A selective tagging method for detecting minor alternatively-localized populations of a protein is used to study a disease-associated transmembrane form of prion protein. The analysis reveals key features of transmembrane prion protein metabolism and one way this is altered by human disease-causing mutants.
Abstract
Proteins are often made in more than one form, with alternate versions sometimes residing in different cellular compartments than the primary species. The mammalian prion protein (PrP), a cell surface GPI-anchored protein, is a particularly noteworthy example for which minor cytosolic and transmembrane forms have been implicated in disease pathogenesis. To study these minor species, we used a selective labeling strategy in which spatially restricted expression of a biotinylating enzyme was combined with asymmetric engineering of the cognate acceptor sequence into PrP. Using this method, we could show that even wild-type PrP generates small amounts of the CtmPrP transmembrane form. Selective detection of CtmPrP allowed us to reveal its N-terminal processing, long half-life, residence in both intracellular and cell surface locations, and eventual degradation in the lysosome. Surprisingly, some human disease-causing mutants in PrP selectively stabilized CtmPrP, revealing a previously unanticipated mechanism of CtmPrP up-regulation that may contribute to disease. Thus, spatiotemporal tagging has uncovered novel aspects of normal and mutant PrP metabolism and should be readily applicable to the analysis of minor topologic isoforms of other proteins.
INTRODUCTION
A characteristic feature of complex organisms is their ability to markedly diversify the protein products generated from a comparatively small number of genetic elements. Through the use of alternative promoter usage, alternative splicing, translational regulation, modification, processing, regulated degradation, and trafficking, a single gene may generate any of numerous potential protein products in a highly context dependent manner. While the major protein product is typically the most extensively studied, it is increasingly appreciated that minor or transiently generated alternative protein species often have critical functional roles.
For example, many proteins are primarily made and reside in an inactive form, with the active form represented by minor or transiently produced products. Hence, the minor species of key factors like steroid hormone receptors (Yudt and Cidlowski, 2002), membrane-bound transcription factors (Sannerud and Annaert, 2009), or caspases (Pop and Salvesen, 2009) are in fact the primary functional product. Even seemingly artifactual minor truncated versions of Glucocorticoid receptor resulting from internal translation initiation seem to be physiologically relevant functional products in vivo (Lu and Cidlowski, 2005; Lu et al., 2007). Thus, understanding the nuances and regulation of complex organism physiology will require an understanding of not only how proteomic complexity is generated, but the functional roles of the various major and minor products.
However, studying the alternative minor species of a protein often poses several technical obstacles depending on its comparative abundance and difference from the major species. In the simplest case, the minor species can be differentiated on the basis of a unique sequence element (e.g., generated by an isoform-specific exon) that can be immunologically detected with high specificity and sensitivity. However, this is not always possible. For example, alternative forms generated via facultative translocation or trafficking can generate minor variants whose main distinguishing feature is its alternative cellular location.
Indeed, proteins with targeting signals for the ER, mitochondria, peroxisomes, and chloroplasts can potentially reside in the cytosol as a minor (or ‘eclipsed’) nontranslocated form (reviewed by Regev-Rudzki and Pines, 2007). While this is sometimes apparently attributable to errors (Anandatheerthavarada et al., 2003; Drisaldi et al., 2003; Rane et al., 2004; Levine et al., 2005), numerous examples have been described where the noncompartmentalized form has a function (see Yogev et al., 2010, for an especially compelling recent example). This species can be generated by alternative splice variants that lack the targeting signal (Tong et al., 2003), alternative translation initiation sites downstream of the targeting signal (Land and Rouault, 1998; Goulet et al., 2004), modifications or processing close to the targeting signal (Addya et al., 1997; Robin et al., 2002; Colombo et al., 2005; Dasari et al., 2006; Boopathi et al., 2008), or just intrinsic inefficiencies in the targeting or translocation process (Stein et al., 1994; Sass et al., 2001; Shaffer et al., 2005; Regev-Rudzki et al., 2005; Naamati et al., 2009; Matthews et al., 2010). Studying these minor, differentially compartmentalized, and often transiently generated species has been challenging. Yet, they are nonetheless important. In cases where they represent mistakes, their quality control and degradation is likely to help maintain protein homeostasis in the cytosol. In cases where they have functional relevance, delineating their function requires their selective identification and analysis.
One example where alternative species are thought to be important is mammalian Prion protein (PrP). While the function of this widely expressed cell surface GPI-anchored glycoprotein remains to be clearly elucidated (Westergard et al., 2007; Aguzzi et al., 2008), it is best known for causing various fatal neurodegenerative diseases when misfolded in certain ways (Prusiner, 1998; Aguzzi and Calella, 2009). Recently, several cell biological studies have focused on the possibility that mislocalization of PrP can be detrimental to the cell and perhaps be a contributing factor in the pathogenesis of at least some of the associated diseases (reviewed in Chakrabarti et al., 2009). One source of PrP mislocalization stems from a slight inefficiency in the function of its N-terminal signal sequence (Kim et al., 2002; Rane et al., 2004; Levine et al., 2005). This typically results in ∼10% failed translocation into the ER (Rane et al., 2004; Levine et al., 2005), generating a minor cytosolic form of PrP termed cyPrP (Ma and Lindquist, 2002; Drisaldi et al., 2003; Rane et al., 2004). However, signal sequence inefficiency can have another outcome. If, after targeting to the translocon, the signal sequence fails to initiate translocation of the N terminus, an internal hydrophobic domain (HD, at residues ∼112–135) can engage the translocon and direct membrane insertion of PrP (Kim et al., 2001; Kim et al., 2002; Kim and Hegde, 2002; see Supplemental Figure S1). This transmembrane form, termed CtmPrP, has the N terminus in the cytosol and C terminus in the ER lumen (Hegde et al., 1998). Importantly, mutations that increase hydrophobicity of the hydrophobic domain result in increased CtmPrP production (Hegde et al., 1998; Kim et al., 2001; Stewart and Harris, 2001, 2003).
Studies in transgenic mice manipulating PrP translocation in several ways have illustrated that increased generation of either cyPrP or CtmPrP can cause neurodegeneration (Hegde et al., 1998, 1999; Ma et al., 2002; Stewart et al., 2005; Rane et al., 2008). Of note, several human disease mutations are in the central hydrophobic domain that, based on in vitro assays, result in increased CtmPrP (Hegde et al., 1998; Stewart and Harris, 2001; Kim and Hegde, 2002; Rane et al., 2010). One of these, A117V, has also been modeled in transgenic mice and shown to cause a late-onset neurodegeneration (Hegde et al., 1999; Yang et al., 2009; Rane et al., 2010) and increased CtmPrP (Hegde et al., 1999; Rane et al., 2010). As predicted from the above description of how cyPrP and CtmPrP are generated, improving signal sequence efficiency resulted in reduced generation of these forms in vitro (Kim and Hegde, 2002) and in transgenic mice (Rane et al., 2010), and rescued mice from neurodegeneration (Rane et al., 2010). Thus, CtmPrP and cyPrP, while minor species of PrP, may be of considerable importance to the pathogenesis of at least some diseases. However, these forms, particularly CtmPrP, are especially difficult to detect and follow in biochemical analyses, and this has severely limited insights into its biosynthesis, trafficking, and metabolism.
