Abstract
Background
Malaria, an Anopheles-borne parasitic disease, remains a major global health problem causing illness and death that disproportionately affects developing countries. Despite the incidence of malaria, which remains one of the most severe infections of human populations, there is no licensed vaccine against this life-threatening disease. In this context, we decided to explore the expression of Plasmodium vaccine antigens fused to the granule bound starch synthase (GBSS), the major protein associated to the starch matrix in all starch-accumulating plants and algae such as Chlamydomonas reinhardtii.
Methods and Findings
We describe the development of genetically engineered starch granules containing plasmodial vaccine candidate antigens produced in the unicellular green algae Chlamydomonas reinhardtii. We show that the C-terminal domains of proteins from the rodent Plasmodium species, Plasmodium berghei Apical Major Antigen AMA1, or Major Surface Protein MSP1 fused to the algal granule bound starch synthase (GBSS) are efficiently expressed and bound to the polysaccharide matrix. Mice were either immunized intraperitoneally with the engineered starch particles and Freund adjuvant, or fed with the engineered particles co-delivered with the mucosal adjuvant, and challenged intraperitoneally with a lethal inoculum of P. Berghei. Both experimental strategies led to a significantly reduced parasitemia with an extension of life span including complete cure for intraperitoneal delivery as assessed by negative blood thin smears. In the case of the starch bound P. falciparum GBSS-MSP1 fusion protein, the immune sera or purified immunoglobulin G of mice immunized with the corresponding starch strongly inhibited in vitro the intra-erythrocytic asexual development of the most human deadly plasmodial species.
Conclusion
This novel system paves the way for the production of clinically relevant plasmodial antigens as algal starch-based particles designated herein as amylosomes, demonstrating that efficient production of edible vaccines can be genetically produced in Chlamydomonas.
Introduction
Malaria is a mosquito-borne disease and remains a major global health problem causing illness and death that disproportionately affects developing countries. The worldwide incidence of malaria is estimated by the Word Health Organization to be approximately 300 to 500 million clinical cases annually, with one million deaths, the majority of which are young children [1]–[4]. Of particular concern, the emergence of insecticide-resistant mosquito vectors and multi-drug resistant parasites has contributed to resurgences of the disease. Therefore, malaria control is a continuous battle that requires long-term sustainability and commitment. The development of a vaccine that reduces morbidity and mortality would be a valuable new tool in the fight against malaria. Plasmodium falciparum causes the most severe form of the disease [5], [6]. Infection begins when malaria sporozoites are injected by mosquito into the host and within minutes parasites invade hepatocytes, where they multiply and differentiate into the next stage. The emerging merozoites invade red blood cells leading to clinical illness [7]. The most advanced vaccine candidate, designated RTS,S/AS02A, is based on the major sporozoite surface antigen. However, this candidate vaccine, currently in Phase 3 clinical trials, has shown only 30–65% efficiency in field studies [8] and a vaccine with higher levels of protection is still sought. Over time, people living in malaria-endemic areas develop immunity to clinical disease caused by P. falciparum and IgG from immune adults has been shown to reduce parasite density and clinical symptoms when administered to children with clinical malaria [9]–[11]. Thus, proteins expressed during the blood-stage of the life cycle are good candidates for inclusion in a vaccine [12], [13], as a blood-stage vaccine would reduce or prevent severe illness and complications of the disease.
In this context, we decided to explore the expression of Plasmodium vaccine antigens fused to the granule bound starch synthase (GBSS), the major protein associated to the starch matrix in all starch accumulating plants and algae [14], [15]. Starch-bound proteins are known to remain stable for years within the polysaccharide matrix purified from plants and algae. Starch can be easily purified from plants and algae by straightforward sedimentation procedures that in some systems do not even require centrifugation. In addition, cereal starch, with its role in human and animal diets, represents an approved source for the production of glucose for injection in humans. As a first approach to the use of recombinant polysaccharide particles for vaccine production, we focussed our efforts on the production of transgenic starch from chloroplasts of the green algae Chlamydomonas reinhardtii. We chose this system because of the ease and unparalleled speed with which constructs can be introduced and proteins expressed and correctly targeted to the chloroplast and polysaccharide granule. In addition, expression of recombinant vaccine antigens into starch granules localized into the Chlamydomonas chloroplasts would avoid protein N-glycosylation, a post-translational modification that seems to be generally absent in Plasmodium falciparum. Moreover, starch metabolism has been investigated in great detail in this system through genetic dissection of mutants allowing optimization of starch granule protein content and polysaccharide structure [16]–[18]. To evaluate this novel system, we have chosen two well-studied Plasmodium vaccine candidates, MSP1 [19], [20] and AMA1 [21], [22], which are thought to be involved in invasion of human red blood cells. The biosynthesis, purification, characterization, and immunologic properties of starch-stored clinically relevant antigens produced in Chlamydomonas reinhardtii chloroplast are described.
