We functionally characterized RhoS/RSA-14-44 as a new member of Rho GTPase subfamily in spermatogenesis, which provides a direct link between Rho family GTPase and the proteasome biogenesis.
Abstract
Most Rho family GTPases serve as key molecular switches in a wide spectrum of biological processes. An increasing number of studies have expanded their roles to the spermatogenesis. Several members of Rho family have been confirmed to be essential for mammalian spermatogenesis, but the precise roles of this family in male reproduction have not been well studied yet. Here we report a surprising function of an atypical and testis-specific Rho GTPase, RSA-14-44 in spermatogenesis. Featured by unique structural and expressional patterns, RSA-14-44 is distinguished from three canonical members of Rho cluster. Thus, we define RSA-14-44 as a new member of Rho GTPases family and rename it RhoS (Rho in spermatogenic cells). RhoS associates with PSMB5, a catalytic subunit of the proteasome, in a series of stage-specific spermatogenic cells. More importantly, RhoS does not directly modulate the cellular proteasome activity, but participates in regulating the stability of “unincorporated” PSMB5 precursors. Meanwhile, our data demonstrate that the activation of RhoS is prerequisite for negatively regulating the stability of PSMB5 precursors. Therefore, our finding uncovers a direct and functional connection between the Rho GTPase family and the pathway of proteasome biogenesis and provide new clues for deciphering the secrets of spermatogenesis.
INTRODUCTION
Mammalian spermatogenesis is a paradigm for development, during which the genetic information from male germ stem cells is reedited, reorganized, and finally distributed into spermatozoa, along with a dramatic metamorphosis of germ cells (Kierszenbaum, 1994; de Kretser et al., 1998; Trasler, 2009). This intricate process is strictly regulated by a system that is constructed from a sophisticated and well-coordinated program of gene expression (Schultz et al., 2003; Rolland et al., 2008; Lui and Cheng, 2008). The most notable function of this program was performed by numerous testis and/or germ cell–specific genes in spermatogenesis (Eddy, 2002; Sha et al., 2002; White-Cooper, 2010). Thus, identification and further characterization of these specific genes is of great value to the determination of the mechanism of spermatogenesis.
The mammalian Rho GTPases consist of over 20 distinct members that are homologous in evolution, forming a subgroup of the Ras super family (Bustelo et al., 2007). Represented by CDC42, RAC1, and RhoA, most members of this super subfamily have been shown to act as key molecular switches by cycling between an active GTP-bound state and an inactive GDP-bound state (Etienne-Manneville and Hall, 2002). They are known for their pivotal roles in a wide variety of cellular functions, including cytoskeleton organization, cell polarity, microtubule dynamics, membrane transport pathways, and transcription factor activity (Mackay and Hall, 1998; Ridley, 2001; Boureux et al., 2007; Vega and Ridley, 2008). Recently, several members of this subfamily including Rnd2, Cdc42, Rac1, Rac2, and RhoB, coupled with a group of their regulator proteins, have been identified in the testis. Some of them have been proved to respectively participate in regulating Sertoli-germ cell tight junctions, germ cell movement, or cell division (Freeman et al., 2002; Naud et al., 2003; Lui et al., 2005; Sarkar et al., 2007; Adly and Hussein, 2010; Wong and Cheng, 2009). However, to date, much more work remains to be done to thoroughly understand the physiological roles of Rho family GTPases in the testis. In particular, exploring the putative testis and/or germ-specific Rho-like GTPases and their related pathways will contribute greatly to our knowledge of spermatogenesis.
The ubiquitin-proteasome system (UPS) takes the central stage in nonlysosomal protein degradation in eukaryotic cells (Tanaka et al., 1992; Rivett, 1993). As a core component of the UPS, the proteasome works as a protease with high efficiency and specificity in various cellular processes including cell cycle progression, transcriptional regulation, signal transduction, and cell fate determination (Zwickl, 2002; Adams, 2003; Hendil and Hartmann-Petersen, 2004). The highly conserved structure of the 26S proteasome is based on two subcomplexes, namely, a 20S catalytic core particle (CP) and a 19S regulatory particle (RP; Walz et al., 1998; Cheng, 2009). In eukaryotes, the CP is composed of two identical outer α-rings and two identical inner β-rings, each consisting of seven homologous subunits (α1–7 and β 1–7), that stack to form a catalytic cylinder (; Groll et al., 2002; Unno et al., 2002a,b). All of catalytic β-subunits (β1, β2, β5) are expressed in immature precursor forms, thereby preventing self-assembly of the catalytic subunits and active site exposure. Thus, a major confusion in understanding the biogenesis of proteasome is how de novo–synthesized β-subunits precursors are restricted in the assembly of proteasome to avoid being diffused into untargeted subcellular regions, where these precursors might be incorrectly activated by nonspecific cleavages.
On the other hand, recent studies have revealed a dynamic and heterogeneous nature in the proteasome biogenesis. Mammals encode four major additional catalytic β-subunits (β1i, β2i, β5i, and β5t), which are incorporated upon induction in place of the corresponding β-subunits (β1, β2, and β5). This variation in β-subunits composition results in different proteasome subtypes, that is, immunoproteasomes for proteasomes containing β1i, β2i, and β5i; and thymoproteasomes for those containing β5t (Aki et al., 1994; Heink et al., 2005; Ostrowska et al., 2006; Tomaru et al., 2009). Interestingly, some reports had described alternative or unique proteasome subunits and its associated proteins in the testes of various species, whose notable functions are related to the reproductive ability (Khor et al., 2006; Belote and Zhong, 2009; Zhong and Belote, 2007; Rivkin et al., 2009;). However, the mechanisms underlying the proteasome biogenesis in mammalian development processes, especially in spermatogenesis, are poorly understood.
Here, we identified a novel testis-specific small GTPase, RhoS, in spermatogenesis. In terms of the structural and enzymatic features of RhoS, we classified it into the Rho small GTPase subfamily. Surprisingly, instead of canonical regulators or effectors of Rho GTPases, a core subunit of the 20S proteasome, PSMB5, was identified as a novel RhoS-associated protein in spermatogenesis. Notably, we found that the primary functional role of RhoS is to work as a switch for the degradation of unincorporated PSMB5 precursors, instead of directly attenuating the proteasome activity. Collectively, these findings present the first piece of evidence for a direct link between Rho family GTPases and the proteasome biogenesis, expanding the catalogue of the functions of Rho GTPases in spermatogenesis.
MATERIALS AND METHODS
Expression Constructs
cDNA spanning the full-length open reading frame of RhoS/RSA-14-44 or PSMB5 were obtained from rat testis by RT-PCR with corresponding primers (for RhoS/RSA-14-44, the forward primer was 5′-ACCAT GGCTG CCATC CGGAA GAAAC TGGTG ATCGT GGGAG-3′ and the reverse primer was 5′-TCAAA AGACA AAGCA ACCAG TCTTT TTCTT CACTC GATTCG-3′; for PSMB5, the forward primer was 5′-ATGGC GCTGG CTAGC GTGTT-3′ and the reverse primer was 5′-GATAT CGGGA CAGAT ACACT ACTG-3′). The PCR products were cloned into a pGEM-T1 vectors (Invitrogen, Carlsbad, CA), which were confirmed by sequencing. Deletion mutants and wild type of RhoS were generated by PCR using the pGEM-T1-RhoS as the template and then were subcloned into a pcDNA6/myc-His B vector (Invitrogen) at KpnI/ApaI sites with N-terminal Flag tag or N-terminal HA tag (pcDNA6-Flag-RhoS or pcDNA6-HA-RhoS) for expressing Flag-RhoS or HA-RhoS. Point mutations of RhoS were introduced into pcDNA6-Flag-RhoS using a PCR method for expressing Flag-RhosD13T, RhosG14V, RhosT19N, RhosQ63L, or RhoS C190S, respectively. The coding region of PSMB5 was subcloned into pcDNA6/myc-His B with or without N-terminal Flag-tag at EcoRI/XhoI sites for expressing PSMB5-Myc or Flag-PSMB5-Myc and was also subcloned into pcDNA4/TO/myc-His B at the same sites for the inducible expression of PSMB5-Myc. The C-terminally 3×Flag-tagged PSMB5 was constructed by inserting PSMB5's coding region into a p3×FLAG-CMV-14 vector (Sigma-Aldrich, St. Louis, MO) at HindIII/Xba I sites.
The coding regions of RhoA, RhoB, and RhoC were amplified from mouse tissue cDNAs by PCR with following primers (forward and reverse): RhoA, 5′-GTCGG ATCCA CCATG GCTGC CATCC GGAAG AAACT G-3′ and 5′-CGCGA ATTCT CACAA GACAA GGCAC CCAG-3′; RhoB, forward primer as for RhoA and 5′-CGTCT AGATC ATAGC ACCTT GCAGC AGTTG ATGCA GCCAT TCTGA GATCC G-3′; and RhoC, forward primer as for RhoA and 5′-CGCGA ATTCT CATCA GAGAA TGGGA CAGCC CCTC-3′. The PCR products were cloned into pcDNA6/myc-His B vector individually at BamHI/EcoRI sites with N-terminal Flag tag, except for RhoB at BamHI/XbaI sites.
For the prokaryote expression vectors, the entire coding sequence of RhoS/RSA-14-44 was inserted into pGEX-4T-3 (GE Healthcare, Little Chalfont and Buckinghamshire, United Kingdom) and pET-30a (Novagen, Madison, WI) at EcoRI/XhoI sites.
Northern Blot Analysis
A normalized Northern blot was used to identify RhoS/RSA-14-44 transcripts in different rat tissues. Total RNA isolated from the indicated tissues of adult rats using the Trizol reagent (Invitrogen) was separated and blotted onto a positively charged nylon membrane (Boehringer Mannheim, Ingelheim on Rein, DE). The membrane was probed sequentially with a γ-32P-labeled RhoS/RSA-14-44 cDNA probe and then with a γ-32P-labeled β-actin probe. After hybridization, blots were washed at high stringency and exposed to x-ray film.