The difficulty in detecting and analyzing the minor cyPrP and CtmPrP forms is primarily an issue of signal to noise. In the case of CtmPrP, the polypeptide is identical (or nearly so) to the major fully translocated species: both are glycosylated and GPI-anchored, migrate similarly on SDS-PAGE, and are composed of the same polypeptide sequence (Hegde et al., 1998; Stewart et al., 2001; Kim and Hegde, 2002; Stewart and Harris, 2003). Thus, selective antibody detection is difficult. While conformation-specific antibodies are a theoretical possibility, this poses substantial technical hurdles given that the specific conformations are not easy to generate in high amounts as a source of antigens. Thus, to study CtmPrP, a method to selectively mark this minor species is needed. Here we have explored the possibility of using compartment-restricted and site-specific biotinylation as a method for selective tagging of CtmPrP. We characterize this method for use in mammalian cells and use it to delineate basic but key features of CtmPrP biosynthesis and metabolism. Illustrating the power of this approach, we show that, unexpectedly, CtmPrP metabolism is selectively perturbed by disease-associated mutants in PrP previously unknown to affect PrP topology.
MATERIALS AND METHODS
DNA Constructs
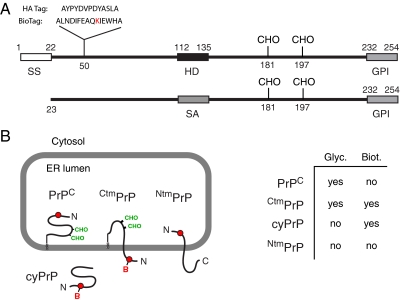
Wild-type Hamster PrP and the various derivatives from it [ΔSS-PrP, SA-PrP, Prl-PrP, PrP(G123P), PrP(AV3), Prl-PrP(G123P)] have been described (Hegde et al., 1998; Kim et al., 2002; Rane et al., 2004; Chakrabarti and Hegde, 2009). Wild-type Human PrP and its various disease-associated point mutants have been described (Ashok and Hegde, 2009). In each case, the BioTag or an HA epitope tag was inserted using synthetic oligos into the unique Bsu36I site at codon 50. Both epitopes were flanked by an Alanine on both sides. BirA was cloned by PCR from total E. coli K-12 DNA. An N-terminal FLAG tag was incorporated into the 5′ primer and the product inserted by standard methods into a mammalian (pCDNA3.1-based) expression vector. BirA for bacterial expression lacked the FLAG tag and was inserted into the His-tag containing pRSET-A vector. SS-BirA-KDEL was made by PCR amplifying the BirA open reading frame with the FLAG epitope and KDEL signal encoded in the 5′ and 3′ primers, respectively, and ligating to a mammalian expression plasmid containing the Prl signal sequence (Kim et al., 2002). For the purposes of topology analysis, we have not observed any differences between Hamster PrP and Human PrP. Human PrP was used in all of the experiments shown except in Figure 1 and Figure 2, where Hamster constructs were used.
Figure 1.
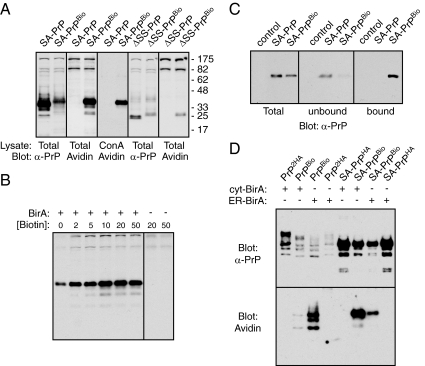
Experimental strategy for compartment-restricted biotinylation. (A) Diagram of PrP (top) and SA-PrP (bottom) indicating key elements: SS – signal sequence; HD – hydrophobic domain; CHO – glycosylation site; GPI – signal for glycosylphosphatidylinositol anchor attachment; SA – signal anchor (from Asialoglycoprotein receptor). In addition, the BioTag and control HA sequences are shown at their site of insertion. The Lysine that becomes biotinylated is in red. SA-PrP lacks an N-terminal signal sequence and is instead targeted to and inserted via the SA domain. The flanking charges ensure its type II orientation, resulting in the C terminus being translocated into the ER lumen. Numbering is for Human PrP. (B) Diagram of the four potential topologic forms that can be achieved by PrP, with the glycans indicated in green, and the BioTag in red. On introduction of BirA to the cytosol, cyPrP and CtmPrP become biotinylated (‘B’). A chart summarizing the modifications is shown.
Figure 2.
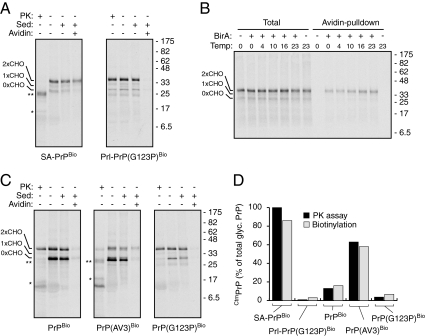
Biotinylation assay for PrP topology in vitro. (A) SA-PrPBio and Prl-PrP(G123P)Bio were in vitro translated and inserted into ER-derived microsomes using reticulocyte lysate and pancreatic microsomes. After translation, the samples were incubated for 1 h on ice with BirA and subjected to proteinase K (PK) digestion, sedimentation to isolate the microsomes, and/or pulldown using immobilized Avidin as indicated. The products were analyzed by SDS-PAGE and autoradiography. The positions of PrP containing 0, 1, or 2 glycans (CHO) are indicated. The double and single asterisks indicates the positions of glycosylated and unglycosylated C-terminal fragments generated by partial PK digestion of CtmPrP (Hegde et al., 1998). Note that SA-PrPBio is quantitatively in the CtmPrP topology as judged by PK digestion, and is efficiently biotinylated as judged by Avidin-pulldown. By contrast, Prl-PrP(G123P)Bio is quantitatively PK-protected, and fails to be biotinylated by BirA, both indicative of its complete translocation into the lumen of the microsomes. (B) Biotinylation reactions of in vitro translated and membrane inserted SA-PrPBio at various temperatures for 30 min, followed by Avidin-pulldowns as in panel A. (C) PK digestion and biotinylation assays for PrPBio, PrP(AV3)Bio, and PrP(G123P)Bio as in panel A. (D) Quantification of results from panels A and C. Plotted are the proportions of fully glycosylated PrP that is PK-accessible (black bars), or that is recovered by immobilized Avidin (gray bars), both of which are indicators of CtmPrP.
