM6, eve-lacZ females. For each of these single crosses, the lethality was scored and a balanced GE11015-excision/SM6, eve-lacZ or, if viable homozygous, stock was established. Breakpoints were determined molecularly by genomic PCR and sequencing.
Embryo fixation
Embryos were collected on agar in Petri dishes, dechorionated for 2.5 min in 100% bleach (DanKlorix, Colgate-Palmolive), washed with H2O for 2 min and fixed by shaking for 20–30 min in 800 µl 5X Buffer B (50 mM K-Phosphate pH6.8, 225 mM KCl, 75 mM NaCl, 65 mM MgCl2), 800 µl formaldehyde, 2.5 ml H2O and 8 ml heptane. After removing the vitelline membrane by shaking the embryos 30 sec in methanol and two washes in methanol embryos were stored at 4°C until staining.
In situ hybridization
4 µg of the RE33150 EST clone of lms, obtained from the Riken embryo library, were digested with EcoRI, gel purified with the QIAquick Gel Extraction Kit (Qiagen) and resuspended in 30 µl of DEPC-H2O. 10 µl of this preparation were used for a 2 h transcription reaction at 37°C using 3 µl of T3 RNA Polymerase in 30 µl final volume. 0.5 µl of the reaction was tested on a gel and the remaining probe was treated with 10 µl of 5X Carbonate Buffer (300 mM Na2CO3, 200 mM NaHCO3, pH 10.2) and 20 µl DEPC-H2O for 20 min at 65–70°C. 50 µl of 2X Stop Solution (0.2 M NaAc, pH 6.0) were added. The probe was precipitated after addition of 15 µl of 4 M LiCl, 2 µl of tRNA (50 mg/ml) and 300 µl of 100% EtOH, washed with 500 µl of 70% EtOH in DEPC-H2O, dissolved in 70 µl of Hybridization Solution (50% formamide, 5X SSC, 50 µg/ml heparin, 100 µg/ml tRNA, 100 µg/ml ssDNA, 0.1% Tween, pH 6.5) and used at 1∶2000 dilution for in situ hybridization.
Immunohistochemistry
Whole mount embryo in situ hybridization, double fluorescent in situ hybridization and antibody labeling were performed as described in [9] and detected with Fast Red reagent (Sigma-Aldrich) or tyramide reagents (PerkinElmer). Immuno-stainings of larval imaginal discs were performed as described in [56]. In situ hybridization on larval discs was performed according to [57]. The stainings were either scanned with a Leica confocal microscope or recorded by using a Zeiss Apotome microscope. Ultramicroscopic images of adult flies were generated according to [44], [45]. Images were assembled using Photoshop.
The following primary antibodies were used: rabbit anti-βgalactosidase (Cappel/MP Biomedicals, CA); rabbit anti-GFP (Molecular Probes, OR); rat anti-Tropomyosin (Babraham, UK), mouse anti-Cut (Developmental Studies Hybridoma Bank, DSHB), rabbit anti-Vg (gift from S Carroll; HHMI/Univ. of Wisconsin, Madison, WI), rabbit anti-myosin (D. Kiehart, Duke Univ., NC), anti-β3-Tubulin (R. Renkawitz-Pohl, Univ. Marburg), mouse anti-Lbe ([58]), rabbit anti-Kr (1∶2000; gift from P. Carrera and G. Vorbrüggen, MPI Göttingen). For fluorescent detection, FITC-, Cy3-, or Cy5-conjugated secondary antibodies were used. Secondary antibodies were obtained from Jackson Laboratories.
Ultramicroscopy
Ultramicroscopy (UM) is a microscopy technique allowing three-dimensional reconstructions of up to cm-sized specimen with micrometer resolution using a laser light sheet [44], [45]. Using this technique, we performed three-dimensional reconstructions of the inner anatomy of chemically cleared entire Drosophila flies [59]. Flies were anaesthetised by ether, fixed in 4% paraformaldehyde overnight, and dehydrated in an ascending ethanol series (50%, 70%, 96%, 2×100% for 6 h). Afterwards, they were incubated in a clearing solution consisting of 2 parts benzyl benzoate and 1 part benzylalcohol (Sigma-Aldrich, Germany) for 3 days. Imaging the flies' inner morphology was performed using autofluorescence of Drosophila muscles. Autofluorescence was excited at 488 nm laser light sheet having a beam diameter of approx. 3.2 µm at the focus. Images were recorded with a 10X objective (NA 0.30) using a CCD camera (CoolSnap K4, Roper Scientific) as described in [44]. Two individuals from each genotype were analyzed (yw, lmsS95/Df(2R)exu2, lmsS119/Df(2R)exu2) and the mutant flies utilized showed strong held-out wing phenotypes.