Semiquantitative Relative RT-PCR
With Trizol reagent, total RNAs were isolated from tissues of adult mice and from testes of mice at various ages, ranging from 1 to 10 wk after birth, respectively. Isolated total RNA, 1 μg, was converted to cDNA with reverse transcription system (Promega, Madison, WI). cDNA samples were subjected to PCR amplification using following target and reference gene primers: RhoA, 5′-CGGAA TGACG AGCAC ACGAG ACGG-3′ and 5′-CAAGA TGAGG CACCC AGA-3′; RhoB, 5′-GCGCA GCGAC GAGCA TGTCC GCAC-3′ and 5′-TAGCA CCTTG CAGCA GTTGA TG-3′; RhoC, 5′-GAGGC AAGATGAGCA TACCA GGAGA-3′ and 5′-GAGAA TGGGA CAGCC CCTCC GGCG-3′; RhoS/RSA-14-44, 5′-CGGAA TGACT TCTAC ACGAT ACAA-3′ and 5′-AAAGA CAAAG CAACC AGT-3′; and GAPDH, 5′-AGCGA GATCC CTCCA AAATC-3′ and 5′-GGCAG AGATG ATGAC CCTTT-3′. Cycling conditions were 1 cycle at 95°C for 2 min, followed by 30 cycles of denaturation at 95°C for 15 s, and annealing/extension at 56°C for 15 s.
Tissues and Cells
The tissue samples were prepared from adult male BALB/c mice for analyzing the spatial expression pattern of RhoS by RT-PCR, whereas testes tissues for studying the temporal expression pattern were obtained from male BALB/c mice at different ages (from 1 to 10 wk after birth). The tissues samples for Northern blot were isolated from adult male Sprague-Dawley rats. All of the experimental and surgical procedures were approved by the Animal Ethics Committee of National Research Institute for Family Planning Beijing.
Human embryonic kidney (HEK) 293T, MCF-7, and Hela cells were maintained in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% fetal bovine serum (FBS) at 37°C with 5% CO2. HEK293T cells were transfected with Vigorous transfection reagent (Vigorous, Beijing, CN). MCF-7 and Hela cells were transfected with Fugene HD transfection reagent (Roche, Basel, CH).
For establishing PSMB5 inducible expression stable cell line with Tet-on system, Hela cells were transfected with combined pcDNA4-TO-PSMB5 and pcDNA6-TR at a ratio of 6:1 and then selected for the stable clones with the mixture of Zeocin and blasticidin according to the manual of the products (Invitrogen). The cell lines with lower background expression were selected for the protein stability assay.
Protein Expression and Purification
Recombinant glutathione S-transferase (GST)- and 6×His-tagged RhoS were produced in Escherichia coli BL21 (DE3) cells and purified with glutathione Sepharose 4B (GE Healthcare, Piscataway, NJ) or Ni-NTA Sepharose (Qiagen, Hilden, DE) according to the manufacturer's instructions. Finally, the purified proteins were dialyzed against buffer A (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA, and 1 mM dithiothreitol) plus 20% glycerol.
Assay of Rho GTPase Activity
The GTPase activity of purified GST-RhoS was measured using a RhoGAP assay Biochem Kit (BK105; Cytoskeleton, Denver, CO). All procedures were performed according to the manufacturer's protocol.
Active Rho GTPase Pulldown Assay
HEK 293T cells were transfected with the indicated plasmids. Twenty-four hours after transfection, the activation of Rho GTPase was investigated using a Rho Activation Assay Kit (STA-403; Cell Biolabs, San Diego, CA) according to the manufacturer's protocol.
Antibodies
A mouse polyclonal antibody against purified recombinant 6xHis-tagged RSA-14-44/RhoS was raised and purified by the chromatography-affinity method. Other antibodies as follows were purchased from the corresponding companies: anti-Flag (clone M2; Sigma-Aldrich, St. Louis, MO), anti-Myc (mouse mAb; Abmart, Shanghai, CN), anti-Myc (rabbit polyclonal antibody; MBL, Woburn, MA), anti-PSMB5 (Enzo, Plymouth Meeting, PA), anti-PSMA2 (Cell Signaling, Danvers, MA), anti-GFP (MBL, Woburn, MA), anti-GAPDH (Santa Cruz Biotechnology, Santa Cruz, CA), anti-LaminA+C (Abcam, Cambridge, United Kingdom), anti-Calnexin (Santa Cruz Biotechnology), and anti-POMP (Sigma-Aldrich).
2.9. Immunohistochemical Analysis
Rat testes were fixed in buffered paraformaldehyde at 4°C and embedded in paraffin. Deparaffinized sections (7 μm) were incubated in phosphate-buffered saline (PBS) containing 3% H2O2 to quench endogenous peroxidase activity. Sections were then blocked in species-specific normal sera for 30–60 min to reduce nonspecific staining and subsequently were incubated with primary antibodies or preimmune sera followed by horseradish peroxidase–conjugated secondary antibody. The signals were detected using a 3,3′-diaminobenzidine substrate working solution until the desired staining was achieved. Nuclei were counterstained with hematoxylin.
2.10. Immunofluorescence Analysis
The separation of germ cells was performed as previously described (Qiao et al., 2004). Freshly prepared spermatogenic cell suspensions were smeared on slides. After fixing with methanol and blocking nonspecific sites, the cells were subjected to double-fluorescent staining. The cells were incubated with mouse anti-RhoS antibody plus rabbit anti-PSMB5 antibody followed by FITC-conjugated anti-mouse goat IgG and rhodamine-conjugated anti-rabbit goat IgG. For the control samples, the primary antibodies were replaced by the IgGs from the same species. Nuclei were counterstained with DAPI (1 μg/ml) (Invitrogen). The signals were detected by Leica TCS NT laser confocal microscopy (Deerfield, IL).
For MCF-7 cells, after fixing in 4% paraformaldehyde, the cells were permeabilized and blocked with PBS containing 0.3% Triton X-100 and 5% FBS. Subsequently, they underwent double-fluorescent staining according to the same protocols as described above except that the primary antibodies were replaced by an anti-Flag antibody and an anti-Myc polyclonal antibody.
2.11. Immunoprecipitation and Immunoblot (IB)
For immunoprecipitation (IP), HEK293T cells were lysed with ice-cold buffer B (50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 10 mM MgCl2, 2 mM ATP, and 1 mM dithiothreitol) containing 0.5% Triton X-100 and centrifuged at 13,000 rpm for 10 min at 4°C. The supernatant (∼1 mg) was added to M2-agarose (Sigma-Aldrich) and incubated for 1 h at 4°C. The immunoprecipitates were washed several times with ice-cold buffer A containing 0.2% Triton X-100 and then boiled in SDS sample buffer with beta-mercaptoethanol.
For immunoblotting, the samples were separated by 12% SDS-PAGE (Rottinger et al., 2006) and transferred to a PVDF membrane (Millipore, Bedford, MA). The membranes were immersed in 5% BSA overnight at 4°C and incubated with primary antibody. The bound antibodies were detected using corresponding horseradish peroxidase–conjugated secondary antibodies (anti-mouse, goat IgG; anti-rabbit, goat IgG; Santa Cruz Biotechnology) and ECL reagents (Engreen, Beijing, CN).
Determination of PSMB5 Protein Stability
Twenty-four hours after transfection with indicated plasmids, cells were incubated with 100 μg of cycloheximide (CHX) for effectively blocking the protein synthesis. The concentration of PSMB5, including both the precursor and mature forms, was monitored by Western blotting at various time points after CHX addition.
For measuring the PSMB5 protein level in steady state using the Tet-on expression system, the selected stable cell lines with low level of leaky expression were transfected with the indicated plasmids. Twenty-four hours after transfection, the cells was induced by replacing with tetracycline (1 μg/ml) containing media for 4 h. To suppress the expression of PSMB5, the cells were then cultured in Tet-free medium for another 1 h before harvest at different time points. The protein level of PSMB5 was analyzed by Western blot.
Cellular Proteasome Activity Assay
The HEK293T cells were transfected with corresponding plasmids and homogenized on ice in buffer B devoid of sodium. The cell lysates were clarified by centrifugation, and the supernatants were used for the determination of protein concentration and the subsequent assay for proteasome activity using the fluorogenic substrate succinyl LLVY-7amc. The detailed procedures were as same as described previously (Hirano et al., 2005).
Glycerol Gradient Analysis
After transfection, HEK 293T cells were lysed in buffer B containing 0.5% NP-40. The lysates were clarified by centrifugation at 13,000 rpm at 4°C, and the supernatants were subjected to 10–40% (vol/vol) linear glycerol density gradient centrifugation at 100,000 × g for 12 h, as described previously (Qiu et al., 2006).
RESULTS
RhoS/RSA-14-44 Is Specifically Expressed in Testis
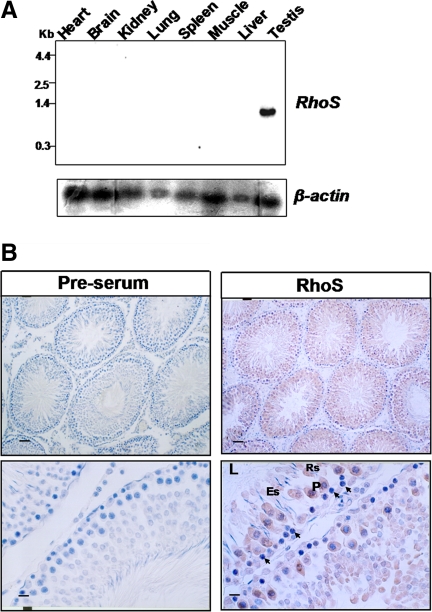
In previous studies, we had identified a series of factors in spermatogenesis through a system combining laser capture microdissection (LCM) and suppressive subtractive hybridization (Zhang et al., 2003; Liang et al., 2004; Chen et al., 2008). Based on this technique, a new gene was isolated and cloned from rat testis cDNA library and assigned the name of RSA-14-44 (GenBank ID: 297173). To elucidate the expression pattern of RSA-14-44, a Northern blot analysis was applied to a panel of rat tissues, the result of which showed its expression is restricted to the testis (Figure 1A). To further determine the distribution of RSA-14-44 in the testis, we raised a specific antibody against RSA-14-44 and used it to probe the frozen section of rat testis (Figure 1B). All positive signals were exclusively found in seminiferous tubules, and notably, much stronger stains were observed in an array of spermatogenic cell lineages at defined stages of spermatogenesis, including panchytene spermatocytes, round spermatids, and elongated spermatids. Thus, RSA-14-44 is a testis-specific gene with putative roles at some specific stages of spermatogenesis.