Antibodies
Anti-PrP antibodies used in this study were either PrP-A, a rabbit polyclonal against the PrP N terminus from residues 23–38 (Ashok and Hegde, 2008; Rane et al., 2008), or 3F4 (Covance, Princeton, NJ), a mouse monoclonal whose epitope is at residues 109–112. PrP-A recognizes all mammalian species of PrP and the SA-PrP construct. 3F4 recognizes Hamster and Human PrP, but not SA-PrP. These were used interchangeably with indistinguishable results, except in experiments with SA-PrP, in which case PrP-A was used. The TRAPα antibody has been described (Fons et al., 2003).
Cell Culture
Unless otherwise indicated, all experiments were performed in Neuro 2a (N2a) cells obtained from ATCC. Key results were confirmed to be reproducible in HeLa, HEK293, and HT1080 cells (unpublished results). To facilitate much of the routine characterization, N2a cells stably expressing BirA were produced by isolating and selecting for clones from single cells using standard methods. However, transient cotransfection with BirA produced identical results and stable expression was not essential. In instances where BirA was cotransfected, it occupied between one-sixth and one-fifth of the total DNA. Transfection was with either Lipofectamine 2000 or Effectene with similar results. Cells were assayed between 24 and 48 h after transfection. In nearly all experiments, a small amount (typically one-fifth of the total amount of the DNA) of a CFP or GFP expression plasmid was cotransfected to verify that efficiency of transfection was at least 70%. Although not shown, this was often used as an expression control. Experiments were typically performed on cells at ∼70–90% confluent. Pulse-chase studies were performed on cells in six-well dishes as described (Rane et al., 2004; Ashok and Hegde, 2008; Ashok and Hegde, 2009). Although adding exogenous biotin to the media was not absolutely necessary, the experiments here contained 10 uM biotin in the culture media that was typically added at the time of transfection.
In Vitro Biotinylation Assays
In vitro transcription, translation in reticulocyte lysate, and translocation using canine pancreatic rough microsomes has been described (Kim et al., 2002; Fons et al., 2003; Sharma et al., 2010). The protease protection assay for topology was as before (Kim et al., 2002). For biotinylation, translation reactions were placed on ice and recombinant BirA added to a final concentration of 50 ng/ul, which was determined in preliminary experiments to be the lowest concentration needed to give saturating biotinylation. Biotin was added to 50 uM, and the reaction was allowed to proceed on ice for 1 h. Experiments at various temperatures showed that higher temperatures were unnecessary and provided no additional modification. The reaction was then placed onto a sucrose cushion (of 0.5 M sucrose in PSB: 100 mM KAc, 50 mM HEPES, pH 7.4, 2 mM MgAc2) and the microsomes isolated by sedimentation (Kim et al., 2002). This was necessary to remove the free biotin, which otherwise competed in the subsequent avidin pulldown. The microsomes were solubilized in 1% SDS, 0.1M Tris, pH 8, denatured by boiling, diluted 10-fold in IP buffer (1% Triton X-100, 50 mM HEPES, pH 7.4, 100 mM NaCl), and incubated with 10 μl (packed volume) of immobilized Avidin (Pierce) that was prewashed in IP buffer. After incubation for 2 h with end-over-end rotation, the beads were washed three times in IP buffer and the bound products eluted by boiling in SDS-PAGE sample buffer.
Analysis of Biotinylated Products from Cells
For most analyses (unless specifically indicated), cells were washed twice in PBS and lysed by addition of 1% SDS, 0.1 M Tris, pH 8.0, and immediately denatured by boiling. The DNA was sheared by repeated boiling and vortexing until the viscosity was reduced. Typically, one well of a six-well dish was harvested in 150 μl. This method of rapid denaturation ensured that no post-lysis biotinylation could occur, as was confirmed by mixing experiments (Supplemental Figure S3). Almost all experiments used this strategy, except where detergent solubility of PrP products was being examined (e.g., Figure 7, A and E). In this case, cells were washed in PBS and lysed in ice-cold detergent buffer (typically 1 ml per well of a six-well dish) composed of 100 mM NaCl, 50 mM HEPES, pH 7.5, 0.5% Triton X-100, 0.5% Deoxycholate. Again, mixing experiments verified that under these conditions, post-lysis biotinylation was not detectable in the time frame of our experiments. These samples were then homogenized by repeated pipetting and/or passage through 23–25 gauge needles, and spun at maximum speed in a cold-room microfuge for 30 min. The supernatant and pellet were then solubilized in SDS-PAGE sample buffer. For avidin pulldowns, samples were adjusted with IP buffer to reduce the SDS concentration to 0.1%. Any insoluble material was sedimented for 10 min in a microfuge, and the supernatant incubated for 1 h at 4°C with plain sepharose beads to pre-clear the lysate. The supernatant from this pre-clearing step was then incubated with avidin-agarose (10 μl, pre-equilibrated in IP buffer) for 2 h at 4°C with constant mixing. The beads were washed four times in IP buffer, once in IP buffer containing 2 M urea (to reduce background), and eluted in SDS-PAGE sample buffer by boiling. Avidin blots used Avidin-HRP, using biotin-free purified BSA (3%) as a blocking reagent. Preliminary experiments were used to identify the optimal dilution of the Avidin-HRP to minimize background. To verify that recovery was uniform in all of the avidin pulldown samples (e.g., in a pulse-chase), each sample was spiked before the pulldown with a known quantity (typically 1 μg) of biotinylated BSA. This was insufficient to compete for binding to the beads but allowed confirmation of equal recovery and loading of all samples upon coomassie staining of the gel.
Figure 7.
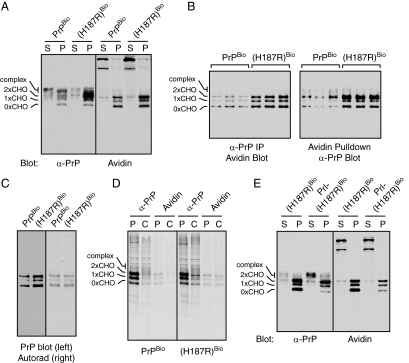
A disease-causing PrP mutant selectively alters CtmPrP metabolism. (A) BirA-expressing N2a cells were transfected with either PrPBio or PrP(H187R)Bio, lysed under nondenaturing conditions (with 0.5% Triton X-100 and 0.5% Deoxycholate), and separated into a detergent soluble fraction (S) and insoluble pellet (P). The samples were probed with anti-PrP or Avidin-HRP. (B) Total cell lysates in triplicate expressing BirA and the indicated construct were either immunoprecipitated with anti-PrP and blotted with Avidin-HRP (left) or pulled down with Avidin and blotted with anti-PrP (right). In both cases, the amount of biotinylated PrP was consistently higher for PrP(H187R)Bio. (C) BirA-expressing N2a cells were transfected with either PrPBio or PrP(H187R)Bio, pulse-labeled for 30 min, harvested, and pulled down with Avidin to isolate biotinylated products. The samples were separated by SDS-PAGE and transferred to nitrocellulose membrane. The membrane was probed with anti-PrP antibodies to detect total biotinylated products (left). After the blot, the membrane was dried and exposed to x-ray film to detect newly synthesized (i.e., radiolabeled) biotinylated products (right). (D) Pulse-chase analysis of PrPBio and PrP(H187R)Bio. Pulse labeling was for 30 min, and chase was for 7 h. Samples were divided in two and either immunoprecipitated with anti-PrP or pulled down with immobilized Avidin. (E) PrP(H187R)Bio and Prl-PrP(H187R)Bio were analyzed as in panel A.