Results and Discussion
Genetic engineering of Chlamydomonas vectors expressing transgenic starch bound plasmodial antigens
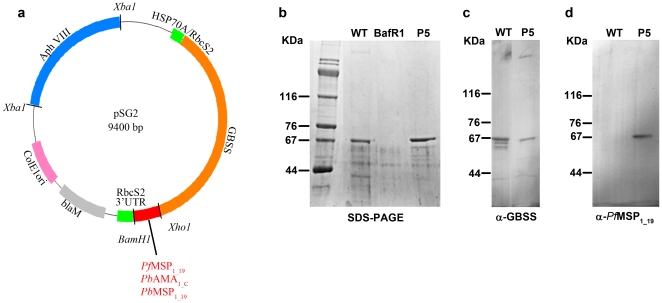
We constructed a Chlamydomonas expression vector containing the gene encoding a GBSS protein carrying a deletion that yielded a product truncated for 130 amino acids at the C-terminus (Figure 1a and Figure S1 online). We have previously shown that the absence of this C-terminal tail does not impair stability and targeting into the chloroplast [15]. A synthetic gene that encoded the 19 kDa C-terminal peptide of P. falciparum MSP1 (PfMSP1-19) was designed taking into account the GC-rich codon bias of Chlamydomonas (Figure S2). This synthetic gene was fused to the truncated GBSS gene followed by the paromomycin resistance gene for selection of algal transformants (Figure 1a). The expression of GBSS-PfMSP1-19 fusion protein is under the control of a strong chimeric rubisco RBCS2 and HSP70A promoter (Figure 1a). The Chlamydomonas BafR1 mutant strain with a complete gene deletion at the STA2 locus that encodes GBSS [18] was transformed with the GBSS-PfMSP1-19 expression vector. The transformants developed normally during vegetative growth and were indistinguishable from wild type algae. Starch granules from Chlamydomonas transformants were purified using French press disruption followed by sedimentation and Percoll gradient centrifugation. The starch bound proteins were analyzed on SDS-PAGE and Coomassie blue staining. Out of twenty clones analyzed in a pilot experiment, three Chlamydomonas transformants strongly expressed the GBSS-PfMSP1-19 fusion, as determined by their expected electrophoretic mobility. One positive Chlamydomonas transformant, designated P5, showed levels of starch-associated GBSS-PfMSP1-19 protein that are slightly higher than those of the standard GBSS of wild type reference algae (Figure 1b , lanes P5 and WT). This increase of GBSS-PfMSP1-19 in transgenic algae compared to the wild type GBSS is probably due to both the chromosomal insertion site insertion and the strength of the chimeric HSP70A/RbcS2 promoter used in this context. In addition, the promoter of the construct might not respond in an identical fashion to circadian clock control, which is known to operate on the wild-type GBSS gene [23]. Western blots using rabbit-specific polyclonal antibodies confirmed that the fusion protein of P5 transgenic algae contains the Chlamydomonas starch bound GBSS fused to the P. falciparum MSP1-19 antigen (Figure 1c and 1d).
Figure 1. Anti-malaria vaccine strategy and antigen expression in C. reinhardtii.
(a) Map of the vector used for C. reinhardtii transformation and expression. The plasmid of 9.4 kb, designated pSG2, carries the genomic region of 3 kb containing all introns and exons necessary for coding the C. reinhardtii granule bound starch synthase (GBSS). The XhoI and BamHI restriction sites at the end of the truncated GBSS gene have been utilized for cloning in-frame cDNAs encoding the C-terminal domain of P. berghei major surface protein 1 (MSP1-19); P. berghei apical major antigen (AMA1-C) and P. falciparum MSP1-19. Expression of Chlamydomonas GBSS-Plasmodium fusion protein is driven by a strong chimeric Rubisco RBCS2 and HSP70A promoter. (b) High-level of GBSS-P. falciparum MSP1-19 protein expression in one representative Chlamydomonas transformant (P5) was obtained by co-transformation of the BafR1 mutant strain lacking the GBSS gene. Samples representing 1 mg of purified starch after French press disruption and Percoll-gradient centrifugation were resuspended in SDS-βmercaptoethanol loading buffer, separated by SDS-PAGE and stained by Coomassie blue. Note that minor protein bands accidently trapped in the polysaccharide matrix are common to wild type WT, P5 and mutant BafR1 algae lacking the GBSS gene. The starch protein extracts from WT and P5 algae were also blotted to nitrocellulose and incubated with rabbit polyclonal antibodies specific to C. reinhardtii GBBS (c), or rabbit anti-P. falciparum MSP1-19 (d).The molecular weights of protein markers are given in kDa.
Demonstration that genetically engineered starch particles stored malaria antigens inside the chloroplast of Chlamydomonas
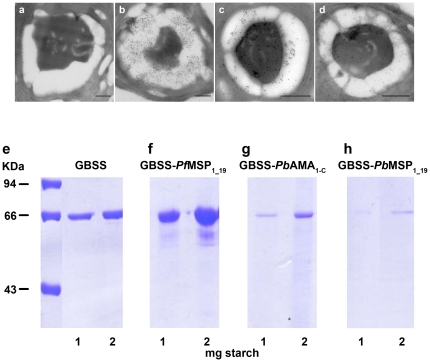
While we explored the expression of P. falciparum MSP1-19 in Chlamydomonas, we also extended our system to production of other malaria candidate vaccine antigens. AMA1 and MSP1 C-terminal domains derived from P. berghei, designated GBSS-PbAMA1-C and GBSS-PbMSP1-19 (Figure S2), were investigated in detail, as the murine malaria parasite is frequently used for animal vaccine formulation and testing. Chlamydomonas transformed with these constructs were screened by visualising the starch bound proteins among a sample of 20 to 200 transformants. The presence of transgenic starch containing GBSS-PbAMA1-C and GBSS-PbMSP1-19 was demonstrated by ultrastructural-immunogold labelling using two positive algal clones and specific polyclonal antibodies (Figure 2c and 2d). As expected, there was no gold-staining observed by electron microscopy on the starch of wild type algae (Figure 2a), while strongly positive starch granules containing P. falciparum GBSS-PfMSP1-19 were detected in transgenic algae (Figure 2b). The data suggest a stronger accumulation of P. falciparum GBSS-PfMSP1-19 protein in the transgenic algae (Figure 2b) compared to P. berghei GBSS-PbAMA 1-C and GBSS-PbMSP1-19 fusion proteins (Figure 2c and 2d). The differences in levels of P. berghei GBSS-PbAMA 1-C and GBSS-PbMSP1-19 fusion proteins relative to that of P. falciparum GBSS-PfMSP1-19 are likely due mainly to the influence of random site integration events in the chromosomes of transgenic Chlamydomonas. Hence, all GBSS-Chlamydomonas transformants expressed the starch-associated GBSS-parasite antigens in transgenic algae chloroplasts, as illustrated by full-ultrastructural section (Figure S3). Generally, a significant accumulation of GBSS-PfMSP 1-19, GBSS-PbAMA1-C and GBSS-PbMSP1-19 was detected within the starch sheath around the single chloroplast pyrenoid (P) (Figure 2b-d and Figure S3), which defines a protein agglomerate of mostly Rubisco protein. However, gold particles were also observed over the pyrenoid structure itself. The immunogold signal over the pyrenoid was not detected in untransformed controls (Figure 2a), suggesting the presence of small fragments of transgenic starch granules surrounding the pyrenoid. The total recombinant GBSS-parasite fusion protein content was determined in all transgenic transformants after starch purification. The highest concentrations were confirmed in starch-associated GBSS-PfMSP1-19 (Figure 2f), demonstrating the presence of higher levels of starch-bound protein in the strains expressing GBSS-PfMSP1-19 in comparison to those expressing wild type GBSS alone (Figure 2e), GBSS-PbAMA1-C (Figure 2g), or GBSS-PbMSP1_19 (Figure 2h). These results indicate that levels of starch associated GBSS-parasite fusion protein estimated at about 0.2 to 1.0 µg of protein per 1.0 mg of purified starch can be obtained, validating the usefulness of C. reinhardtii as an expression platform for complex recombinant parasite proteins within starch granules.