Figure 1.
The expression of RhoS/RSA-14-44 is restricted to germ cells. (A) Tissue distribution of RhoS/RSA-14-44 mRNA in adult rats was analyzed by Northern blot. Each lane contains 20 μg of total RNA. (B) The spermatogenic cell–specific expression pattern of RhoS. Immunohistochemistry was carried out using anti-RhoS polyclonal antibodies (right) to clarify the localization of the protein in rat testis. A negative control was performed using normal rabbit sera (left). P, pachytene spermatocytes; Rs, round spermatids; Es, elongated spermatids; L, lumen of seminiferous. Bar, 100 μm in top panels and 20 μm in bottom panels.
RhoS/RSA-14-44 Is Classified into the Rho GTPases Family
Based on our in silico analysis utilizing the Pfam (http://www.sanger.ac.uk/resources/databases/pfam.html) and InterProScan (http://www.ebi.ac.uk/Tools/InterProScan) databases, RSA-14-44 was predicted to contain a Rho-type structure (amino acids 1-182) and a CAAX motif (amino acids 190-193; C is cysteine, A is an aliphatic amino acid, and X is variable; Figure 2A). These two conserved parts are actually the typical structural symbols of Rho family GTPases. In addition, there was an “insert region” (amino acids 123-136) in RSA-14-44 (Figure 2A) that was similar in some degree with the “insert loop” of Rho subfamily GTPases. Collectively, these results raised a high possibility that RSA-14-44 belongs to the Rho GTPases family, and thus we renamed it RhoS (Rho in spermatogenic cells).
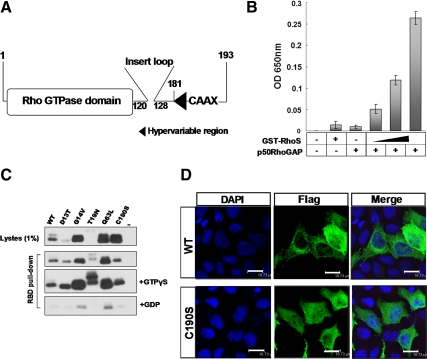
Figure 2.
The conservation of RhoS in the structure and enzymatic activity as a Rho GTPase. (A) A schematic structural model of RhoS. Numbers indicate corresponding amino acids in sequence. (B) GTP hydrolysis activity assay. Enzymatic activity was measured by GTP hydrolysis using purified GTPase protein. The phosphate generated by hydrolysis of GTP was measured by the addition of CytoPhos (Cytoskeleton) reagent and reading the absorbance at 650 nm. Results are presented as the mean ± SD from three experiments. (C) Activated Rho GTPase (GTP-bound) protein pulldown assay. Extracts were prepared from HEK293T cells transiently expressing Flag-tagged wild-type or mutated RhoS including RhoS(D13T), RhoS(G14V), RhoS(T19N), RhoS(Q63L), and RhoS(C190S). After a 1-h incubation with GST-RBD glutathione beads, the bound proteins were analyzed by Western blotting using anti-Flag antibody. (D) A conserved C-terminal site (C190) is essential for the proper localization of RhoS. Subcellular distributions of wild-type and C190S mutant were analyzed by immunofluorescence. Twenty-four hours after transfection, MCF-7 cells transfected with the indicated plasmids were prepared for immunofluorescence analysis using anti-Flag antibody. Bar, 18.73 μm.
To determine whether RhoS could hydrolysis GTP in the same enzymatic way as other Rho family GTPases, we performed GTP hydrolysis analysis based on a well-developed in vitro assay using purified GST-RhoS protein (Supplemental Figure 1). The intrinsic GTPases activity is generally very low in the case of the Ras superfamily of GTPases, including Rho family. This hydrolysis can be accelerated by RhoGAP proteins like p50RhoGAP, with distinct specificities for their respective GTP-binding proteins (Zhang and Zheng, 1998). As shown in Figure 2B, p50RhoGAP significantly elevated the GTP hydrolysis reaction catalyzed by RhoS, whereas the intrinsic activity of RhoS remained at a considerably low level when p50RhoGAP was absent in the reaction system. Therefore, we characterized RhoS as a Rho type GTPase.
We then made sequence alignment of RhoS with three Rho GTPase isoforms (RhoA, RhoB, and Rho C). The result indicates that RhoS shared high similarity with these members in the Rho GTPase domain (amino acids 1-121) and the CAAX motif (Figure 3A), represented by some well-conserved residues such as D13, G14, T19, Q63, and C190. Previous studies confirmed that the mutations in these specific residues can genetically modify the activation of Rho family GTPases. For example, D13T or T19N mutation occurred in RhoA results in a dominant negative form of GTPase (Zallen et al., 2000), whereas G14V or Q63L mutant is constitutively active (Bourne et al., 1991). Moreover, most Rho family members undergo CAAX-terminal posttranslational modification at C190 site, which is critical for the membrane association and biological activity of Rho GTPases (Hori et al., 1991; Ihara et al., 1998; Solski et al., 2002). Therefore, to further verify the correlation of the Rho type structure of RhoS to its GTPase activity, we introduced respective mutations into RhoS to assess their effects on the activation of the GTPase through a pulldown assay, in which activated Rho GTPase could be isolated through selectively binding to RBD (Rho-binding domain) fused beads (Knaus et al., 2007). As shown in Figure 2C, all the mutants we generated behaved in ways similar to those described in previous reports on other Rho family members. Higher efficiency in pulldown of activated GTPase was exhibited in the G14V and Q63L groups compared with that of wild-type protein, whereas low levels of positive signals were detected in D13T, T19N, and C190S groups. Meanwhile, we compared the cellular localization of the C190S mutant and wide-type protein by IF (Figure 2D). In contrast to the perinuclear centered pattern of the wild-type protein, C190S mutant was dispersed in both nuclear and cytoplasmic regions, which is similar to the case of RhoAC190S (Benetka et al., 2006). This dramatic difference occurred in the C190S mutant was also confirmed by the Western blot analysis (Supplemental Figure 2). These results indicate that Rho type structure of RhoS provides a dynamic base for its biological activity, thus confirming RhoS is a member of the Rho GTPases family. On the other hand, this experiment validated a group of RhoS activity–related mutants that became powerful tools in our subsequent work for exploring the biological functions of RhoS.
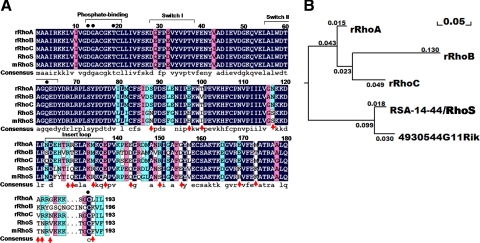
Figure 3.
The structural features distinguish RhoS from canonical Rho GTPases. (A) Alignment of the amino acid sequence of RhoS and three classical Rho GTPases (RhoA, RhoB, and RhoC). The alignment was performed by DNAMAN (Lynnon, Quebec, Canada). Homology levels are highlighted in different colors. Black: 100%; Pink: 75%; Blue: 50%. Arrows: residues conserved between RhoS and 4930544G11Rik (mRhoS) but significantly distinct from RhoA/B/C; Points: conserved resides for regulating the activity of Rho type GTPases. (B) Phylogenetic tree analysis of three Rho isoforms, RhoS, and 4930544G11Rik. Data generated from alignment (Figure 3A) were loaded to reconstruct the rooted neighbor-joining phylogenetic tree by the maximum likelihood method. Numbers indicate branch length. r, rat.
Characterization of RhoS as a New Member of the Rho GTPases Family
Despite a high level of similarity between RhoS and the canonical Rho family members (RhoA, RhoB, and RhoC), there are substantial differences between these GTPases, not only in their detailed structures but also in their expression patterns. Many divergences in the protein sequences of Rho GTPases occur in the hypervariable region in the C-terminal structure (Figure 2A). In addition, the insert region also contributes to forming the structural and functional distinction in different Rho GTPases. Actually, in RhoS, there are evolutionally conserved variations in these two symbolic structures compared with RhoA, RhoB, and RhoC (red arrows in Figure 3A and Supplemental Figure 3). Further phylogenetic tree analysis confirmed that these differences significantly lead to the separation of RhoS from other members of Rho subfamily (Figure 3B).
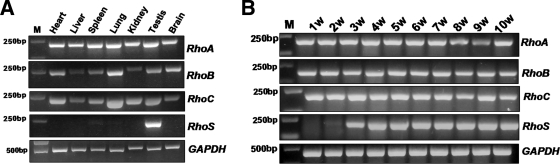
Even though, we still wonder whether RhoS serves as an unconventional Rho GTPase with unique biological roles. For this purpose, we chased the spatial and temporal expression patterns of RhoS. RT-PCR analysis showed that the expression of RhoS is restricted to mouse testis, whereas RhoA, RhoB, and RhoC are expressed ubiquitously in all of tissues examined (Figure 4A). This result is consistent with the previous reports on the distribution of RhoA, RhoB, and RhoC (Liu et al., 2001; Ducummon and Berger, 2006; Mitchell et al., 2007). Further analysis was performed on the testes obtained from mice at different times after birth. Previous studies confirmed that for the mice at the age of 1 wk, most of the cells in testis belong to somatic population, and the proportion of germ cells sequentially increases as spermatogenesis proceeds. As showed in Figure 4B, only the expression of RhoS showed dynamic variation in the whole process of establishing spermatogenesis. Lowest level of expression appeared in the second week, and the level began to increase after 3 wk, reaching the peak at about 4 or 5 wk. These data provide another piece of evidence for the germ cell–specific expression pattern of RhoS, distinguished from the cases of canonical Rho GTPases (RhoA, RhoB, and RhoC). Therefore, we conclude that RhoS is a new member of Rho GTPases family that might play unique roles in mammalian spermatogenesis.