Analysis of CtmPrP N-Terminal Processing
Cells in a six-well dish were pulse-labeled for 30 min with [35S]methionine (Ashok and Hegde, 2008) and harvested in denaturing conditions with 1% SDS, 0.1M Tris, pH 8. After boiling the sample, the material from three wells were pooled and diluted 10-fold in IP buffer. This was incubated with 100 μl of packed ConA-sepharose for 1.5 h with constant gentle mixing. The beads were washed twice in IP buffer and twice with IP buffer containing 0.5 M NaCl. The washed beads were resuspended in TEV digestion buffer (as per the manufacturer's directions) and incubated with TEV protease overnight at 4°C. The released products were separated from the beads and subjected to pulldown with Avidin-sepharose as above. The final products were analyzed by SDS-PAGE next to in vitro translated markers. To generate these markers, the relevant region of the PrPBio/TEV construct was PCR amplified with primers encoding an SP6 promoter in the 5′ primer and stop codon in the 3′ primer. The PCR product was then translated in reticulocyte lysate (Sharma et al., 2010). As a positive control to monitor the TEV digestion reaction, the above steps were performed but using immobilized anti-PrP antibodies (the 3F4 monoclonal; Covance) instead of ConA. After the digestion reaction on the IP samples, SDS-PAGE sample buffer was added and the total products analyzed directly.
Miscellaneous Biochemistry
Immobilized ConA was from GE Biosciences (Piscataway, NJ) and was used at 10 μl per 1-ml sample. Incubation was for 2 h at 4°C in IP buffer as for the Avidin pulldowns. Immunoprecipitations were as before and used the anti–PrP-A antibody (Ashok and Hegde, 2008) or 3F4 antibody (Covance). EndoH and PNGase treatments and analysis of PrP solubility in nondenaturing detergents was as before (Rane et al., 2004; Ashok and Hegde, 2008; Ashok and Hegde, 2009). BirA was expressed in E. coli BL21(DE3) pLysS cells with IPTG (1 mM) induction at midlog phase growth. Expression was for 3 h, after which the cells were harvested in 300 mM NaCl, 50 mM Tris, pH 8 and lysed by sonication. The His-tagged BirA was purified by immobilized Ni+2 or Co+2 affinity chromatography (using Chelating-Sepharose from GE), dialyzed against PBS, and stored in aliquots at −80°C. SDS-PAGE was on 12% Tris-Tricine gels, except the experiments in Figure 6, which were on 15% Tris-Tricine gels.
Figure 6.
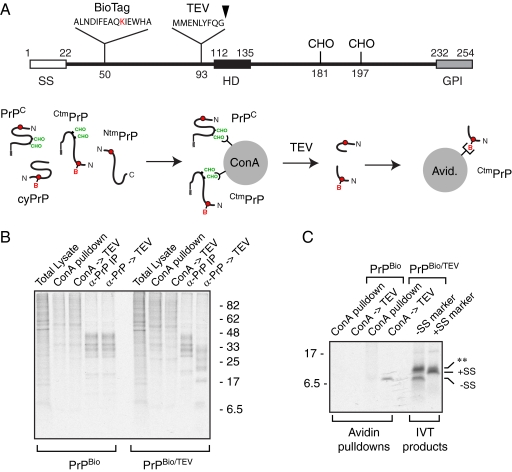
The signal sequence has been processed on CtmPrP. (A) Experimental strategy to isolate the N-terminal domain selectively from CtmPrP. Diagram of the TEV site inserted into PrPBio. The two Methionines preceding the TEV site are to allow labeling; the cleavage site is indicated with an arrowhead. The fate of the four topologic forms of PrP upon ConA pulldown, TEV elution, and Avidin pulldown are shown. The final fragment recovered should necessarily be from CtmPrP because it was selected for both glycosylation and biotinylation. (B) Characterization of the ConA pulldown and TEV cleavage steps. Pulse-labeled cell lysates expressing PrPBio without or with the TEV site were analyzed directly (total lysate), after ConA pulldown, after TEV cleavage of ConA-selected products (ConA→TEV), after anti-PrP immunoprecipitation (α-PrP IP), and after TEV cleavage of the PrP IP products (α-PrP→TEV). The products were visualized by autoradiography. Note that no digestion occurs in the absence of the TEV site, while all of the PrP products are digested efficiently by TEV. PrP is barely visible in the ConA-selected products, which contains many endogenous cellular glycoproteins. Analysis was on 15% Tricine gels to maximize separation of small products. (C) Samples treated as in panel B were further selected by Avidin pulldowns and visualized by autoradiography. Markers for PrP from residue 1 up to the TEV site (+SS) or residue 23 up to the TEV site (−SS) were produced by in vitro translation and analyzed on the same gel. Note that an unidentified background translation product was seen in both samples (asterisks). The source of this product is unknown, although the nuclease digested translation system likely contains numerous mRNA fragments, some of which may be translated to generate small truncated products not normally visualized at the bottom of a gel. Analysis of this experiment was on 15% Tricine gels.
Quantification
Autoradiographs were quantified by phosphorimaging on a Typhoon system, while images for the figures were produced by exposure of the same gel to film and scanning in Photoshop. Comparative quantification of western blots were by comparing samples to serial dilutions of standards that were run on the same blot.
RESULTS AND DISCUSSION
Experimental Strategy
To selectively identify and monitor CtmPrP, we exploited its unique topology relative to the other PrP forms: cytosolically exposed N-terminal domain with C-terminal domain in the ER lumen. Because the C-terminal domain has acceptor sites for N-linked glycosylation, its residence in the lumen can be readily monitored. To generate a similar topologic marker for the cytosolically exposed N terminus, we took advantage of orthogonal biotinylation of an artificial short sequence by the bacterial BirA biotin ligase (Schatz, 1993). This enzyme, which covalently attaches biotin to its substrate proteins, has been found by screening and refinement to biotinylate a short artificial acceptor sequence of 13 residues (Schatz, 1993; Beckett et al., 1999; hereafter termed the BioTag; see Figure 1A). This BioTag, which contains an acceptor lysine for biotinylation, is efficiently modified by BirA, is not found in any natural proteins, and is not recognized by mammalian biotinylating enzymes (Schatz, 1993; Beckett et al., 1999; Howarth et al., 2005; Kulman et al., 2007). This makes the BirA-BioTag system orthogonal, allowing the generation of a highly specific and easily detectable modification.