Supporting Information
lms[S119] homozygous flies. Non-flying behaviour of homozygous lmsS119 flies in open dish.
(1.90 MB MOV)
yw control inside. Scan of yw control fly from the inside (anterior to the right) (see also Fig. 8H).
(0.80 MB MP4)
lms[S95]/Df(2R)exu2 inside. Scan of lmsS95/Df(2R)exu2 fly from the inside (anterior to the right) (see also Fig. 8J).
(0.95 MB MP4)
lms[S119]/Df(2R)exu2 outside. Scan of lmsS119/Df(2R)exu2 fly from the outside (anterior to the left) (see also Fig. 8M).
(0.98 MB MP4)
Acknowledgments
We acknowledge Claudia Obermeier for technical support.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: National Institutes of Health (HD30832) to MF (http://www.nichd.nih.gov), Hertie-Foundation to HUD (http://www.ghst.de/en/index.php). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Bate M. The mesoderm and its derivatives. In: Bate M, Martinez-Arias A, editors. The development of Drosophila melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.; 1993. pp. 1023–1090. [Google Scholar]
- 2.Beckett K, Baylies MK. The development of the Drosophila larval body wall muscles. Int Rev Neurobiol. 2006;75:55–70. doi: 10.1016/S0074-7742(06)75003-6. [DOI] [PubMed] [Google Scholar]
- 3.Maqbool T, Jagla K. Genetic control of muscle development: learning from Drosophila. J Muscle Res Cell Motil. 2007;28:397–407. doi: 10.1007/s10974-008-9133-1. [DOI] [PubMed] [Google Scholar]
- 4.Dohrmann C, Azpiazu N, Frasch M. A new Drosophila homeobox gene is expressed in mesodermal precursor cells of distinct muscles during embryogenesis. Genes Dev. 1990;4:2098–2111. doi: 10.1101/gad.4.12a.2098. [DOI] [PubMed] [Google Scholar]
- 5.Bourgouin C, Lundgren S, Thomas J. Apterous is a Drosophila LIM domain gene required for the development of a subset of embryonic muscles. Neuron. 1992;9:549–561. doi: 10.1016/0896-6273(92)90192-g. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz-Gomez M, Romani S, Hartmann C, Jäckle H, Bate M. Specific muscle identities are regulated by Krüppel during Drosophila embryogenesis. Development. 1997;124:3407–3414. doi: 10.1242/dev.124.17.3407. [DOI] [PubMed] [Google Scholar]
- 7.Jagla T, Bellard F, Lutz Y, Dretzen G, Bellard M, et al. ladybird determines cell fate decisions during diversification of Drosophila somatic muscles. Development. 1998;125:3699–3708. doi: 10.1242/dev.125.18.3699. [DOI] [PubMed] [Google Scholar]
- 8.Crozatier M, Vincent A. Requirement for the Drosophila COE transcription factor Collier in formation of an embryonic muscle: transcriptional response to Notch signalling. Development. 1999;126:1495–1504. doi: 10.1242/dev.126.7.1495. [DOI] [PubMed] [Google Scholar]
- 9.Knirr S, Azpiazu N, Frasch M. The role of the NK-homeobox gene slouch (S59) in somatic muscle patterning. Development. 1999;126:4525–4535. doi: 10.1242/dev.126.20.4525. [DOI] [PubMed] [Google Scholar]
- 10.Balagopalan L, Keller CA, Abmayr SM. Loss-of-function mutations reveal that the Drosophila nautilus gene is not essential for embryonic myogenesis or viability. Dev Biol. 2001;231:374–382. doi: 10.1006/dbio.2001.0162. [DOI] [PubMed] [Google Scholar]
- 11.Fujioka M, Wessells RJ, Han Z, Liu J, Fitzgerald K, et al. Embryonic even skipped-dependent muscle and heart cell fates are required for normal adult activity, heart function, and lifespan. Circ Res. 2005;97:1108–1114. doi: 10.1161/01.RES.0000191546.08532.B2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Azpiazu N, Frasch M. tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 1993;7:1325–1340. doi: 10.1101/gad.7.7b.1325. [DOI] [PubMed] [Google Scholar]
- 13.Nose A, Isshiki T, Takeichi M. Regional specification of muscle progenitors in Drosophila: the role of the msh homeobox gene. Development. 1998;125:215–223. doi: 10.1242/dev.125.2.215. [DOI] [PubMed] [Google Scholar]