Figure 4.
RhoS is a new member of Rho GTPases family closely related to mammalian spermatogenesis. (A) RT-PCR analysis of the expressions of RhoS, RhoA, RhoB and RhoC in a panel of adult male mice tissues. M indicates the maker for DNA electrophoresis analysis. (B) Expression analysis of RhoS, RhoA, RhoB, and RhoC in testes from the mice at different ages after birth. W: week; each number represents the age of mice after birth.
RhoS Associates with PSMB5 in Spermatogenesis
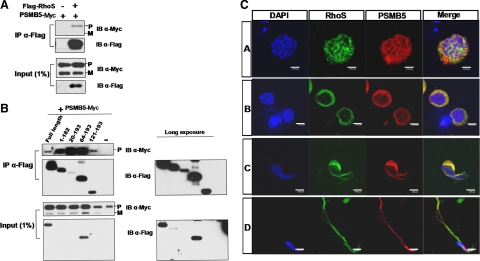
To elucidate the biological roles of RhoS in the spermatogenesis, we tried to seek its associated proteins in the testis though yeast two-hybridization screening. PSMB5, a key catalytic subunit of the 20S proteasome, was identified as a novel candidate protein. CoIP of PSMB5 with RhoS provided the further evidence to confirm the association between these two proteins (Figure 5A). To define the region in RhoS responsible for the association with PSMB5, several deletion mutants of RhoS were generated. Different constructs respectively expressing the full-length or deletion mutant RhoS were cotransfected into HEK293T cells with PSMB5. As shown in Figure 5B, different RhoS proteins exhibited significant variation in the affinity for PSMB5. The highest affinity was observed in the phosphate-binding loop deletion (20–193) group, followed by the switch I loop deletion (64–193) and the C-terminal deletion (1–182) groups; the GTPase domain deletion (121–193) and full-length groups had much lower affinity for PSMB5. These results indicate that RhoS associates with PSMB5 mainly through its GTPase domain (1–120), particularly the region covering the switch I and switch II loops (20–120). It is noted that the switch I and switch II loops not only are involved in Rho GTPase effectors' binding, but also define the specificities of Rho GTPases to their effectors (Bishop and Hall, 2000). So, we proposed that PSMB5 might be a novel target protein of RhoS.
Figure 5.
RhoS associates with PSMB5. (A) CoIP of RhoS with PSMB5. HEK293T cells were transfected with Flag-tagged plasmid encoding RhoS and Myc-tagged plasmid encoding PSMB5 as indicated. The immunoprecipitates were immunoblotted for Flag and Myc epitopes. P, precursor; M, matured. (B) Mapping the region in RhoS interacting withPSMB5. Wild-type RhoS and its deletion mutants were coexpressed with PSMB5 in HEK293T cells respectively. The following IP and immunoblotting were performed as described in A. (C) RhoS physiologically associates with PSMB5 in spermatogenesis. Isolated spermatogenic cell population were fixed and used for immunocytochemical analysis with anti-PSMB5 and anti-RhoS antibodies. The data were collected and analyzed using confocal microscopy. The nucleus was stained with DAPI. Bar, 8 μm. A, spermatocytes; B, round spermatid; C, elongated spermatid and D, spermatozoa.
GTPases of Rho family generally act as switches that convert extracellular signals into multiple intracellular effects, which is mediated by various target proteins. To clarify whether PSMB5 is a physiological effector of RhoS, we tracked the expression of RhoS and PSMB5 in the whole process of the spermatogenesis. It was turned out that the distributions of RhoS and PSMB5 were dynamic and highly overlapped in multiple types of spermatogenic cells in the spermatogenesis (Figure 5C). In spermatocytes, both of these two proteins were restricted to a cytoplasmic region near the nucleus. In round spermatids, both of them exhibited a scattered distribution surrounding the nucleus. However, in elongated spermatids, these two proteins were seemed to be translocated into a space around the post-acrosome area, where RhoS was predominantly localized in the vicinity of the posterior area of nucleus, whereas PMSB5 mainly resided in the cytoplasmic sect peripheral to RhoS. Finally, in spermatozoa, representing the end of spermiogenesis, the distributions of RhoS and PSMB5 underwent another remodeling process, during which they were relocated to the acrosome and midpiece parts of spermatozoa. Notably, an earlier description of the ubiquitin-proteasome system in the spermatogenesis had shown similar results as that from this observation (Berruti and Martegani, 2005; Tengowski et al., 2007). Therefore, we propose that PSMB5 is an important physiological partner of RhoS in spermatogenesis.
RhoS Is Not a Modulator Directly Regulating the Cellular Proteasome Activity
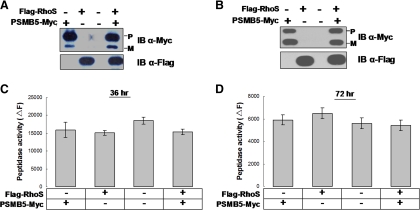
PSMB5 is one of three catalytic β subunits undertaking the function of proteasomes in protein degradation (Voges et al., 1999). As RhoS associates with PSMB5 in spermatogenesis, we simply hypothesized that RhoS might directly regulate the proteasome activity as described in the reports on the proteasome-associated proteins (Kleijnen et al., 2000; Liu et al., 2006). To verify this hypothesis, the proteasome activity was measured in the cells with transient expressions of target proteins. We found unexpectedly that the cellular proteasome activity was not altered significantly under either coexpression of RhoS and PSMB5 or single expression of each protein in a confined time course, namely, from 36 to 72 h (Figure 6). So, in our hands, there was no evidence that RhoS directly regulate the cellular proteasome activity.
Figure 6.
Transient expression of RhoS has little effect on the cellular proteasome. HEK293T cells were transfected with indicated plasmids. The Western blot analysis of target proteins, coupled with a cellular proteasome activity assay, were performed on the cells harvested at different time points, i.e., A and C, 36 h and B and D, 72 h after transfection. Data are presented as the mean ± SD of three independent experiments. P, precursor; M, matured.
RhoS Associates with Only “Unincorporated” PSMB5 Precursors
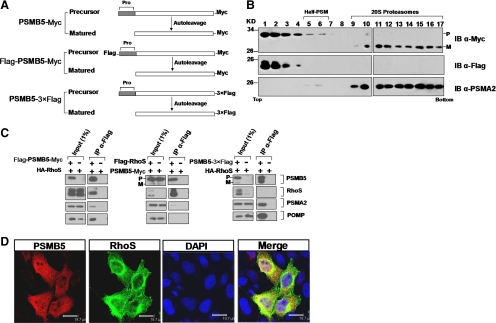
The negative results from the proteasome activity assay led us to look for alternative ways to explore the functions of the association between RhoS and PSMB5. Similar to many other proteases, the active β subunits of the proteasome including PSMB5 are synthesized as precursor form of proteins with N-terminal propeptides. On cleavage of the propeptide, these β subunits are activated in the assembled proteasomes (Figure 7A). Following this clue, we planned to define which type of PSMB5 associates with RhoS in the cells. First, we performed a glycerol density gradient–based assay to analysis the dynamic distribution of RhoS and PSMB5 in different steps of the proteasome assembly process. Assembly of the 20S proteasome starts with the formation of a half-proteasome intermediate that contains one full α-ring and one β-ring containing incorporated but unprocessed precursor forms of subunits. At the late stage of the assembly, the dimerization of half-proteasomes leads to the maturation of 20S proteasome, during which the proteolytic β subunits become active through autocleavage (Tanaka, 2009). The α-subunits are usually taken as “trackers” to reflect the whole assembly process, because they reside in both assembly intermediates and mature protesome. Thus, we chose PSMA2 to track the proteasome assembly process. As shown in Figure 7B, the distribution of PSMA2 defined two types of proteasomes in the assembly process: one was located in fractions 5–6, representing the half-proteasome (half-PSM) and the other in fractions 9–17, standing for the mature 20S proteasome. All of the mature PSMB5 existed exclusively in the 20S proteasome fractions; in contrast, PSMB5 precursor was absent from matured 20S proteasome. Except for only small part of the PSMB5 precursors located in the assembly intermediates, most of them stayed in the floating factions (1–4), to which RhoS also was restricted. This result implies that RhoS associates with only unincorporated PSMB5 precursors and is unlikely to impede the steps after the formation of the half-proteasome. Second, we designed a series of IP experiments to compare the composition of the PSMB5 precursor containing complexes isolated from the cells according to different strategies (Figure 7C). PSMA2 and a proteasome assembly chaperone, POMP, both existed in the complex coimmunoprecipitated with the PSMB5 precursor, whereas they were totally disappeared in the complex coimmunoprecipitated with RhoS. Based on this result, we clarified two important facts: 1) the transiently expressed PSMB5 is able to participate in the proteasome assembly pathway, thus verifying the model we used; and 2) the PSMB5 precursor associating with RhoS remains an “unincorporated” state in the cells, which is consistent with our novel finding in the preceding glycerol gradient analysis (Figure 7B). Finally, we found that in transfected MCF-7 cells, RhoS was just partially colocalized with PSMB5 in a perinuclear region, seemed to be the endoplasmic reticulum (ER), where the proteasome assembly is thought to take place (Fricke et al., 2007; Figure 7D). Collectively, these data testify that RhoS associates with only the unincorporated precursor of PSMB5 and suggest that it is mainly involved in the earlier steps of the proteasome biogenesis.