We therefore reasoned that PrP BioTagged within its N-terminal domain could allow selective CtmPrP detection (Figure 1B). When this Biotagged PrP (PrPBio) is expressed in cells containing BirA in the cytosol, only CtmPrP should be both biotinylated and glycosylated. The majority of PrP would be fully translocated into the ER lumen cotranslationally, thereby escaping biotinylation (but not glycosylation). By contrast, cytosolic PrP would be biotinylated but not glycosylated, and NtmPrP would be neither glycosylated nor biotinylated. Thus, using two topologically restricted modifications allows, in principle, the four potential forms of PrP to be discriminated (Figure 1B). In practice however, any potential inefficiencies in the modification reactions could make the lack of a modification difficult to interpret with certainty, especially when dealing with low abundance species. For this reason, and because of its clear physiological importance, we focused on CtmPrP, whose double modification can be interpreted with a high degree of confidence.
Our overall strategy was to develop and validate this compartment-restricted tagging strategy in vitro and in cells using variants of PrP with well-defined and uniform (or nearly uniform) topology. Once validated, we sought to then apply the method to wild-type PrP so that basic but critical unanswered questions about CtmPrP could be addressed. And finally, we investigated whether inherited mutants in human PrP that cause neurodegeneration might influence CtmPrP metabolism.
Analysis of Compartment-Restricted Biotinylation in Vitro
The 13-residue BioTag, with a flanking Alanine on either side, was inserted into PrP at codon 50, between the flexible N-terminal 27 residues and the octarepeat region. Insertion of a short tag in this region is not expected to have any effect on PrP based on earlier studies inserting either an epitope tag or GFP (Negro et al., 2001; Lorenz et al., 2002; Chakrabarti and Hegde, 2009; Ashok and Hegde, 2009). Thus, throughout this study, all constructs were modified at this site and the modified constructs indicated by a superscript ‘Bio’ (or ‘HA’ for the HA epitope tag) in the name. Two versions of PrP that are uniformly (or very nearly so) in the CtmPrP or fully translocated topologic forms were BioTagged and tested in an in vitro system (Figure 2A).
As characterized before (Chakrabarti and Hegde, 2009), SA-PrPBio translated in rabbit reticulocyte lysate in the presence of canine pancreatic rough microsomes was inserted efficiently into the membrane as judged by its glycosylation, cosedimentation with microsomes, protease accessibility of its N terminus, and protease protection of its C terminus. By contrast, Prl-PrP(G123P)Bio (containing the highly efficient signal sequence from Prolactin fused to PrP with a Gly to Pro change in the potential membrane-spanning region) was fully translocated into the lumen based on its glycosylation and complete protection from protease digestion (Figure 2A). These two constructs therefore serve as the two extremes in topology, representing the greatest and least amount of CtmPrP possible.
When these translation products were incubated with recombinant BirA, SA-PrP Bio but not Prl-PrP(G123P) Bio could be efficiently recovered with immobilized Avidin (Figure 2A). Importantly, capture with Avidin was dependent on adding BirA, required the BioTag, and could be competed by excess biotin (Figure 2B and unpublished data). Thus, BirA-dependent biotinylation of the BioTag can occur on CtmPrP, and this product could be selectively captured with immobilized Avidin. Quantification showed that if the extent of SA-PrP Bio capture by Avidin was defined as 100%, capture of Prl-PrP(G123P) Bio was ∼3%. Because this was roughly the same amount of translation product recovered in the absence of BirA, it presumably represents the background of this assay generated by low level nonspecific sticking of PrP to the Avidin resin. Relative to these standards, wild-type PrPBio showed ∼13% biotinylation of the glycosylated product, consistent with the fact that ∼16% of this band was found to be in the CtmPrP form by protease-protection assays (Figure 2, C and D). Importantly, PrP(AV3)Bio and PrP(G123P)Bio, mutants that respectively increase and decrease CtmPrP generation (Hegde et al., 1998), led to corresponding changes in biotinylation (Figure 2, C and D). Thus, biotinylation of the BioTag by cytosolic BirA, when combined with glycosylation, can be used as a specific, sensitive, and quantitative topologic reporter of CtmPrP.
Analysis of Compartment-Restricted Biotinylation in Vivo
To evaluate the suitability of using biotinylation as a means to detect CtmPrP in vivo, we coexpressed SA-PrPBio with BirA in N2a cells and analyzed total cell lysates for biotinylated products by blotting with Avidin-HRP. In preliminary experiments, we initially confirmed that cells expressing BirA without a BioTagged construct, or a BioTagged construct without BirA, showed the same weakly staining bands as observed in GFP-transfected control cells (data not shown). This indicated that BirA has no detectable targets endogenous to mammalian cells, and that the BioTag is not modified by endogenous enzymes. Similar results were seen in multiple cell types (data not shown), where the main endogenous products detected by Avidin-HRP were two high molecular weight bands characterized before as mitochondrial biotin-containing enzymes (Howarth et al., 2005).
When SA-PrPBio was introduced into BirA-expressing cells, it was clearly visualized on blots with both anti-PrP antibodies and Avidin-HRP (Figure 3A). SA-PrP biotinylation was entirely dependent on the BioTag, because SA-PrP lacking the BioTag was not detected by Avidin-HRP despite expression at high levels as judged by its detection with anti-PrP. The presence of biotin in the media maximized biotinylation of SA-PrPBio, but was not absolutely required (Figure 3B). Presumably, low level biotin in the serum and endogenous cellular biotin pools are sufficient. The biotinylated SA-PrPBio was confirmed to be glycosylated by its recovery with immobilized ConA (Figure 3A). Controls with glycosylated and unglycosylated PrP verified the glycan specificity of ConA binding under the conditions used in this study (Supplemental Figure S2). Furthermore, blotting of lysates before and after incubation with immobilized Avidin illustrated that most of the SA-PrPBio was biotinylated (Figure 3C). The residual amount remaining in the unbound fraction was likely due to inefficiency in Avidin binding using batch-pulldown. Thus, a construct expected to be in the CtmPrP form can be site-specifically and efficiently biotinylated in vivo with cytosolic BirA. That this biotinylated species is also glycosylated confirms its CtmPrP topology.
Figure 3.