- 14.Clark IB, Boyd J, Hamilton G, Finnegan DJ, Jarman AP. D-six4 plays a key role in patterning cell identities deriving from the Drosophila mesoderm. Dev Biol. 2006;294:220–231. doi: 10.1016/j.ydbio.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 15.Duan H, Zhang C, Chen J, Sink H, Frei E, et al. A key role of Pox meso in somatic myogenesis of Drosophila. Development. 2007;134:3985–3997. doi: 10.1242/dev.008821. [DOI] [PubMed] [Google Scholar]
- 16.Hooper JE. Homeotic gene function in the muscles of Drosophila larvae. Embo J. 1986;5:2321–2329. doi: 10.1002/j.1460-2075.1986.tb04500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Michelson A. Muscle pattern diversification in Drosophila is determined by the autonomous function of homeotic genes in the embryonic mesoderm. Development. 1994;120:755–768. doi: 10.1242/dev.120.4.755. [DOI] [PubMed] [Google Scholar]
- 18.Enriquez J, Boukhatmi H, Dubois L, Philippakis AA, Bulyk ML, et al. Multi-step control of muscle diversity by Hox proteins in the Drosophila embryo. Development. 2010;137:457–466. doi: 10.1242/dev.045286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmann C, Landgraf M, Bate M, Jäckle H. Krüppel target gene knockout participates in the proper innervation of aspecific set of Drosophila larval muscles. EMBO J. 1997;16:5299–5309. doi: 10.1093/emboj/16.17.5299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Junion G, Bataille L, Jagla T, Da Ponte JP, Tapin R, et al. Genome-wide view of cell fate specification: ladybird acts at multiple levels during diversification of muscle and heart precursors. Genes Dev. 2007;21:3163–3180. doi: 10.1101/gad.437307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bate M, Rushton E, Currie D. Cells with persistent twist expression are the embryonic precursors of adult muscles in Drosophila. Development. 1991;113:79–89. doi: 10.1242/dev.113.1.79. [DOI] [PubMed] [Google Scholar]
- 22.Roy S, VijayRaghavan K. Muscle pattern diversification in Drosophila: the story of imaginal myogenesis. Bioessays. 1999;21:486–498. doi: 10.1002/(SICI)1521-1878(199906)21:6<486::AID-BIES5>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 23.Roy S, VijayRaghavan K. Metamorphosis and the Formation of the Adult Musculature. In: Sink H, editor. Drosophila Muscle Development. Georgetown, TX: Landes; 2005. [Google Scholar]
- 24.Fernandes J, Celniker S, Lewis E, VijayRaghavan K. Muscle development in the four-winged Drosophila and the role of the Ultrabithorax gene. Curr Biol. 1994;4:957–964. doi: 10.1016/s0960-9822(00)00219-0. [DOI] [PubMed] [Google Scholar]
- 25.Roy S, VijayRaghavan K. Homeotic genes and the regulation of myoblast migration, fusion, and fibre-specific gene expression during adult myogenesis in Drosophila. Development. 1997;124:3333–3341. doi: 10.1242/dev.124.17.3333. [DOI] [PubMed] [Google Scholar]
- 26.Rivlin PK, Gong A, Schneiderman AM, Booker R. The role of Ultrabithorax in the patterning of adult thoracic muscles in Drosophila melanogaster. Dev Genes Evol. 2001;211:55–66. doi: 10.1007/s004270000126. [DOI] [PubMed] [Google Scholar]
- 27.Maqbool T, Soler C, Jagla T, Daczewska M, Lodha N, et al. Shaping leg muscles in Drosophila: role of ladybird, a conserved regulator of appendicular myogenesis. PLoS ONE. 2006;1:e122. doi: 10.1371/journal.pone.0000122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soler C, Daczewska M, Da Ponte JP, Dastugue B, Jagla K. Coordinated development of muscles and tendons of the Drosophila leg. Development. 2004;131:6041–6051. doi: 10.1242/dev.01527. [DOI] [PubMed] [Google Scholar]
- 29.Ghazi A, Anant S, VijayRaghavan K. Apterous mediates development of direct flight muscles autonomously and indirect flight muscles through epidermal cues. Development. 2000;127:5309–5318. doi: 10.1242/dev.127.24.5309. [DOI] [PubMed] [Google Scholar]