Figure 7.
Only unincorporated PSMB5 precursors associate with RhoS. (A) Schematic models showing the structures and autocleavage of different constructs used in this work. Pro, the propeptide in the precursor proteins. (B) Glycerol gradient analysis of the association between RhoS and PSMB5 in the biogenesis of proteasomes. Extracts were prepared from HEK293T cells cotransfected with Flag-Rhos and PSMB5-Myc. Two milligrams of cell lysate were then resolved on a linear glycerol gradient (10–40%), and the fractions were analyzed by Western blot using anti-Flag, anti-Myc, and anti-PSMA2 antibodies. P, precursor; M, matured. (C) Comparing the composition of the protein complexes associated with the RhoS, PSMB5 precursors alone, or both precursor and matured PSMB5 to define which type of PSMB5 associates RhoS. The proteins complex were immunoprecipitated from the indicated cells using anti-Flag antibody and resolved for detecting the existence of RhoS, PSMB5 precursor, PSMA2, and POMP individually. (D) Partial colocalization of RhoS and PSMB5 in the cells. MCF-7 cells coexpressing Flag-RhoS and PSMB5-Myc were fixed and probed with anti-Flag and anti-Myc antibodies. Nuclei were stained with DAPI. Bar, 19.7 μm.
3.7. RhoS Down-Regulates the Stability of PSMB5 Precursors
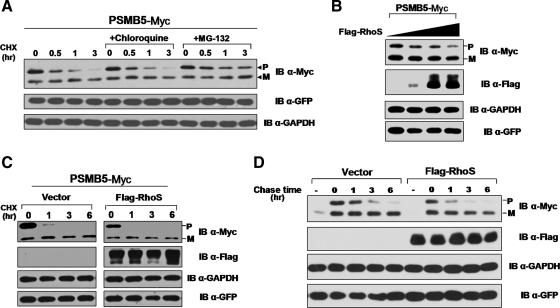
In previous studies, we noticed an intriguing fact that the protein level of PSMB5 precursors is remained very low in mammalian cells, compared with the mature form of PSMB5 (Hirano et al., , 2005, 2006). Despite the contribution of rapid turnover from the precursor to the mature proteins (Chen and Hochstrasser, 1996; Ramos et al., 1998), in our opinion, a lower level of PSMB5 precursors seems to be partially a result of its protein stability, significantly lower than the matured PSMB5. To verify this notion, we checked the stability of a PSMB5 precursor using CHX to block the protein synthesis in the cells. The PSMB5 precursor was obviously unstable, but could be stabilized only with MG-132, an inhibitor of the proteasome activity, whereas the mature form of PSMB5 behaved conversely (Figure 8A). In particular, the loss of the precursor was much greater than the increase in the mature protein, implying it is the proteasome-mediated protein degradation that plays major roles in decreasing the level of PSMB5 precursor. Most of the transiently expressed PSMB5 precursor stays in an unincorporated state and RhoS associates with this part of precursor in the cells (Figure 7). Thus, we wondered whether RhoS affects the protein stability of PSMB5 precursor. To this end, we first observed the effect of increased amounts of RhoS on the level of PSMB5. The level of PSMB5 precursor decreased significantly as a result of the increased dose of RhoS; in contrast, no disturbance was observed in the level of mature PSMB5 (Figure 8B). This result indicates that RhoS is involved in regulating the protein level of PSMB5 precursor rather than its autocleavage, which is consistent with the notion that it associates with only unincorporated part of the PSMB5 precursor. Second, we measured the stability of PSMB5 precursor under transient expression of RhoS. Overexpressed RhoS dramatically down-regulated the stability of the PSMB5 precursor and exhibited no significant effect on the stability of the mature PSMB5 (Figure 8C). To avoid any artifact interference from inhibiting protein synthesis, the stability of PSMB5 precursor was also measured in “steady state” based on an inducible expression system. As shown in Figure 8D, the expression of PSMB5 was efficiently repressed before adding the tetracycline and was very sensitive to the inducer, indicating the successful set-up of a system to study the stability of PSMB5. We found that the half-life of PSMB5 precursor was still short, even shorten in further under the overexpression of RhoS. These results suggest that RhoS down-regulates the stability of PSMB5 precursor.
Figure 8.
RhoS modulates the protein stability of PSMB5 precursors. (A) The PSMB5 precursor is a short-life protein, whose degradation is mediated by proteasomes. Thirty-six hours after transfection, HEK293T cells transiently expressing PSMB5-Myc were incubated with cycloheximide (CHX; 100 μg/ml) for the indicated time. For valuating the putative effect of the lysosome- and proteasome-mediated protein degradation on this process, the lysosome inhibitor, chloroquine (100 μM) and proteasome inhibitor, MG132 (25 μM) were included in the drugs treatment, respectively. The variation in the levels of both precursor and matured PSMB5 was measure by Western blot using anti-Myc antibody. P, precursor; M, matured. (B) The effect of increased expression of RhoS on the level of coexpressed PSMB5. Increasing amounts of RhoS were coexpressed with PSMB5-Myc. The cells were homogenized and analyzed 72 h after transfection. (C) Overexpression of RhoS accelerates the degradation of PSMB5 precursors. GFP and GAPDH serving as transfection efficiency and loading controls. (D) Observation of the effect of RhoS on the protein stability of PSMB5 precursors in a steady state. The selected Hela stable cell line capable of expressing PSMB5-Myc in response to the inducer with low background expression was used. After induction with tetracycline (Erschbamer et al., 2005; 1 μg/ml) for 4 h and prolonged Tet-free culture for 1 h, the cells were harvested at the indicated time points and subjected to Western blot analysis. The uninduced controls were included for monitoring the repression of background expression.
The PSMB5 Precursor Serves as a Unconventional Effector of RhoS
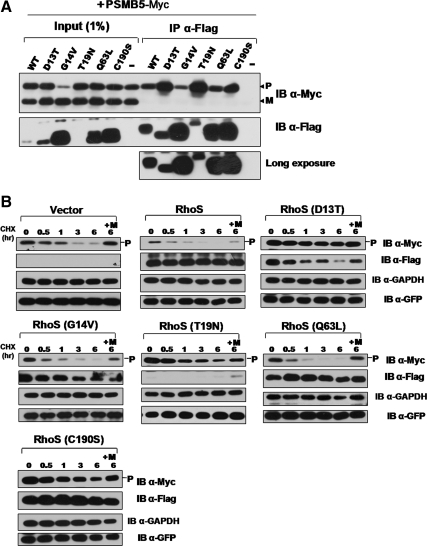
Most Rho GTPases act as signaling gates, switching on when bound to GTP (activated) and switching off when bound to GDP (inactivated). After specifically binding to the activated GTPases, various effector proteins could recognize and discriminate the differences in the conformation of GTPases with GTP loading (Bishop and Hall, 2000). To determine whether the PSMB5 precursor belongs to this kind of effectors of RhoS, we investigated the effect of GTP binding on the interaction between RhoS and the PSMB5 precursor. All of the dominant active or dominant negative mutants used in this part had already been validated in preceding works (Figure 2C). Strikingly, either dominant active (G14V and Q63L) or dominant negative RhoS mutants (D13T, T19N, and C190S) could be coIPed with PSMB5 precursors. However, there was a significant discrimination in the efficiency in coIP of the PSMB5 precursor between dominant negative and dominant active RhoS, suggesting the activation of RhoS must have something to do with the association between these two proteins (Figure 9A). Next, we changed our focus on testing the effect of RhoS activation on the stability of PSMB5 precursor. Intriguingly, all dominant negative RhoS mutants not only neutralized the effect of wild-type RhoS on PSMB5, but also somehow elevated the stability of PSMB5 precursor compared with the control group (empty vector). In contrast, compared with wide-type protein, all dominant active RhoS mutants exerted more vigorous, at least equal effect on the stability of PSMB5 precursor, indicating the GTP loading is a critical switch-on signal for modulating the PSMB5 precursor stability by RhoS (Figure 9B). Considered together, these observations suggest that the PSMB5 precursor serves as an unconventional effector of RhoS, thereby providing important complements to the classical regulation networks of Rho family GTPases.
Figure 9.
PSMB5 serves as a novel effector of RhoS. (A) GTP loading is not essential for the association between RhoS and PSMB5. The efficiency of RhoS and its mutants in coimmunoprecipitating of PSMB5 precursors were compared to evaluate the effect of GTP binding on the association between RhoS and PSMB5. (B) The activation of RhoS is a prerequisite for its activity in down-regulating the protein stability of PSMB5 precursors. The same group of RhoS mutants used in A was introduced into the protein stability assay based on CHX treatment as described above. Total lysates, 10 μg, were subjected to the Western blot analysis with GFP and GAPDH as transfection efficiency and loading controls. M, MG132.
DISCUSSION
Identification and characterization of differentially expressed genes in testis has provided us additional insight into the mechanisms of spermatogenesis (Lui and Cheng, 2008; White-Cooper, 2010).