Figure 2. Malarial antigen accumulation in starch of transgenic C. reinhardtii.
Pre-immune rabbit serum used as control shows no gold particles either on starch grains or over the pyrenoid matrix (a). Transverse sections of transgenic algae visualized by electron microscopy and immunogold labelling with rabbit polyclonal antibodies specific to P. falciparum MSP1-19 (b), P. berghei AMA1-C (c) and P. berghei MSP1-19 (d). Note the presence of positive gold particles (black dots) on the starch grains (white) surrounding the pyrenoid matrix in the middle, which also contained a significant amount of GBSS-parasite fusion protein. The bar represents 500 nm. The total starch-bound protein content of purified granules containing wild type GBSS (e) was compared to those of GBSS-PfMSP1-19 (f), GBSS-PbAMA1-C (g) and GBSS-Pf MSP1-16 (h). One and two milligrams of starch purified from transformants and wild type algae were analyzed by SDS-PAGE and stained by Coomassie blue. The molecular weights of three protein markers are given in kDa.
Transgenic starch-bound antigens elicit immune responses and protection against P. berghei challenge
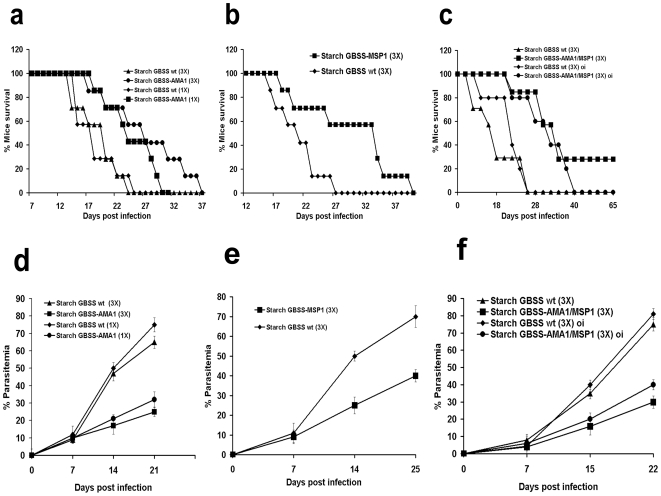
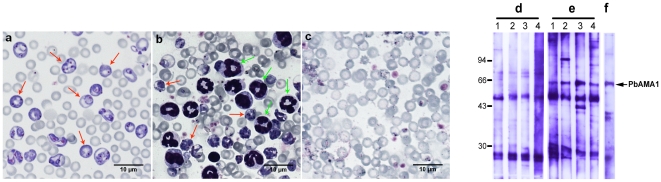
To investigate whether the levels of transgenic starch-parasite antigens achieved were sufficient to elicit protective immune responses, we tested mice immunized with purified starch containing GBSS-PbAMA1-C, or GBSS-PbMSP1-19. After immunization, mice were challenged with lethal doses (104 infected red blood cells) of the highly virulent P. berghei ANKA strain. A single dose of intraperitoneal immunization with starch containing GBSS-PfAMA1-C showed a significant and reproducible delay of mortality in P. berghei-challenged mice, compared to mice immunized with wild type starch GBSS alone (Figure 3a). When the animals were immunized with three doses of starch containing GBSS-PbAMA1-C prior to P. berghei challenge, a longer delay in mortality was observed (Figure 3a). Similarly, mice immunized with three doses of starch containing GBSS-PbMSP1-19 displayed significant protection and delay in mortality after parasite challenge (Figure 3b). Consistent with the delay in mortality, progression in the proportion of red blood cells infected (% parasitemia) in immunized and challenged mice was slower in comparison to control animals (Figure 3d and 3e). Moreover, we showed that the sera from immunized mice contain specific immunoglobulins (IgG) generated against the native P. berghei AMA1 (PbAMA1) using Western blot analyses (Figure 4, panel e). When mice were simultaneously immunized with both starch bound GBSS-PbAMA1-C and GBSS-PbMSP1-19, a delay in mice mortality persisted for a longer period of time (Figure 3c) and reduction in parasitemia (Figure 3f) was also observed. Interestingly, 25–30% of mice challenged with P. berghei survived (Figure 3c) after two months post-challenge even though few infected red blood cells can be visualized (Figure 4, panel c compare to panels a and b). We confirmed that the remaining infected red blood cells were not viable parasites because they were not able to develop any disease symptoms and intra-erythrocytic development in naive mice. At this time-point, the duration of the induced protection was also investigated by re-challenging vaccinated mice with lethal doses of P. berghei. Vaccinated mice displayed sterile protection, since no infected red blood cells or any mortality was observed. There was also a significant delay in mortality when mice were fed purified starch containing both GBSS-PbAMA1-C and GBSS-PbMSP1-19 (Figure 3c).
Figure 3. Starch-bound antigens elicit protection against P. berghei challenge.