- 30.Sudarsan V, Anant S, Guptan P, VijayRaghavan K, Skaer H. Myoblast diversification and ectodermal signaling in Drosophila. Dev Cell. 2001;1:829–839. doi: 10.1016/s1534-5807(01)00089-2. [DOI] [PubMed] [Google Scholar]
- 31.Bernard F, Lalouette A, Gullaud M, Jeantet AY, Cossard R, et al. Control of apterous by vestigial drives indirect flight muscle development in Drosophila. Dev Biol. 2003;260:391–403. doi: 10.1016/s0012-1606(03)00255-0. [DOI] [PubMed] [Google Scholar]
- 32.Bernard F, Dutriaux A, Silber J, Lalouette A. Notch pathway repression by vestigial is required to promote indirect flight muscle differentiation in Drosophila melanogaster. Dev Biol. 2006;295:164–177. doi: 10.1016/j.ydbio.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 33.Tomancak P, Beaton A, Weiszmann R, Kwan E, Shu S, et al. Systematic determination of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2002;3:research0088.0081–0088.0014. doi: 10.1186/gb-2002-3-12-research0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith ST, Jaynes JB. A conserved region of engrailed, shared among all en-, gsc-, Nk1-, Nk2- and msh-class homeoproteins, mediates active transcriptional repression in vivo. Development. 1996;122:3141–3150. doi: 10.1242/dev.122.10.3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Capovilla M, Kambris Z, Botas J. Direct regulation of the muscle-identity gene apterous by a Hox protein in the somatic mesoderm. Development. 2001;128:1221–1230. doi: 10.1242/dev.128.8.1221. [DOI] [PubMed] [Google Scholar]
- 36.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 37.Liotta D, Han J, Elgar S, Garvey C, Han Z, et al. The Him gene reveals a balance of inputs controlling muscle differentiation in Drosophila. Curr Biol. 2007;17:1409–1413. doi: 10.1016/j.cub.2007.07.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lawrence PA. Cell lineage of the thoracic muscles of Drosophila. Cell. 1982;29:493–503. doi: 10.1016/0092-8674(82)90166-0. [DOI] [PubMed] [Google Scholar]
- 39.Fernandes J, Bate M, Vijayraghavan K. Development of the indirect flight muscles of Drosophila. Development. 1991;113:67–77. doi: 10.1242/dev.113.1.67. [DOI] [PubMed] [Google Scholar]
- 40.Rebeiz M, Reeves NL, Posakony JW. SCORE: a computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Site clustering over random expectation. Proc Natl Acad Sci U S A. 2002;99:9888–9893. doi: 10.1073/pnas.152320899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roy S, VijayRaghavan K. Patterning muscles using organizers: larval muscle templates and adult myoblasts actively interact to pattern the dorsal longitudinal flight muscles of Drosophila. J Cell Biol. 1998;141:1135–1145. doi: 10.1083/jcb.141.5.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Benzer S. Genetic dissection of behavior. Sci Am. 1973;229:24–37. doi: 10.1038/scientificamerican1273-24. [DOI] [PubMed] [Google Scholar]
- 43.Dutta D, VijayRaghavan K. Metamorphosis and the Formation of the Adult Musculature. In: Sink H, editor. Drosophila Muscle Development. Georgetown, TX: Landes; 2005. [Google Scholar]
- 44.Becker K, Jährling N, Kramer ER, Schnorrer F, Dodt HU. Ultramicroscopy: 3D reconstruction of large microscopical specimens. J Biophotonics. 2008;1:36–42. doi: 10.1002/jbio.200710011. [DOI] [PubMed] [Google Scholar]
- 45.Dodt HU, Leischner U, Schierloh A, Jährling N, Mauch CP, et al. Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat Methods. 2007;4:331–336. doi: 10.1038/nmeth1036. [DOI] [PubMed] [Google Scholar]
- 46.Miller A. The internal anatomy and histology of the imago of Drosophila melanogaster. In: Demerec M, editor. Biology of Drosophila. New York: Wiley; 1950. pp. 420–480. [Google Scholar]