In the present study, we identified a spermatogenesis stage–specific gene RSA-14-44 from rat germ cells. This gene encodes a protein containing a well-conserved Rho type structure (Figure 2A); thus we renamed it RhoS (Rho in spermatogenic cells). Enzymatic analysis of RhoS shows that it could catalyze the GTP hydrolysis like other members of Rho GTPase family (Figure 2B). Though RhoS shares high amino acid similarity with the canonical members of Rho GTPase family, it still reasonable for us to suspect that it might be a new member of this family, because sharing high structural similarity is never a surprise for the members of this family, especially for the Rho cluster (RhoA, RhoB, and RhoC; Takai et al., 2001). The most structural divergences in the members of Rho GTPase family focus two “hotspots”: one is in the “hypervariable region” and the other locate in the “insert loop” (Michaelson et al., 2001; Wheeler and Ridley, 2004). We observed significant and evolutionally conserved variations within these two structures in RhoS, distinct from the corresponding parts in the classical Rho isoforms (Figure 3, A and B; Supplemental Figure 3). However, in this family with “high similarity,” the more solid discrimination for each member relies on the differences in their expression patterns or their cellular activities. RT-PCR analysis showed that RhoS is a spermatogenic cell–specific gene, seriously distinct form the conventional Rho isoforms (Figure 4, A and B). Interestingly, when looking for the putative homologues of RhoS in other species by searching the NCBI database, we found that an uncharacterized gene, 4930544G11Rik, is considered to be the ortholog of RhoS in mice (mRhoS; Okazaki et al., 2002). Microarray data from the Genomics Institute of the Novartis Research Foundation (La Jolla, CA; Su et al., 2004) confirmed that the expression of 4930544G11Rik is also restricted to the testis. These results provide vigorous supports for us to propose that RhoS is a new and spermatogenesis-specific member of Rho GTPases family. Even though, much works needs to be done for thoroughly understanding the biological role of RhoS as a GTPase. For example, identifying the regulators like GEFs (guanine nucleotide exchange factors), GAPs (GTPase-activating proteins) and GDIs (guanine nucleotide dissociation inhibitors) specific for RhoS should be helpful to explore its related signaling pathways and regulation mechanisms.
Screening the RhoS-associated proteins in the testis led us to identify PSMB5, a catalytic subunit of the proteasome. Surprisingly, only the PSMB5 precursor associates with RhoS (Figure 9A) and is merely subject to the regulation of activated RhoS (Figure 9B). This is a novel finding, not only because so far there are few cases describing the direct association between Rho GTPases and proteasome subunits (Dong et al., 2004), but also because such a regulation mechanism is somehow unconventional for both Rho family GTPase and the proteasome biogenesis. However, considering Rho family GTPases as one of the most important molecular switches controlling nearly every basic cellular event, it is not surprising that a unique member of this family, RhoS participate in the proteasome biogenesis pathway. RhoS associates with PSMB5 in a novel pattern that is distinguished from the traditional ones in the cases of typical Rho GTPases and their effectors. In our opinion, there are three possible reasons to understand this phenomenon: 1) it might be a artifact resulted from the forced expressions of target proteins in cells; 2) PSMB5 may not be the direct effector of RhoS, and there should be some unidentified factor (or factors), most likely some enzymes that would be recruited by RhoS, and be capable of discriminating the signal from the activation of RhoS and converting it to the event of regulating the PSMB5 precursor stability; and 3) until now, most of literature on Rho GTPases family paid much more attention to the roles of a few classical members such as RhoA, Rac1, and Cdc42. This preference definitely brings us some interference to thorough understanding the biological activities of uncanonical Rho GTPases like RhoS.
Further evidences indicate that RhoS is involved in the regulation of the stability of PSMB5 precursor. Due to the high similarity between RhoS and other three Rho isoforms, i.e., RhoA, RhoB, and RhoC, we wondered whether this function is unique for RhoS. Actually, we found that except for RhoA, both RhoB and RhoC associate with the PSMB5 precursor and down-regulate its stability in the similar way as observed in RhoS (Supplemental Figures 4 and 5). In our opinion, there are two possibilities for explaining this phenomenon. The first one is that the high level of similarity between these Rho family members led to the functional redundancy. The region in RhoS responsible for its associating with PSMB5 was shown to be located in the conserved Rho GTPase domain (Figure 5B), which is shared by other three Rho isoforms. In fact, most RhoA targets identified so far also interact with RhoB and RhoC (Bryan and D'Amore, 2007). The other possibility is that RhoB and RhoC might be the functional ortholog of RhoS in somatic cells. This idea could be supported by two facts: 1) the protein level of the PSMB5 precursor is low in somatic cells where RhoB and RhoC, instead of RhoS, exist; and 2) RhoA, though sharing higher similarity in the structure with RhoS than RhoB and RhoC, showed less effect on the stability of the PSMB5 precursor.
Assembly of the proteasome is not a straightforward process. The complicated architecture of proteasomes justifies the great amount of energy consumed by the cell to assemble this degradation machinery correctly. Although the fundamental mechanisms of 20S proteasome assembly are well described, there is still at least one question remains to be answered: how to balance the synthesis of β subunits precursors and the proteasome assembly to keep unnecessary energy wastes as low as possible? Our finding provides some possible mechanisms for this challenge. First, the proteasome-mediated degradation of β subunit precursors (Figure 8A) would establish a feedback mechanism to realize the self-control of the proteasome level in cells. Second, the rapid autocleavage of β subunit precursors and the slow metabolism of assembled proteasomes make it possible that a low level of β subunit precursors is enough for maintaining the proper level of proteasomes. That is exactly what has been observed in this and previous studies (Figure 7B). Finally, the rapid and controlled degradation of the β subunit precursor could extremely minimize the accumulation of the unincorporated precursors, even the proteasome assembly was inhibited for some reason.
The composition and organization of the proteasome complex is highly dynamic (Matias et al., 2010). Recent studies indicate that a variety of additional or alternative subunits could be assembled into proteasomes resulting in a series of subtype proteasomes (Dahlmann et al., 2000; Merforth et al., 2003; Schmidt et al., 2006). These proteasome subtypes were mostly isolated from different tissues. For example, the β5t-containing subtype was isolated from the thymus (Murata et al., 2007). On the other hand, a series of specific events are interwoven in spermatogenesis, such as reduction of the chromosomal number from diploid to haploid in meiosis, remodeling of the nucleosomes by replacing histones with protamines, and formation of acrosomes (Trasler 2009). These processes definitely require highly efficient and dynamic protein metabolism, most of which is mediated by various types of proteasomes. We believe that the organization pattern of proteasomes in the spermatogenesis must be unique and dynamically adaptive to the complicated regulation networks. This notion is also supported by recent findings (Tengowski et al., 2007; Zhong and Belote, 2007). The question then is how to balance the formation of the specific proteasome subtypes such as immunoproteasome and the formation of the constitutive ones. Our data suggest that at least in mammalian spermatogenesis, RhoS serves as a switch for sequestering the unincorporated PSMB5 precursor from the proteasome assembly pathway. Once activated by intracellular or extracellular signals, RhoS could lower the pool of PSMB5 available for its assembly into the constitutive proteasomes, thus accelerating the formation of different proteasome subtypes. Intriguingly, some reports showed that extracellular stimuli such as IFN-γ could activate Rho family members (Badr et al., 2010; Utech et al., 2005). In addition, our unpublished data also support this model by demonstrating that the organization of proteasomes in the testis is indeed distinct from that in other tissues.
In summary, our present study provides evidence that a spermatogenesis specific Rho GTPase, RhoS serves as a switch for modulating the stability of a proteasome β subunit (PSMB5) precursor, hence participating in the regulation of proteasome biogenesis pathway. Further studies are needed to define the mechanism for this regulation and clarify its related signal networks.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the National Program for the Important Research Plan (2006 CB944002) and National Program for the Key Basic Research Project (2006 CB504001) from the Ministry of Science and Technology of China and by Grant 30721063 for the Creative Research Group from the National Nature Science Foundation of China and National Laboratory Special Fund (2060204).
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-04-0310) on October 27, 2010.