All mice survival experiments were performed using a group of 10 Balb/c mice. (a) Life expectancy of a group of mice vaccinated after a single, or three doses of starch containing GBSS-PbAMA1-C. Each dose contains 10 mg of purified starch derived GBSS-PbAMA1-C and Freund's adjuvant. As a negative control, a group of mice was vaccinated with a single, or three doses of wild type (WT) starch containing GBSS alone. After immunization, mice were challenged with 104 (lethal dose) of P. berghei ANKA strain. Four independent experiments have been performed (n = 4) and P<0.001. (b) Life expectancy of a group of mice immunized by three doses of starch containing GBSS-PbMSP1-19. Each dose contains 10 mg of purified starch derived GBSS-PbMSP1-19 and Freund's adjuvant. As a negative control, a group of mice was vaccinated with three doses of wild type (WT) starch containing GBSS alone. After immunization, the vaccinated mice were challenged with P. berghei and analyzed as above. Three independent experiments have been performed (n = 3) and P<0.05. (c) Life expectancy of a group of mice vaccinated by three doses of starch containing a mixture of 5 mg of GBSS-PbAMA1-C and 5 mg of GBSS-PbMSP1-19 with Freund's adjuvant. As a negative control, a group of mice was also vaccinated with three doses of wild type (WT) starch containing GBSS alone. After vaccination, mice were challenged with P. berghei and analyzed as above. Three independent experiments have been performed (n = 3) and P<0.001. A group of mice were also fed three times with both starches GBSS-PbAMA1-C and GBSS-PbMSP1-19 mixed with the B-subunit enterotoxin mucosal adjuvant. As a negative control, a group of mice was fed with wild type (WT) starch containing GBSS alone. After oral immunization, mice were challenged with 104 (lethal dose) of P. berghei and analyzed as above. n = 3 and P<0.001. For all experiments, life expectancy and parasitemia were monitored daily. The panels d, e and f represent parasitemia profiles corresponding to mice vaccinated and challenged in experiments shown in panels a, b and c, respectively. Data represent mean values +/− s.d. and are derived from at least three or four independent experiments with similar results (P<0.001).
Figure 4. Starch-bound antigens elicit immune responses and reduce parasitemia after P. berghei challenge.
The Giemsa stained thin smears of red blood cells isolated from mice vaccinated with starch containing wild type GBSS (negative controls), challenged with P. berghei ANKA and analyzed after 3 weeks post infection. The red arrows indicate that the mice were highly infected with P. berghei (a). The Giemsa stained thin smears of red blood cells isolated from mice vaccinated with starch containing both GBSS-PbAMA1-C and GBSS-PbMSP1-19, challenged with P. berghei ANKA and analyzed after 3 weeks post infection (b). The Giemsa stained thin smear of red blood cells isolated from mice vaccinated with starch containing both GBSS-PbAMA1-C and GBSS-PbMSP1-19, challenged with P. berghei ANKA and analyzed after 6 weeks post-infection (c). Note the presence of numerous leukocytes (probably neutrophils, green arrows) and fewer infected red blood cells (red arrows) in the vaccinated mice after 3 weeks post-infection (b). Both leukocytes and P. berghei infected red blood cells were not detected in mice, which survived after 6 weeks post-infection (c). Western blots of total extracts of antigens prepared from red blood cells infected by P. berghei. The immunoblots were incubated with the immune sera isolated from 4 mice immunized with starch containing wild type GBSS (panel d, lanes 1-4). Blots probed with immune sera of 4 mice immunized with starch containing GBSS-PbAMA1-C (panel e, lanes 1–4). Lane f corresponds to the blots incubated with the positive rabbit polyclonal antibodies specific to P. berghei AMA1 produced in E. coli.
Immune sera and purified IgG specific to starch bound antigen block red blood cell entry by P. falciparum
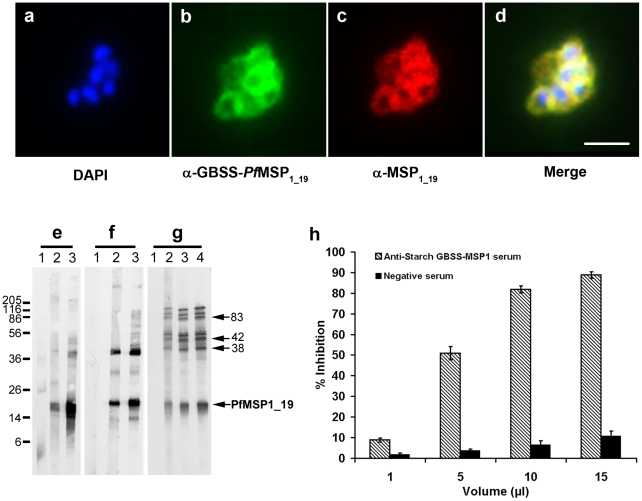
Humans are the natural host for P. falciparum and the paucity of animal models for infections with this Plasmodium species makes the discovery of vaccine candidates challenging. PfMSP1 is a leading anti-blood stage malaria vaccine candidate [24], [25] that undergoes proteolytic processing during merozoite maturation resulting in four major fragments of 83, 30, 38 and 42 kDa [26]. Before erythrocyte entry, the 42-kDa fragment undergoes a secondary proteolytic cleavage, leaving the C-terminal 19-kDa fragment (MSP1-19) associated with the merozoite [24]–[26]. Antibodies that are specific for MSP1-19 in naturally exposed individuals have potent inhibitory activities against P. falciparum growth in vitro [24]. To explore whether antibodies elicited by starch-associated antigens could inhibit in vitro growth of P. falciparum, GBSS-PbMSP1-19 starch was used to immunize a group of ten mice. Indirect immunofluorescence assays were used to demonstrate that antibodies, which were raised against purified starch containing GBSS-PfMSP1-19 recognized specifically P. falciparum merozoites within mature schizonts, as shown by co-localization with the signal of native MSP1 detected with rabbit specific antibodies (Figure 5a-d). In addition, the anti-GBSS-PfMSP1-19 antibodies recognized all PfMSP1 processed products including the major 19-kDa merozoite surface antigen (Figure 5e-5g). Importantly, anti-GBSS-PfMSP1-19 specific sera displayed very strong inhibition (circa 95%) of red blood cell invasion by P. falciparum (Figure 5h). A dose-dependent inhibition of red blood cell invasion by P. falciparum was also seen with IgG purified from sera raised against starch-bound GBSS-PfMSP1-19 (Figure S4). Our data indicate that 50% of growth inhibition of P. falciparum HB3 strain can be achieved with approximately 7+/−0.5 µg/ml (P<0.005) of purified IgG while a weaker inhibition was observed with the 3D7 strain. These results demonstrate that the biological activities of the antibodies produced against starch associated GBSS-PfMSP1-19 can be obtained in a parasite strain-specific manner [27].