Biotinylation assay for PrP topology in vivo. (A) SA-PrP and ΔSS-PrP lacking or containing the BioTag were transfected into BirA-expressing N2a cells and analyzed 24 h later. Total cell lysates or ConA-bound products were separated by SDS-PAGE, blotted, and probed with either anti-PrP antibodies (α-PrP) or Avidin-HRP as indicated. Note that low levels of endogenous PrP are visualized at ∼35 kDa in the ΔSS-PrP samples probed with α-PrP. (B) SA-PrPBio was transfected into N2a cells expressing or lacking BirA and cultured in the indicated concentrations of biotin for 24 h. Total cell lysates were analyzed for biotinylated products by blotting with Avidin-HRP. (C) N2a cells expressing BirA were transfected with either a GFP-expressing plasmid (control), SA-PrP, or SA-PrPBio. Total cell lysates were incubated with immobilized Avidin to pulldown the biotinylated proteins. Samples of the total lysate, unbound fraction, and bound fraction were analyzed by immunoblotting with anti-PrP. (D) The indicated constructs were transfected into N2a cells expressing either cytosolic BirA or SS-BirA-KDEL (ER-BirA). Total cell lysates were analyzed by blotting and probed for either total PrP (α-PrP) or biotinylated products (with Avidin-HRP). ‘2HA’ indicates a construct containing two tandem HA tags.
As with SA-PrPBio, a cytosolic version of PrP lacking its ER targeting signal (ΔSS-PrPBio) was also biotinylated by BirA in a BioTag-dependent manner (Figure 3A). This is noteworthy because nontargeted cytosolic PrP is rapidly degraded, and the ability to detect its biotinylation illustrates that even short-lived species that are likely chaperone-associated can be tagged by BirA.
We also examined the consequences of directing BirA to the ER lumen with an N-terminal signal sequence and C-terminal KDEL sequence (SS-BirA-KDEL). In principle, this should preclude biotinylation of a cytosolically exposed BioTag, while permitting biotinylation of a BioTag in the ER lumen. As expected, biotinylation of SA-PrPBio was clearly decreased (Figure 3D). In contrast, wild-type PrPBio, the majority of which is expected to be translocated completely into the ER lumen, was biotinylated to a greater extent by ER-targeted BirA than by cytosolic BirA (Figure 3D). While generally consistent with expectations, biotinylated and glycosylated SA-PrPBio was still detectable in cells expressing SS-BirA-KDEL. This could be due to either residual SS-BirA-KDEL in the cytosol (i.e., due to translocation inefficiency), small amounts of SA-PrPBio being fully translocated into the ER lumen, or post-lysis biotinylation.
We favor the first explanation for two reasons. First, sensitive reporter assays have previously shown that signal-containing proteins are never translocated with complete efficiency, leaving at least a few percent in the cytosol (Levine et al., 2005). Second, mixing experiments of cell lysates from BirA-expressing cells with lysates from PrPBio-expressing cells showed no detectable post-lysis biotinylation under either the denaturing or nondenaturing lysis conditions we used (Supplemental Figure S3). This is presumably because upon washing and lysis of the cells, the concentrations of BirA, substrate, and biotin are reduced many orders of magnitude below their cellular levels, making the reaction very inefficient even if nondenaturing conditions are used. Thus, we conclude that while the majority of SS-BirA-KDEL is translocated and resides in the ER lumen [where it is apparently quite active, as observed before (Mize et al., 2008)], sufficient amounts of it are mislocalized to the cytosol where it can partially biotinylate a cytosolically exposed BioTag. Because SS-BirA-KDEL does not represent an absolute compartment-restricted enzyme, we focused the remainder of our efforts on using cytosolic BirA.
Detection of CtmPrP Generated by Wild-Type PrP
The SA-PrP model protein illustrated that biotinylation combined with glycosylation provides a rigorous means to detect CtmPrP since each modification is strictly restricted to different compartments. We sought to apply this method to unambiguously address whether CtmPrP is a normal product of wild type PrP biosynthesis. Although CtmPrP is generated from wild-type PrP in vitro, it is a minor product that some have argued represents inefficiencies or mistakes of an in vitro reaction comprising heterologous components (such as reticulocyte lysate and microsomes from pancreas). Whether CtmPrP is generated in vivo has been debatable because the assays for its detection have limited sensitivity and rely on either cumbersome biochemical fractionation (such as isolation of microsomes) or relatively crude limited protease digestion assays.
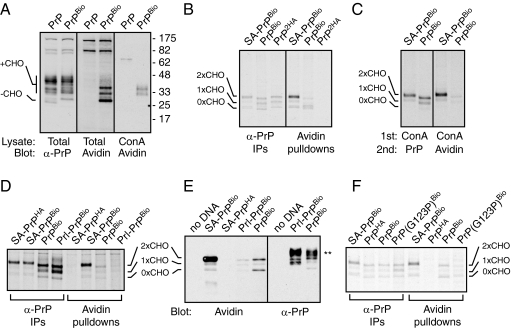
To address this issue, PrPBio was coexpressed with cytosolic BirA, and the products were analyzed by various methods. PrP, whether it contains or lacks the BioTag, is expressed as a heterogeneously glycosylated set of products, with a small amount of unglycosylated material (Figure 4A). Detection of the biotinylated species with Avidin-HRP showed several BioTag-dependent PrP species (Figure 4A). The smallest of these products comigrated with unglycosylated PrP and was verified as such by its lack of binding to immobilized ConA. Furthermore, this band was the most efficiently biotinylated PrP product relative to its abundance in the total PrP blot, suggesting a high degree of cytosolic exposure. These observations suggest this species represents cyPrP, although it may also include CtmPrP that failed to receive N-linked glycans. Among glycosylated PrP species, a small proportion (∼5–10%) was detected with Avidin-HRP. Most of these products could be pulled down with immobilized ConA and visualized by Avidin-HRP, confirming its dual modification with biotin and N-glycan, consistent with the CtmPrP topology.
Figure 4.
The biotinylation assay reveals CtmPrP generated by wild-type PrP. (A) Wild-type Human PrP constructs lacking or containing the BioTag were transfected into BirA-expressing N2a cells and analyzed 24 h later. Total cell lysates or ConA-bound products were separated by SDS-PAGE, blotted, and probed with either anti-PrP antibodies (α-PrP) or Avidin-HRP as indicated. The positions of glycosylated (+CHO) and unglycosylated (-CHO) PrP are indicated. (B) The indicated constructs were transfected into BirA-expressing N2a cells and the cells pulse-labeled with [35S]methionine for 30 min. Total cell lysates were then immunoprecipitated with anti-PrP antibodies or pulled down with immobilized Avidin, and the products visualized by SDS-PAGE and autoradiography. (C) An experiment as in panel B, except that the samples were first pulled down with immobilized ConA, after which the bound products were either immunoprecipitated with anti-PrP antibodies or pulled down with immobilized Avidin. (D) An experiment as in panel B was performed on the indicated constructs. (E) BirA-expressing N2a cells were transfected with the indicated constructs, and total cell lysates were analyzed by blotting with anti-PrP or Avidin HRP. The positions of the unglycosylated and core-glycosylated products are indicated. The double-asterisk indicates complex glycosylated PrP species. (F) An experiment as in panel B was performed on the indicated constructs.