REFERENCES
- Adams J. The proteasome: structure, function, and role in the cell. Cancer Treat. Rev. 2003;29(Suppl 1):3–9. doi: 10.1016/s0305-7372(03)00081-1. [DOI] [PubMed] [Google Scholar]
- Adly M. A., Hussein M. R. Immunohistological profile of the Ras homologous B protein (RhoB) in human testes showing normal spermatogenesis, spermatogenic arrest and Sertoli cell only syndrome. Pathol. Oncol. Res. 2010;16:427–433. doi: 10.1007/s12253-009-9232-3. [DOI] [PubMed] [Google Scholar]
- Aki M., Shimbara N., Takashina M., Akiyama K., Kagawa S., Tamura T., Tanahashi N., Yoshimura T., Tanaka K., Ichihara A. Interferon-gamma induces different subunit organizations and functional diversity of proteasomes. J. Biochem. 1994;115:257–269. doi: 10.1093/oxfordjournals.jbchem.a124327. [DOI] [PubMed] [Google Scholar]
- Badr G., Saad H., Waly H., Hassan K., Abdel-Tawab H., Alhazza I. M., Ahmed E. A. Type I interferon (IFN-alpha/beta) rescues B-lymphocytes from apoptosis via PI3Kdelta/Akt, Rho-A, NFkappaB and Bcl-2/Bcl(XL) Cell Immunol. 2010;263:31–40. doi: 10.1016/j.cellimm.2010.02.012. [DOI] [PubMed] [Google Scholar]
- Belote J. M., Zhong L. Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis. Heredity. 2009;103:23–31. doi: 10.1038/hdy.2009.23. [DOI] [PubMed] [Google Scholar]
- Benetka W., Koranda M., Maurer-Stroh S., Pittner F., Eisenhaber F. Farnesylation or geranylgeranylation? Efficient assays for testing protein prenylation in vitro and in vivo. BMC Biochem. 2006;7:6. doi: 10.1186/1471-2091-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berruti G., Martegani E. The deubiquitinating enzyme mUBPy interacts with the sperm-specific molecular chaperone MSJ-1, the relation with the proteasome, acrosome, and centrosome in mouse male germ cells. Biol. Reprod. 2005;72:14–21. doi: 10.1095/biolreprod.104.030866. [DOI] [PubMed] [Google Scholar]
- Bishop A. L., Hall A. Rho GTPases and their effector proteins. Biochem. J. 2000;348(Pt 2):241–255. [PMC free article] [PubMed] [Google Scholar]
- Boureux A., Vignal E., Faure S., Fort P. Evolution of the Rho family of ras-like GTPases in eukaryotes. Mol. Biol. Evol. 2007;24:203–216. doi: 10.1093/molbev/msl145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991;349:117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Bryan B. A., D'Amore P. A. What tangled webs they weave: Rho-GTPase control of angiogenesis. Cell Mol. Life Sci. 2007;64:2053–2065. doi: 10.1007/s00018-007-7008-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustelo X. R., Sauzeau V., Berenjeno I. M. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P., Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- Chen X., Hu T., Liang G., Yang M., Zong S., Miao S., Koide S. S., Wang L. A novel testis protein, RSB-66, interacting with INCA1 (inhibitor of Cdk interacting with cyclin A1) Biochem. Cell Biol. 2008;86:345–351. doi: 10.1139/o08-072. [DOI] [PubMed] [Google Scholar]
- Cheng Y. Toward an atomic model of the 26S proteasome. Curr. Opin. Struct. Biol. 2009;19:203–208. doi: 10.1016/j.sbi.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlmann B., Ruppert T., Kuehn L., Merforth S., Kloetzel P. M. Different proteasome subtypes in a single tissue exhibit different enzymatic properties. J. Mol. Biol. 2000;303:643–653. doi: 10.1006/jmbi.2000.4185. [DOI] [PubMed] [Google Scholar]
- de Kretser D. M., Loveland K. L., Meinhardt A., Simorangkir D., Wreford N. Spermatogenesis. Hum. Reprod. 1998;13(Suppl 1):1–8. doi: 10.1093/humrep/13.suppl_1.1. [DOI] [PubMed] [Google Scholar]
- Dong J., Chen W., Welford A., Wandinger-Ness A. The proteasome alpha-subunit XAPC7 interacts specifically with Rab7 and late endosomes. J. Biol. Chem. 2004;279:21334–21342. doi: 10.1074/jbc.M401022200. [DOI] [PubMed] [Google Scholar]
- Ducummon C. C., Berger T. Localization of the Rho GTPases and some Rho effector proteins in the sperm of several mammalian species. Zygote. 2006;14:249–257. doi: 10.1017/S0967199406003790. [DOI] [PubMed] [Google Scholar]
- Eddy E. M. Male germ cell gene expression. Recent Prog. Horm. Res. 2002;57:103–128. doi: 10.1210/rp.57.1.103. [DOI] [PubMed] [Google Scholar]
- Erschbamer M. K., Hofstetter C. P., Olson L. RhoA, RhoB, RhoC, Rac1, Cdc42, and Tc10 mRNA levels in spinal cord, sensory ganglia, and corticospinal tract neurons and long-lasting specific changes following spinal cord injury. J. Comp. Neurol. 2005;484:224–233. doi: 10.1002/cne.20471. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A. Rho GTPases in cell biology. Nature. 2002;420:629–635. doi: 10.1038/nature01148. [DOI] [PubMed] [Google Scholar]
- Freeman E. A., Jani P., Millette C. E. Expression and potential function of Rho family small G proteins in cells of the mammalian seminiferous epithelium. Cell Commun. Adhes. 2002;9:189–204. doi: 10.1080/15419060216016. [DOI] [PubMed] [Google Scholar]
- Fricke B., Heink S., Steffen J., Kloetzel P. M., Kruger E. The proteasome maturation protein POMP facilitates major steps of 20S proteasome formation at the endoplasmic reticulum. EMBO Rep. 2007;8:1170–1175. doi: 10.1038/sj.embor.7401091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groll M., Nazif T., Huber R., Bogyo M. Probing structural determinants distal to the site of hydrolysis that control substrate specificity of the 20S proteasome. Chem. Biol. 2002;9:655–662. doi: 10.1016/s1074-5521(02)00144-8. [DOI] [PubMed] [Google Scholar]
- Heink S., Ludwig D., Kloetzel P. M., Kruger E. IFN-gamma-induced immune adaptation of the proteasome system is an accelerated and transient response. Proc. Natl. Acad. Sci. USA. 2005;102:9241–9246. doi: 10.1073/pnas.0501711102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendil K. B., Hartmann-Petersen R. Proteasomes: a complex story. Curr. Protein Pept. Sci. 2004;5:135–151. doi: 10.2174/1389203043379747. [DOI] [PubMed] [Google Scholar]
- Hirano Y., Hayashi H., Iemura S., Hendil K. B., Niwa S., Kishimoto T., Kasahara M., Natsume T., Tanaka K., Murata S. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol. Cell. 2006;24:977–984. doi: 10.1016/j.molcel.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Hirano Y., Hendil K. B., Yashiroda H., Iemura S., Nagane R., Hioki Y., Natsume T., Tanaka K., Murata S. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature. 2005;437:1381–1385. doi: 10.1038/nature04106. [DOI] [PubMed] [Google Scholar]
- Hori Y., Kikuchi A., Isomura M., Katayama M., Miura Y., Fujioka H., Kaibuchi K., Takai Y. Post-translational modifications of the C-terminal region of the rho protein are important for its interaction with membranes and the stimulatory and inhibitory GDP/GTP exchange proteins. Oncogene. 1991;6:515–522. [PubMed] [Google Scholar]
- Ihara K., Muraguchi S., Kato M., Shimizu T., Shirakawa M., Kuroda S., Kaibuchi K., Hakoshima T. Crystal structure of human RhoA in a dominantly active form complexed with a GTP analogue. J. Biol. Chem. 1998;273:9656–9666. doi: 10.1074/jbc.273.16.9656. [DOI] [PubMed] [Google Scholar]
- Khor B., et al. Proteasome activator PA200 is required for normal spermatogenesis. Mol. Cell. Biol. 2006;26:2999–3007. doi: 10.1128/MCB.26.8.2999-3007.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kierszenbaum A. L. Mammalian spermatogenesis in vivo and in vitro: a partnership of spermatogenic and somatic cell lineages. Endocr. Rev. 1994;15:116–134. doi: 10.1210/edrv-15-1-116. [DOI] [PubMed] [Google Scholar]
- Kleijnen M. F., Shih A. H., Zhou P., Kumar S., Soccio R. E., Kedersha N. L., Gill G., Howley P. M. The hPLIC proteins may provide a link between the ubiquitination machinery and the proteasome. Mol. Cell. 2000;6:409–419. doi: 10.1016/s1097-2765(00)00040-x. [DOI] [PubMed] [Google Scholar]
- Knaus U. G., Bamberg A., Bokoch G. M. Rac and Rap GTPase activation assays. Methods Mol. Biol. 2007;412:59–67. doi: 10.1007/978-1-59745-467-4_5. [DOI] [PubMed] [Google Scholar]
- Liang G., Zhang X. D., Wang L. J., Sha Y. S., Zhang J. C., Miao S. Y., Zong S. D., Wang L. F., Koide S. S. Identification of differentially expressed genes of primary spermatocyte against round spermatid isolated from human testis using the laser capture microdissection technique. Cell Res. 2004;14:507–512. doi: 10.1038/sj.cr.7290254. [DOI] [PubMed] [Google Scholar]
- Liu A. X., Rane N., Liu J. P., Prendergast G. C. RhoB is dispensable for mouse development, but it modifies susceptibility to tumor formation as well as cell adhesion and growth factor signaling in transformed cells. Mol. Cell. Biol. 2001;21:6906–6912. doi: 10.1128/MCB.21.20.6906-6912.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Huang W., Li C., Li P., Yuan J., Li X., Qiu X. B., Ma Q., Cao C. Interaction between c-Abl and Arg tyrosine kinases and proteasome subunit PSMA7 regulates proteasome degradation. Mol. Cell. 2006;22:317–327. doi: 10.1016/j.molcel.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Lui W. Y., Cheng C. Y. Transcription regulation in spermatogenesis. Adv. Exp. Med. Biol. 2008;636:115–132. doi: 10.1007/978-0-387-09597-4_7. [DOI] [PubMed] [Google Scholar]
- Lui W. Y., Mruk D. D., Cheng C. Y. Interactions among IQGAP1, Cdc42, and the cadherin/catenin protein complex regulate Sertoli-germ cell adherens junction dynamics in the testis. J. Cell. Physiol. 2005;202:49–66. doi: 10.1002/jcp.20098. [DOI] [PubMed] [Google Scholar]
- Mackay D. J., Hall A. Rho GTPases. J. Biol. Chem. 1998;273:20685–20688. doi: 10.1074/jbc.273.33.20685. [DOI] [PubMed] [Google Scholar]
- Matias A. C., Ramos P. C., Dohmen R. J. Chaperone-assisted assembly of the proteasome core particle. Biochem. Soc. Trans. 2010;38:29–33. doi: 10.1042/BST0380029. [DOI] [PubMed] [Google Scholar]