Figure 5. Inhibition of red blood cell entry by P. falciparum.
Immunofluorescence assay (IFA) of erythrocyte-containing P. falciparum schizont with merozoites incubated with a pool of immune sera generated against purified starch GBSS-PfMSP1-19 (b). The nuclei of merozoites were stained by DAPI (a). The same schizont was also stained with rabbit polyclonal antibodies specific to native MSP1-19 (c). Note the perfect overlap of fluorescence signals (d, yellow signal), which corresponds to merged pictures in panels a, b and c) of anti-starch GBSS-PfMSP1-19 (b, green signal) and that of the positive control sera (red, c). The bar represents 5 µm. Western blots of total extract antigens from ring (e), trophozoite (f) and schizont (g) obtained after synchronization of P. falciparum culture with sorbitol and separated by polyacrylamide gel, transferred to nitrocellulose and probed with antibodies. Lane 1, a pool of non immune serum used as negative control. Lanes 2 and 3, a pool of immune sera from two independent experiments in which a group of mice were immunized with starch GBSS-PfMSP1-19. Lane 4 in panel g is from a third independent experiment. The proteolytically-processed fragments of 83, 42, 38 and 19 kDa derived from the 185–205 kDa PfMSP1 were shown by arrows. (h) Comparison of in vitro inhibitory effect of immune sera obtained after immunization with starch containing GBSS-PfMSP1-19 with that of immune sera generated against starch containing wild type GBSS only using the deadly human P. falciparum HB3 strain. Histograms represent mean values +/− s.d. of separate experiments (n = 3). Results were confirmed in two additional independent experiments (P<0.001).
Conclusion
We have engineered high levels of starch-bound GBSS fused to three distinct malaria vaccine candidates using the unicellular green algae C. reinhardtii. Our animal vaccine studies suggest that the levels of transgenic starch antigens that accumulate in the chloroplast are sufficient to confer substantial protection against lethal P. berghei in mice. Additionally, the functional activity of antibodies elicited to P. falciparum was demonstrated by inhibition of parasite growth in vitro and we ascribed this to blockade of erythrocyte invasion. C. reinhardtii thus provides an ideal expression system for the production of recombinant antigens associated with the starch polysaccharide matrix. In addition, algae can be grown on scales ranging from a few millilitres to 500,000 liters. Starch metabolism has been demonstrated to be identical in C. reinhardtii to that found in vascular plants so that results obtained are likely to be immediately transposable to crop plants including cereals and potatoes [28], [29]. Aside from the tremendous advantage of producing clinically relevant antigens in starch, transgenic algae can also be generated quickly, requiring only a few weeks from the generation of initial transformants to large scale vaccine production. These attributes, and the fact that unicellular green algae fall into the GRAS (Generally Regarded As Safe) category, make C. reinhardtii a particularly attractive alternative for the expression of recombinant antigens. Moreover, starch is easy to purify and represents a protective environment for bound proteins because GBSS is known to be exceptionally stable with no detectable loss of activity even after years of storage. In a first approach to genetically engineering starch particles designated amylosomes, we have produced recombinant anti-malaria vaccines in starch purified from C. reinhardtii. This system should also be amenable to the production of virtually any recombinant antigens including vaccine candidates of viruses, bacteria and other protozoan parasites.
Materials and Methods
Plasmid construction and Chlamydomonas transformation
The pKB101 vector used to transform Chlamydomonas for expression of Plasmodium antigens in the chloroplast-starch compartment is derived from the previously described pMS188 plasmid [30]. The bleomycin resistance gene flanked by MscI and BamHI restriction sites, which is under the control of chimeric RBCS2 and HSP70A promoters was replaced by a PCR product corresponding to a truncated version of the genomic sequence (STA2) [15], encoding the GBSS protein lacking the 130 C-terminal amino-acids. The 5′ end of this PCR fragment (3 kb) was cloned blunted while the 3′ end was modified by inserting into the primer both XhoI and BamHI restriction sites. These sites were used to clone in-frame all Plasmodium synthetic cDNAs designed based on the codon bias of C. reinhardtii. The AphVIII gene conferring paromomycin resistance was introduced for efficient selection of nuclear Chlamydomonas transformants. The AphVIII gene was obtained by PCR amplification from the pSL18 plasmid [31]. The different expression vectors used in this study were introduced by nuclear transformation using the glass beads method [32] into the cell wall-less GBSS mutant strain of Chlamydomonas BafR1 [18]. The transformants were selected on TAP medium plates [33] supplemented with paromomycin at 10 µg.mL−1. The strains expressing the chimeric proteins were identified by analyzing the pattern of proteins bound to the starch granule by SDS-PAGE and Coomassie blue staining.