In addition to detection of steady state levels of biotinylated PrP species, we could also selectively recover these forms from pulse-labeled cells (Figure 4B). As with the steady state analysis, capture of pulse-labeled PrP with immobilized Avidin was dependent on the BioTag. The assignment of bands as glycosylated and unglycosylated products was again confirmed using ConA binding (Figure 4C). Comparing the total pulse-labeled PrPBio (recovered using anti-PrP immunoprecipitation) to that recovered with immobilized Avidin showed that unglycosylated PrPBio was biotinylated to higher specific activity than glycosylated PrPBio. Relative to glycosylated SA-PrPBio, glycosylated PrPBio was biotinylated to ∼10-fold lower levels. Given that SA-PrPBio is uniformly in the CtmPrP topology, this indicates that only a small proportion of glycosylated PrPBio is represented by CtmPrP (∼10% in this experiment). Although the precise proportion of PrPBio in the CtmPrP topology varied depending on cell culture conditions, cell type, expression level, labeling time, and other parameters yet to be delineated, it was always many-fold lower than SA-PrP and was always a detectable but minor proportion of total synthesized PrP. These observations are consistent with unglycosylated PrPBio representing primarily cytosolic species, while the glycosylated PrPBio represents a mixture of fully translocated species with minor amounts of CtmPrP.
To further validate this conclusion, we tested whether biotinylation could be decreased by two independent manipulations that each reduce CtmPrP production. In the first strategy, we replaced the PrP signal sequence with that from Prolactin (Prl). Earlier studies have shown that both CtmPrP and cyPrP are dependent on slight inefficiency of the native PrP signal sequence, and that both can be reduced (although not completely eliminated) by the more efficient Prl signal (Kim and Hegde, 2002; Rane et al., 2004; Kang et al., 2006; Rane et al., 2010). Indeed, relative to PrPBio, Prl-PrPBio was reduced in the amount of both glycosylated and unglycosylated biotinylated PrP products recovered from pulse-labeled cells (Figure 4D). When total cell lysates were analyzed by blotting with Avidin-HRP, Prl-PrPBio again showed a substantially reduced biotinylated population compared with PrPBio despite equal or higher overall expression levels (Figure 4E).
In the second strategy, we analyzed PrP(G123P)Bio, which reduces CtmPrP levels by altering the potential membrane spanning region (e.g., Figure 2, C and D and Hegde et al., 1998). As with Prl-PrPBio, we found that biotinylation of PrP(G123P)Bio was reduced approximately threefold, very close to the limits of reliable detection above background. The reduced biotinylation seen with PrP(G123P)Bio is noteworthy because it indicates that little if any biotinylation is occurring during cotranslational protein targeting. If this were the primary source of biotinylated product, PrPBio and PrP(G123P)Bio would have been biotinylated equally since these two proteins are targeted by the same signal sequence with identical kinetics.
Three additional points argue against cotranslational biotinylation. First, SRP binds to the signal sequence immediately after its emergence from the ribosome (Krieg et al., 1986; Kurzchalia et al., 1986), even before the BioTag is exposed, thereby initiating the targeting process before the BioTag is available for modification. Second, earlier work in vitro and in vivo suggests that after the signal emerges from the ribosome, targeting occurs before even ∼30 additional residues are synthesized (Jungnickel and Rapoport, 1995; Goder et al., 2000). During this period in PrP translation, the BioTag would reside inside the ribosomal tunnel, where it cannot be accessed by the large ∼40 kDa BirA enzyme. Third, even if targeting were delayed, steric hindrance by the ribosome and SRP would likely preclude efficient access to the nascent PrP by BirA. For these reasons, both experimental and theoretical, we believe that biotinylation represents a post-translational event that is reporting on the final topologic location of the BioTag.
Thus, on the basis of dual modification by two topologically restricted enzymes (BirA and Oligosaccharyl transferase) and the results of various mutants that influence PrP topology [SA-PrP, ΔSS-PrP, Prl-PrP, and PrP(G123P)], we conclude that biotinylated and glycosylated PrPBio represents CtmPrP. While unglycosylated biotinylated PrP clearly includes cyPrP, it may well be a mixture with CtmPrP that failed to be glycosylated. Our ability to detect CtmPrP generated by wild-type PrP with high specificity at both steady state and by pulse-labeling paved the way to an analysis of this pathologically important form of PrP.
Characterization of CtmPrP Trafficking and Metabolism
With this assay for CtmPrP in hand, we addressed several basic, but still unresolved, questions regarding its normal biosynthesis and metabolism. Earlier studies using highly CtmPrP-favoring mutants [such SA-PrP or L9R-PrP(AV3)] have come to conflicting conclusions about the metabolism and cellular location of CtmPrP, ranging from ER, Golgi, or various other parts of the secretory pathway (Hegde et al., 1998; Stewart et al., 2001; Stewart and Harris, 2005; Chakrabarti and Hegde, 2009). However, these mutants are rather substantially modified, and it is unclear whether conclusions derived from them are entirely applicable. Whether CtmPrP generated from wild-type PrP ever leaves the ER, and if it does, whether it samples the cell surface, is unknown.
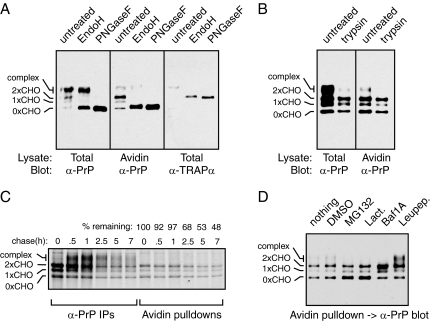
We found that the majority of biotinylated glycosylated PrPBio (i.e., CtmPrP) was sensitive to endoglycosidase H (EndoH) digestion (Figure 5A), indicating that most glycans on CtmPrP have not been processed by Golgi enzymes. However, a small amount of more slowly migrating bands was consistently observed, and these species were resistant to EndoH digestion, only being deglycosylated with PNGase F. The same species was also found to be accessible to extracellular trypsin, while the majority of biotinylated PrP was inaccessible (Figure 5B). When total PrP in the same samples was analyzed by blotting for PrP, the majority of PrP was EndoH resistant and cell surface exposed (Figure 5, A and B). These observations indicate that in contrast to the major population of PrP, which is at the cell surface, CtmPrP is primarily intracellular, possibly in the ER as judged by its EndoH-sensitive glycans. However, at least some CtmPrP clearly leaves the ER and goes to the cell surface as evidenced by an EndoH-resistant population accessible to extracellular trypsin.
Figure 5.