- Merforth S., Kuehn L., Osmers A., Dahlmann B. Alteration of 20S proteasome-subtypes and proteasome activator PA28 in skeletal muscle of rat after induction of diabetes mellitus. Int. J. Biochem. Cell Biol. 2003;35:740–748. doi: 10.1016/s1357-2725(02)00381-3. [DOI] [PubMed] [Google Scholar]
- Michaelson D., Silletti J., Murphy G., D'Eustachio P., Rush M., Philips M. R. Differential localization of Rho GTPases in live cells: regulation by hypervariable regions and RhoGDI binding. J. Cell Biol. 2001;152:111–126. doi: 10.1083/jcb.152.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D. C., Bryan B. A., Liu J. P., Liu W. B., Zhang L., Qu J., Zhou X., Liu M., Li D. W. Developmental expression of three small GTPases in the mouse eye. Mol. Vis. 2007;13:1144–1153. [PMC free article] [PubMed] [Google Scholar]
- Murata S., Sasaki K., Kishimoto T., Niwa S., Hayashi H., Takahama Y., Tanaka K. Regulation of CD8+ T cell development by thymus-specific proteasomes. Science. 2007;316:1349–1353. doi: 10.1126/science.1141915. [DOI] [PubMed] [Google Scholar]
- Naud N., Toure A., Liu J., Pineau C., Morin L., Dorseuil O., Escalier D., Chardin P., Gacon G. Rho family GTPase Rnd2 interacts and co-localizes with MgcRacGAP in male germ cells. Biochem. J. 2003;372:105–112. doi: 10.1042/BJ20021652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okazaki Y., et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- Ostrowska H., Kruszewski K., Kasacka I. Immuno-proteasome subunit LMP7 is up-regulated in the ischemic kidney in an experimental model of renovascular hypertension. Int. J. Biochem. Cell Biol. 2006;38:1778–1785. doi: 10.1016/j.biocel.2006.04.011. [DOI] [PubMed] [Google Scholar]
- Qiao Y., Yang J. X., Zhang X. D., Liu Y., Zhang J. C., Zong S. D., Miao S. Y., Wang L. F., Koide S. S. Characterization of rtSH3p13 gene encoding a development protein involved in vesicular traffic in spermiogenesis. Cell Res. 2004;14:197–207. doi: 10.1038/sj.cr.7290220. [DOI] [PubMed] [Google Scholar]
- Qiu X. B., Ouyang S. Y., Li C. J., Miao S., Wang L., Goldberg A. L. hRpn13/ADRM1/GP110 is a novel proteasome subunit that binds the deubiquitinating enzyme, UCH37. EMBO J. 2006;25:5742–5753. doi: 10.1038/sj.emboj.7601450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos P. C., Hockendorff J., Johnson E. S., Varshavsky A., Dohmen R. J. Ump1p is required for proper maturation of the 20S proteasome and becomes its substrate upon completion of the assembly. Cell. 1998;92:489–499. doi: 10.1016/s0092-8674(00)80942-3. [DOI] [PubMed] [Google Scholar]
- Ridley A. J. Rho family proteins: coordinating cell responses. Trends Cell Biol. 2001;11:471–477. doi: 10.1016/s0962-8924(01)02153-5. [DOI] [PubMed] [Google Scholar]
- Rivett A. J. Proteasomes: multicatalytic proteinase complexes. Biochem. J. 1993;291(Pt 1):1–10. doi: 10.1042/bj2910001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivkin E., Kierszenbaum A. L., Gil M., Tres L. L. Rnf19a, a ubiquitin protein ligase, and Psmc3, a component of the 26S proteasome, tether to the acrosome membranes and the head-tail coupling apparatus during rat spermatid development. Dev. Dyn. 2009;238:1851–1861. doi: 10.1002/dvdy.22004. [DOI] [PubMed] [Google Scholar]
- Rolland A. D., Jegou B., Pineau C. Testicular development and spermatogenesis: harvesting the postgenomics bounty. Adv. Exp. Med. Biol. 2008;636:16–41. doi: 10.1007/978-0-387-09597-4_2. [DOI] [PubMed] [Google Scholar]
- Rottinger E., Croce J., Lhomond G., Besnardeau L., Gache C., Lepage T. Nemo-like kinase (NLK) acts downstream of Notch/Delta signalling to downregulate TCF during mesoderm induction in the sea urchin embryo. Development. 2006;133:4341–4353. doi: 10.1242/dev.02603. [DOI] [PubMed] [Google Scholar]
- Sarkar A., Parikh N., Hearn S. A., Fuller M. T., Tazuke S. I., Schulz C. Antagonistic roles of Rac and Rho in organizing the germ cell microenvironment. Curr. Biol. 2007;17:1253–1258. doi: 10.1016/j.cub.2007.06.048. [DOI] [PubMed] [Google Scholar]
- Schmidt F., Dahlmann B., Janek K., Kloss A., Wacker M., Ackermann R., Thiede B., Jungblut P. R. Comprehensive quantitative proteome analysis of 20S proteasome subtypes from rat liver by isotope coded affinity tag and 2-D gel-based approaches. Proteomics. 2006;6:4622–4632. doi: 10.1002/pmic.200500920. [DOI] [PubMed] [Google Scholar]
- Schultz N., Hamra F. K., Garbers D. L. A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc. Natl. Acad. Sci. USA. 2003;100:12201–12206. doi: 10.1073/pnas.1635054100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sha J., Zhou Z., Li J., Yin L., Yang H., Hu G., Luo M., Chan H. C., Zhou K. Identification of testis development and spermatogenesis-related genes in human and mouse testes using cDNA arrays. Mol. Hum. Reprod. 2002;8:511–517. doi: 10.1093/molehr/8.6.511. [DOI] [PubMed] [Google Scholar]
- Solski P. A., Helms W., Keely P. J., Su L., Der C. J. RhoA biological activity is dependent on prenylation but independent of specific isoprenoid modification. Cell Growth Differ. 2002;13:363–373. [PMC free article] [PubMed] [Google Scholar]
- Su A. I., et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. USA. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y., Sasaki T., Matozaki T. Small GTP-binding proteins. Physiol. Rev. 2001;81:153–208. doi: 10.1152/physrev.2001.81.1.153. [DOI] [PubMed] [Google Scholar]
- Tanaka K. The proteasome: overview of structure and functions. Proc. Jpn. Acad. Ser. B. Phys. Biol. Sci. 2009;85:12–36. doi: 10.2183/pjab.85.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Tamura T., Yoshimura T., Ichihara A. Proteasomes: protein and gene structures. New Biol. 1992;4:173–187. [PubMed] [Google Scholar]
- Tengowski M. W., Feng D., Sutovsky M., Sutovsky P. Differential expression of genes encoding constitutive and inducible 20S proteasomal core subunits in the testis and epididymis of theophylline- or 1,3-dinitrobenzene-exposed rats. Biol. Reprod. 2007;76:149–163. doi: 10.1095/biolreprod.106.053173. [DOI] [PubMed] [Google Scholar]
- Tomaru U., et al. Exclusive expression of proteasome subunit {beta}5t in the human thymic cortex. Blood. 2009;113:5186–5191. doi: 10.1182/blood-2008-11-187633. [DOI] [PubMed] [Google Scholar]
- Trasler J. M. Epigenetics in spermatogenesis. Mol. Cell. Endocrinol. 2009;306:33–36. doi: 10.1016/j.mce.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Unno M., Mizushima T., Morimoto Y., Tomisugi Y., Tanaka K., Yasuoka N., Tsukihara T. Structure determination of the constitutive 20S proteasome from bovine liver at 2.75 A resolution. J. Biochem. 2002a;131:171–173. doi: 10.1093/oxfordjournals.jbchem.a003084. [DOI] [PubMed] [Google Scholar]
- Unno M., Mizushima T., Morimoto Y., Tomisugi Y., Tanaka K., Yasuoka N., Tsukihara T. The structure of the mammalian 20S proteasome at 2.75 A resolution. Structure. 2002b;10:609–618. doi: 10.1016/s0969-2126(02)00748-7. [DOI] [PubMed] [Google Scholar]
- Utech M., Ivanov A. I., Samarin S. N., Bruewer M., Turner J. R., Mrsny R. J., Parkos C. A., Nusrat A. Mechanism of IFN-gamma-induced endocytosis of tight junction proteins: myosin II-dependent vacuolarization of the apical plasma membrane. Mol. Biol. Cell. 2005;16:5040–5052. doi: 10.1091/mbc.E05-03-0193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega F. M., Ridley A. J. Rho GTPases in cancer cell biology. FEBS Lett. 2008;582:2093–2101. doi: 10.1016/j.febslet.2008.04.039. [DOI] [PubMed] [Google Scholar]
- Voges D., Zwickl P., Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- Walz J., Erdmann A., Kania M., Typke D., Koster A. J., Baumeister W. 26S proteasome structure revealed by three-dimensional electron microscopy. J. Struct. Biol. 1998;121:19–29. doi: 10.1006/jsbi.1998.3958. [DOI] [PubMed] [Google Scholar]
- Wheeler A. P., Ridley A. J. Why three Rho proteins? RhoA, RhoB, RhoC, and cell motility. Exp. Cell Res. 2004;301:43–49. doi: 10.1016/j.yexcr.2004.08.012. [DOI] [PubMed] [Google Scholar]
- White-Cooper H. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction. 2010;139:11–21. doi: 10.1530/REP-09-0083. [DOI] [PubMed] [Google Scholar]
- Wong E. W., Cheng C. Y. Polarity proteins and cell-cell interactions in the testis. Int. Rev. Cell. Mol. Biol. 2009;278:309–353. doi: 10.1016/S1937-6448(09)78007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zallen J. A., Peckol E. L., Tobin D. M., Bargmann C. I. Neuronal cell shape and neurite initiation are regulated by the Ndr kinase SAX-1, a member of the Orb6/COT-1/warts serine/threonine kinase family. Mol. Biol. Cell. 2000;11:3177–3190. doi: 10.1091/mbc.11.9.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B., Zheng Y. Regulation of RhoA GTP hydrolysis by the GTPase-activating proteins p190, p50RhoGAP, Bcr, and 3BP-1. Biochemistry. 1998;37:5249–5257. doi: 10.1021/bi9718447. [DOI] [PubMed] [Google Scholar]
- Zhang X., Liu H., Zhang Y., Qiao Y., Miao S., Wang L., Zhang J., Zong S., Koide S. S. A novel gene, RSD-3/HSD-3.1, encodes a meiotic-related protein expressed in rat and human testis. J. Mol. Med. 2003;81:380–387. doi: 10.1007/s00109-003-0434-y. [DOI] [PubMed] [Google Scholar]
- Zhong L., Belote J. M. The testis-specific proteasome subunit Prosalpha6T of D. melanogaster is required for individualization and nuclear maturation during spermatogenesis. Development. 2007;134:3517–3525. doi: 10.1242/dev.004770. [DOI] [PubMed] [Google Scholar]
- Zwickl P. The 20S proteasome. Curr. Top. Microbiol. Immunol. 2002;268:23–41. doi: 10.1007/978-3-642-59414-4_2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.