Electron microscopy, immunofluorescence assays and immunoblot analyses
For immuno-electron microscopy, the pellets of transformed and wild type Chlamydomonas were fixed overnight at 4°C in 8% paraformaldehyde in PBS buffer, thoroughly washed in the same buffer and infused in 2.3 M sucrose containing 20% polyvinyl pyrrolidone 10000 in 0.1 M phosphate buffer. The pellets were mounted on ultracryotome supports and rapidly frozen in liquid nitrogen. Ultrathin sections of about 90 nm were obtained and the grids were incubated with rabbit polyclonal antibodies specific to P. falciparum MSP1, P. berghei MSP1 (generously supplied by Dr Tony Holder, Mill Hill, UK) or P. berghei AMA1 (provided by Dr. Christopher Adda, La Trobe University, Australia). After washing, the sections were incubated at room temperature for 30 min in the corresponding secondary gold conjugates (Jackson ImmunoResearch Laboratories Inc.). After staining with 0.5% uranyl acetate in 1.5% methyl cellulose, the sections were observed on a Hitachi H600 transmission electron microscope.
For immuno-fluorescence assays (IFA), intracellular parasites were permeabilized with 0.1% Triton X-100 in PBS containing 0.1% glycine (PBS-T) for 10 minutes at room temperature. Samples were blocked with 3% BSA in PBS-T buffer and mice immune sera diluted at 1∶1000 were added on parasites in the same buffer for one hour at 37°C. Rabbit secondary antibody coupled to Alexa-488 nm (Molecular Probes) diluted at 1∶1000 was added in addition to DAPI for nucleus staining. For co-localization assay, the rabbit anti-MPS1 serum raised against bacterial expressed recombinant PfMSP1 protein and the goat secondary antibody coupled to Alexa 500 nm were used at the same dilution. Fluorescence was visualized with a ZEISS Axiophot microscope.
For immunoblot analyses, total protein extracts from the synchronized P. falciparum HB3 strain corresponding to ring, trophozoite or schizont stage, respectively were used. The asynchronous P. berghei parasitized red blood cells isolated from mice or purified starch containing GBSS-P. berghei antigens were also analyzed. The samples were boiled in Laemmli's buffer, separated by SDS-PAGE and transferred to Hybond ECL nitrocellulose (Amersham).
In vitro inhibition of red blood cell entry by P. falciparum in the presence of immune sera and purified IgG
The P. falciparum HB3 or 3D7 strain was grown in O+ red blood cells at 6% hematocrit in RPMI 1640 medium supplemented with 10% of AB+ human serum. Cultures were maintained at 37°C and in 90% N2, 5% CO2, 5% O2 atmosphere. Synchronized parasites were obtained by two sorbitol treatments at a 32-hour interval. Synchronized infected red bloods containing schizonts with 1% parasitemia at 2% hematocrit using fresh red blood cells were cultured in complete RPMI medium and invasion assays were performed as previously described [34]. For standardized growth inhibition assays (GIA) validated in the Laboratory of Malaria and Vector Research (NIAID/NIH, USA), IgGs from mice immunized with starch containing GBSS-PfMSP1-19 were purified and tested as previously described [22].
Mouse immunization with transgenic starch and P. berghei challenge
Female BALB/c of 6 weeks of age were purchased and kept in the Pasteur Institute facility. All animals were fed with regular diet and all procedures were in accordance with national regulations on animal experimentation and welfare authorized by the French Ministry of Agriculture and Vetenary committee (N° 59-009145). The Pasteur Institute of Lille and the CNRS review board or ethics committee specifically approved this study. A group of 10 mice were intraperitoneally immunized with complete Freund's adjuvant and three weeks later were challenged twice with incomplete adjuvant in two-week intervals using 10 mg (per mouse) of starch containing GBSS-PbAMA1-C and/or GBSS-PbMSP1-19 or fed three times with the same material in the presence of B-subunit of heat-labile enterotoxin (LTB) from E. coli (Sigma). In some experiments, a single dose of vaccination was performed. Starch containing wild type GBSS alone (10 mg per mouse) was used to immunize intraperitoneally or orally 10 mice for negative vaccination controls. The vaccinated mice were challenged with 10,000 red blood cells parasitized by P. berghei. Parasite growth in vaccinated mice was monitored by Giemsa-stained thin blood smears, and survival of mice was assessed daily.
Statistical analysis
Statistical differences between groups of mice used in this study were evaluated by the Student's t-test. The Mann-Whitney test was used for morphometric Giemsa-staining data and the Log Rank test for survival curves.
Supporting Information
Nucleotide sequences of C. reinhardtii plasmid. The plasmid pKB101 was used to clone P. falciparum and P. berghei genes. This C. reinhardtii expression vector contains the chimeric RBSC2-HSP70A promoter (nucleotide 796 to 1040), GBSS genomic DNA sequence deleted at the 3' end for the 390 last nucleotides (nt 1048 to 4104), blaM Beta-lactamase also named ampicillin resistance cassette (nt 5137 to 5995), ColE1 ori (nt 6155 to 6748), AphVIII or paromomycin resistance cassette (7234-9259).
(TIF)
Parasite nucleotide sequences used for cloning and expression of P. falciparum MSP1-19, P. berghei MSP1-19 and P. berghei AMA1-C. The nucleotide sequences of Plasmodium genes were designed according to the codon bias of C. reinhardtii.
(TIF)
Longitudinal section of C. reinhardtii cell was visualized by electron microscopy. The section of C. reinhardtii cell was probed with polyclonal antibodies specific to P. berghei AMA1. The algae cell has been transformed by a construct expressing P. berghei AMA1-C antigen in the starch localized in the chloroplast. S represents starch grains surrounding the pyrenoid (P) matrix. The gold particles bind to specific anti-AMA1 antibodies present on both starch and the pyrenoid matrix. N, nucleus; C, chloroplast; M, mitochondrion. The bar = 500 nm.
(TIF)
P. falciprum strain-specific growth inhibitory effects in the presence of purified IgGs. A pool of immune sera of mice vaccinated with starch containing GBSS-PfMSP1-19 was used to purify IgGs. The inhibition of red blood cell invasion by either P. falciparum HB3 or 3D7 strain was tested in the presence of purified immune IgGs. Data represent mean values +/- s.d. and are from at least three independent experiments with two different pools of immune sera (P<0.005).