- 47.Kozopas KM, Nusse R. Direct flight muscles in Drosophila develop from cells with characteristics of founders and depend on DWnt-2 for their correct patterning. Dev Biol. 2002;243:312–325. doi: 10.1006/dbio.2002.0572. [DOI] [PubMed] [Google Scholar]
- 48.King DG, Tanouye MA. Anatomy of motor axons to direct flight muscles in Drosophila. J exp Biol. 1983;105:231–239. [Google Scholar]
- 49.Satou Y, Wada S, Sasakura Y, Satoh N. Regulatory genes in the ancestral chordate genomes. Dev Genes Evol. 2008;218:715–721. doi: 10.1007/s00427-008-0219-y. [DOI] [PubMed] [Google Scholar]
- 50.Miyan JA, Ewing AW. How Diptera Move Their Wings: A re-Examination of the Wing Base Articulation and Muscle Systems Concerned with Flight. Philos Trans R Soc Lond, B, Biol Sci. 1985;311:271–302. [Google Scholar]
- 51.Vigoreaux JO. Genetics of the Drosophila flight muscle myofibril: A window into the biology of complex systems. Bioessays. 2001;23:1047–1063. doi: 10.1002/bies.1150. [DOI] [PubMed] [Google Scholar]
- 52.Russell S. The Drosophila dominant wing mutation Dichaete results from ectopic expression of a Sox-domain gene. Mol Gen Genet. 2000;263:690–701. doi: 10.1007/s004380051218. [DOI] [PubMed] [Google Scholar]
- 53.Kehl BT, Cho KO, Choi KW. mirror, a Drosophila homeobox gene in the Iroquois complex, is required for sensory organ and alula formation. Development. 1998;125:1217–1227. doi: 10.1242/dev.125.7.1217. [DOI] [PubMed] [Google Scholar]
- 54.Zikova M, Da Ponte JP, Dastugue B, Jagla K. Patterning of the cardiac outflow region in Drosophila. Proc Natl Acad Sci U S A. 2003;100:12189–12194. doi: 10.1073/pnas.2133156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Isshiki T, Takeichi M, Nose A. The role of the msh homeobox gene during Drosophila neurogenesis: implication for the dorsoventral specification of the neuroectoderm. Development. 1997;124:3099–3109. doi: 10.1242/dev.124.16.3099. [DOI] [PubMed] [Google Scholar]
- 56.Müller D, Kugler SJ, Preiss A, Maier D, Nagel AC. Genetic modifier screens on Hairless gain-of-function phenotypes reveal genes involved in cell differentiation, cell growth and apoptosis in Drosophila melanogaster. Genetics. 2005;171:1137–1152. doi: 10.1534/genetics.105.044453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagaso H, Murata T, Day N, Yokoyama KK. Simultaneous detection of RNA and protein by in situ hybridization and immunological staining. J Histochem Cytochem. 2001;49:1177–1182. doi: 10.1177/002215540104900911. [DOI] [PubMed] [Google Scholar]
- 58.Jagla K, Frasch M, Jagla T, Dretzen G, Bellard F, et al. ladybird, a new component of the cardiogenic pathway in Drosophila required for diversification of heart precursors. Development. 1997;124:3471–3479. doi: 10.1242/dev.124.18.3471. [DOI] [PubMed] [Google Scholar]
- 59.Jährling N, Becker K, Schönbauer C, Schnorrer F, Dodt HU. Three-dimensional reconstruction and segmentation of intact Drosophila by ultramicroscopy. Front Syst Neurosci. 2010;4:1. doi: 10.3389/neuro.06.001.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong YF, Butts T, Holland PW. HomeoDB: a database of homeobox gene diversity. Evol Dev. 2008;10:516–518. doi: 10.1111/j.1525-142X.2008.00266.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lms[S119] homozygous flies. Non-flying behaviour of homozygous lmsS119 flies in open dish.
(1.90 MB MOV)
yw control inside. Scan of yw control fly from the inside (anterior to the right) (see also Fig. 8H).
(0.80 MB MP4)
lms[S95]/Df(2R)exu2 inside. Scan of lmsS95/Df(2R)exu2 fly from the inside (anterior to the right) (see also Fig. 8J).
(0.95 MB MP4)
lms[S119]/Df(2R)exu2 outside. Scan of lmsS119/Df(2R)exu2 fly from the outside (anterior to the left) (see also Fig. 8M).
(0.98 MB MP4)