(TIF)
Acknowledgments
The authors would like to thank Drs Gordon Langsley and Robert Walker for critical reading the manuscript. We gratefully acknowledge the technical assistance of M. Mortuaire, E. Dewailly and T. Duchêne.
Footnotes
Competing Interests: The authors declare no competing financial interests. The CNRS and University of Science and Technology of Lille have filed and registered European and US patents on behalf of inventors D.D., C.D., S.B.G. and S.T. on the use of amylosome to produce parasite vaccine antigens. This does not alter our adherence to all the PLoS ONE policies on sharing data and materials, as detailed online in our guide for authors.
Funding: This research was funded by the Centre National de la Recherche Scientifique (CNRS), Institut Médicale de la Recherche Scientifique (INSERM) and the Institut Pasteur de Lille (IPL). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. WHO, World malaria report, 2008. Available: http://apps.who.int/malaria/wnr2008/malaria2008.pdf.
- 2.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guerra CA, Gikandi PW, Tatem AJ, Noor AM, Smith DL, et al. The limits and intensity of Plasmodium falciparum transmission: implications for malaria control and elimination worldwide. PLos Med. 2008;5:38. doi: 10.1371/journal.pmed.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hay SI, Guerra CA, Gething PW, Patil AP, Tatem AJ, et al. A world malaria map: Plasmodium falciparum endemicity in 2007. PLoS Med. 2009;6:e1000048. doi: 10.1371/journal.pmed.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Severe falciparum malaria. Trans R Soc Trop Med Hyg. 2000;94:S1–S90. [PubMed] [Google Scholar]
- 6.González A, Nicolás JM, Muñoz J, Castro P, Mas J, et al. Severe imported malaria in adults: retrospective study of 20 cases. Am J Trop Med Hyg. 2009;81:595–599. doi: 10.4269/ajtmh.2009.08-0637. [DOI] [PubMed] [Google Scholar]
- 7.Sturm A, Amino R, van de Sand C, Regen T, Retzlaff S, et al. Manupulation of host hepatocytes by the malaria parasite for delivery into liver sinusoids. Science. 2006;313:1245–1246. doi: 10.1126/science.1129720. [DOI] [PubMed] [Google Scholar]
- 8.Ballou WR. The development of the RTS,S malaria vaccine candidate: challenges and lessons. Parasite Immunol. 2009;31:492–500. doi: 10.1111/j.1365-3024.2009.01143.x. [DOI] [PubMed] [Google Scholar]
- 9.Cohen S, McGregor IA, Carrington S. Gamma-globulin and acquired immunity to human malaria. Nature. 1961;192:733–737. doi: 10.1038/192733a0. [DOI] [PubMed] [Google Scholar]
- 10.Bouharoun-Tayoun H, Attanath P, Sabchareon A, Chongsuphajaisiddhi T, Druilhe P. Antibodies that protect humans against Plasmodium falciparum blood stages do not on their own inhibit parasite growth and invasion in vitro but act in cooperation with monocytes. J Exp Med. 1990;172:1633–1641. doi: 10.1084/jem.172.6.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sabchareon A, Burnouf T, Ouattara D, Attanath P, Bouharoun-Tayoun H, et al. Parasitologic and clinical human response to immunoglobulin administration in falciparum malaria. Am J Trop Med Hyg. 1991;45:297–308. doi: 10.4269/ajtmh.1991.45.297. [DOI] [PubMed] [Google Scholar]
- 12.Good MF. Towards a blood-stage vaccine for malaria: are we following all the leads. Nat Rev Immunol. 2001;1:117–125. doi: 10.1038/35100540. [DOI] [PubMed] [Google Scholar]
- 13.Malkin E, Long CA, Stowers AW, Zou L, Singh S, et al. Phase 1 study of two merozoite surface protein 1 (MSP 1 (42) vaccines for Plasmodium falciparum malaria. Plos Clin Trials. 2007;2:e12. doi: 10.1371/journal.pctr.0020012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ball SG, van de Wal MHBJ, Visser RGF. Progress in understanding the biosynthesis of amylose. Trends in Plant Sci. 1998;3:462–467. [Google Scholar]
- 15.Wattebled F, Buléon A, Bouchet B, Ral JP, Liénard L, et al. Granule-bound starch synthase I: a major enzyme involved in the biogenesis of B-crystallites in starch granules. Eur J Biochem. 2002;269:3810–3820. doi: 10.1046/j.1432-1033.2002.03072.x. [DOI] [PubMed] [Google Scholar]
- 16.van de Wal M, D'Hulst C, Vincken JP, Buléon A, Visser R, et al. Amylose is synthesized in vitro by extension of and cleavage from amylopectin. J Biol Chem. 1998;273:22232–22340. doi: 10.1074/jbc.273.35.22232. [DOI] [PubMed] [Google Scholar]
- 17.Van den Koornhuyse N, Libessart N, Delrue B, Zabawinski C, Decq A, et al. Control of starch composition and structure through substrate supply in the monocellular alga Chlamydomonas reinhardtii. J Biol Chem. 1996;271:16281–16287. doi: 10.1074/jbc.271.27.16281. [DOI] [PubMed] [Google Scholar]
- 18.Delrue B, Fontaine T, Routier F, Decq A, Wieruszeski JM, et al. Waxy Chlamydomonas reinhardtii: monocellular algal mutants defective in amylose biosynthesis and granule-bound starch synthase accumulate a structurally modified amylopectin. J Bacteriol. 1992;174:3612–3620. doi: 10.1128/jb.174.11.3612-3620.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellis RD, Martin LB, Shaffer D, Long CA, Miura K, et al. Phase 1 trial of the Plasmodium falciparum blood stage vaccine MSP142-C1/alhydrogel with and without CPG 7909 in malaria naïve adults. Plos One. 2010;5:e8787. doi: 10.1371/journal.pone.0008787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takala SL, Coulibaly D, Thera MA, Dicko A, Smith DL, et al. Dynamics of polymorphism in a malaria vaccine antigen at a vaccine testing site in Mali. Plos Med. 2007;4:e93. doi: 10.1371/journal.pmed.0040093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pizarro JC, Vulliez-Le Normand B, Chesne-Seck ML, Collins CR, Withers-Martinez C, et al. Crystal structure of the malaria vaccine candidate apical membrane antigen 1. Science. 2003;308:408–411. doi: 10.1126/science.1107449. [DOI] [PubMed] [Google Scholar]
- 22.Malkin EM, Diemert DJ, McArthur JH, Perreault JR, Miles AP, et al. Phase 1 clinical trial of apical membrane antigen 1: an asexual blood-stage vaccine for Plasmodium falciparum malaria. Infect Immun. 2005;73:3677–3685. doi: 10.1128/IAI.73.6.3677-3685.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ral JP, Colleoni C, Wattebled F, Dauvillée D, Nempont C, et al. Circadian clock regulation of starch metabolism establishes GBSSI as a major contributor to amylopectin synthesis in Chlamydomonas reinhardtii. Plant Physiol. 2006;142:305–317. doi: 10.1104/pp.106.081885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blackman MJ, Heidrich HG, Donachie S, McBride JS, Holder AA. A single fragment of a malaria merozoite surface protein remains on the parasite during red cell invasion-inhibiting antibodies. J Exp Med. 1990;172:379–382. doi: 10.1084/jem.172.1.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blackman MJ, Whittle H, Holder AA. Processing of the Plasmodium falciparum major merozoite surface protein-1: identification of a 33-kilodalton secondary processing product which is sheed prior to erythrocyte invasion. Mol. Biochem Parasitol. 1991;49:35–44. doi: 10.1016/0166-6851(91)90128-s. [DOI] [PubMed] [Google Scholar]
- 26.Guevara Patiño JA, Holder AA, McBride JS, Blackman MJ. Antibodies that inhibit malaria merozoite surface protein-1 processing and erythrocyte invasion are blocked by naturally acquired human antibodies. J Exp Med. 1997;186:1689–1699. doi: 10.1084/jem.186.10.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyon JA, Angov E, Fay MP, Sullivan JS, Girourd AS, et al. Protection induced by Plasmodium falciparum MSP1 (42) in strain-specific, antigen and adjuvant dependent, and correlates with antibody responses. Plos One. 2008;3:e2830. doi: 10.1371/journal.pone.0002830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Warzecha H, Mason HS, Lane C, Tryggvesson A, Rybicki E, et al. Oral immunogenicity of human papillomavirus-like particles expressed in potato. J Virol. 2003;77:8702–8711. doi: 10.1128/JVI.77.16.8702-8711.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bock R, Warzecha H. Solar-powered factories for new vaccines and antibiotics. Trends in Biotechnol. 2010;28:246–252. doi: 10.1016/j.tibtech.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Schroda M, Beck CF, Vallon O. Sequence elements within an HSP70 promoter counteract transcriptional transgene silencing in Chlamydomonas. Plant Journal. 2002;31:445–455. doi: 10.1046/j.1365-313x.2002.01371.x. [DOI] [PubMed] [Google Scholar]
- 31.Dauvillée D, Chochois V, Steup M, Haebel S, Eckermann N, et al. Plastidial phosphorylase is required for normal starch synthesis in Chlamydomonas reinhardtii. . Plant Journal. 2006;48:274–285. doi: 10.1111/j.1365-313X.2006.02870.x. [DOI] [PubMed] [Google Scholar]
- 32.Kindle K. High-frequency nuclear transformation of Chlamydomonas reinhardtii. Genetics. 1990;87:1228–1232. doi: 10.1073/pnas.87.3.1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris EH. San Diego: Academic Press; 1989. Culture and storage methods. pp. 25–63. In The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use (Harris, E. ed.) [Google Scholar]
- 34.Dzierszinski F, Coppin A, Mortuaire M, Dewailly E, Slomianny C, Ameisen JC, et al. Ligands of the perippheral benzodiazepine receptor are potent inhibitors of Plasmodium falciparum and Toxoplasma gondii in vitro. Antimicrob Agents Chemother. 2002;46:3197–3207. doi: 10.1128/AAC.46.10.3197-3207.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Nucleotide sequences of C. reinhardtii plasmid. The plasmid pKB101 was used to clone P. falciparum and P. berghei genes. This C. reinhardtii expression vector contains the chimeric RBSC2-HSP70A promoter (nucleotide 796 to 1040), GBSS genomic DNA sequence deleted at the 3' end for the 390 last nucleotides (nt 1048 to 4104), blaM Beta-lactamase also named ampicillin resistance cassette (nt 5137 to 5995), ColE1 ori (nt 6155 to 6748), AphVIII or paromomycin resistance cassette (7234-9259).
(TIF)
Parasite nucleotide sequences used for cloning and expression of P. falciparum MSP1-19, P. berghei MSP1-19 and P. berghei AMA1-C. The nucleotide sequences of Plasmodium genes were designed according to the codon bias of C. reinhardtii.
(TIF)
Longitudinal section of C. reinhardtii cell was visualized by electron microscopy. The section of C. reinhardtii cell was probed with polyclonal antibodies specific to P. berghei AMA1. The algae cell has been transformed by a construct expressing P. berghei AMA1-C antigen in the starch localized in the chloroplast. S represents starch grains surrounding the pyrenoid (P) matrix. The gold particles bind to specific anti-AMA1 antibodies present on both starch and the pyrenoid matrix. N, nucleus; C, chloroplast; M, mitochondrion. The bar = 500 nm.
(TIF)
P. falciprum strain-specific growth inhibitory effects in the presence of purified IgGs. A pool of immune sera of mice vaccinated with starch containing GBSS-PfMSP1-19 was used to purify IgGs. The inhibition of red blood cell invasion by either P. falciparum HB3 or 3D7 strain was tested in the presence of purified immune IgGs. Data represent mean values +/- s.d. and are from at least three independent experiments with two different pools of immune sera (P<0.005).
(TIF)