Analysis of CtmPrP metabolism. (A) Cells expressing wild-type PrPBio and BirA were either left untreated or digested with extracellular trypsin. After washing and collecting the intact cells, they were harvested and either analyzed directly, or after capture of biotinylated products with immobilized Avidin. The blot was probed with anti-PrP antibodies. (B) Total lysates from cells expressing PrPBio and BirA were either left untreated or digested with Endoglycosidase H or PNGase F as indicated. The samples were then analyzed directly or after capture of biotinylated products with immobilized Avidin. The blot was probed with anti-PrP antibodies to visualize PrP, or anti–TRAPα, an ER-resident glycoprotein that serves as a control for the glycosidase digestions. (C) Cells expressing PrPBio and BirA were pulse labeled with [35S]methionine for 30 min and chased with unlabeled media for the indicated times. Lysates harvested at each time point were divided in half and immunoprecipitated with anti-PrP or pulled down with immobilized Avidin. The percent of glycosylated biotinylated PrP species remaining at each time point was quantified by phosphorimaging and indicated above the respective lanes. (D) Cells expressing PrPBio and BirA were treated for 16 h with the indicated compounds: 10 uM MG132, 250 nM Bafilomycin 1A, 5 uM Lactacystin, or 0.2 mM Leupeptin. DMSO served as a vehicle control. Total cell lysates were subjected to Avidin pulldowns and the biotinylated products visualized with anti-PrP immunoblotting.
We also noticed in the course of our studies that CtmPrP was more difficult to solubilize in nondenaturing detergents than non-Ctm forms (unpublished observations; see also Figure 7A below). While insolubility could mean aggregation, we do not believe this to be the case because aggregates of a membrane protein would be unlikely to traffic through the secretory pathway and be modified on its glycans en route. Rather, it seems that two modes of membrane anchoring on the same molecule (via a transmembrane segment and GPI anchor) make it more difficult to solubilize. Regardless of the reason, this observation provides a further point of distinction between CtmPrP and the major PrP species. It also emphasizes the need to use efficient solubilization methods (e.g., SDS) to avoid losing subpopulations of PrP in any biochemical analyses. Indeed, standard lysis buffer conditions are insufficient to recover CtmPrP, perhaps explaining some of the earlier conflicting reports on its properties. Thus, unless specifically used for fractionation purposes, all of our experiments were harvested and solubilized in boiling SDS before further analysis to ensure complete recovery of all PrP species.
Using pulse-chase analysis combined with Avidin pulldowns, we estimated the half-life of CtmPrP as being ∼5–7 h (Figure 5C). Consistent with the steady state analysis, most PrP synthesized during the pulse-labeling subsequently matured to heterogeneous complex glycosylated forms, after which it was degraded. By contrast, the majority of CtmPrP remained core-glycosylated. By the pulse-chase method, we found it difficult to reliably detect a biotinylated complex glycosylated species due to a low signal from this diffuse band relative to background. However, the steady state analysis does indicate that some proportion of CtmPrP does undergo complex glycan modification and traffic to the cell surface (Figure 5, A and B). It is worth noting that although most of CtmPrP remains core-glycosylated, its residence in the ER cannot be assumed because glycoproteins can be trafficked through the Golgi without necessarily obtaining further glycan modifications. At this point, we do not know the full itinerary of CtmPrP trafficking, although it does appear to sample much of the secretory and endocytic pathways given its eventual degradation in the lysosome (see below).
We used inhibitors to assess the normal degradation pathway used by CtmPrP. We focused on the ratio of glycosylated biotinylated products (i.e., CtmPrP) to unglycosylated biotinylated PrP (i.e., primarily cyPrP), because the latter is well established to be degraded by the proteasome. Relative to untreated cells (where the CtmPrP and cyPrP bands are roughly equal at steady state), proteasome inhibition (with either MG132 or Lactacystin) clearly results in cyPrP levels exceeding CtmPrP (Figure 5D). By contrast, lysosomal inhibition with either Bafilomycin A1 or Leupeptin resulted in increased CtmPrP relative to cyPrP (Figure 5D). These results indicate that while cyPrP is primarily degraded by a proteasome-dependent pathway, CtmPrP is primarily degraded in lysosomes.
And finally, we wished to assess the status of signal sequence processing on CtmPrP. Although a seemingly esoteric point, this issue is important for two reasons. First, it is quite unusual for a signal sequence containing protein to be generated in a topology where the N terminus is cytosolic. This necessarily means that the polypeptide segment downstream of the signal sequence has failed to be translocated into the ER, a fate inconsistent with signal sequence function. The step at which this fails in vivo has been unclear. If it fails at the step of targeting to or initial gating of the translocon, the signal sequence will remain unprocessed. However, slippage of the polypeptide after initiating translocation could result in signal cleavage if it has transiently accessed the ER lumen where the signal peptidase active site resides. The second reason for assessing this is that because the signal sequence is hydrophobic, its lack of processing on CtmPrP would generate a domain that may be prone to aggregation and/or inappropriate interactions that could play a role in cellular dysfunction caused by CtmPrP. While analysis of a CtmPrP-favoring mutant showed lack of signal sequence processing (Stewart et al., 2001), this construct contains a mutation in the signal sequence itself that may have affected the results.
To address this issue, we initially isolated CtmPrP on the basis of its glycosylation and biotinylation and analyzed its migration relative to signal cleaved and uncleaved standards (data not shown). However, interpretation of this result proved more complicated than expected because of uncertainties about the effect of GPI anchor processing and deglycosylation (which converts Asparagines to Aspartates) on migration. To avoid these complications, we introduced a TEV cleavage site in the N-terminal domain downstream of the BioTag. After binding the glycoproteins (which would include CtmPrP) to immobilized ConA, the N-terminal domain of PrPBio/TEV was liberated by digestion with TEV and the biotinylated N terminus (which necessarily is from CtmPrP) was purified with immobilized Avidin (Figure 6A). This short product was then compared with signal-containing and signal-lacking versions of the fragment generated in vitro.
We first confirmed that TEV would specifically and efficiently cleave immobilized PrP on beads. To do this, PrP was immunoprecipitated with immobilized antibodies and after washing, incubated with TEV protease. PrPBio/TEV, but not PrPBio, was efficiently digested to yield the expected cleavage products (Figure 6B). Parallel samples on immobilized ConA also showed evidence that PrPBio/TEV was being digested similarly (Figure 6B). When the released products from the ConA digestion were further purified on immobilized Avidin, an ∼7-kDa band was specifically recovered in a TEV protease and PrPBio/TEV dependent manner (Figure 6C). Comparison of this product to in vitro translation products of signal-containing and signal-lacking fragments showed that the TEV-cleaved product from in vivo generated CtmPrP comigrates with the signal-lacking marker fragment (Figure 6C). Thus, CtmPrP generated in vivo appears to have its signal sequence removed, suggesting that it is generated by slippage of the N terminus out of the translocon at a step after the polypeptide has transiently accessed the ER lumen. The absence of this hydrophobic peptide may explain why CtmPrP is not recognized as a misfolded substrate for degradation by ER-associated degradation, while earlier studies generating CtmPrP by mutating the signal sequence (which precluded its processing) came to different conclusions (Stewart et al., 2001).
A Disease-Causing PrP Mutant that Alters CtmPrP Metabolism
An increased proportion of PrP is generated in the CtmPrP form when residues in the central hydrophobic domain (HD) are mutated to more hydrophobic residues. This includes artificial mutants such as PrP(AV3), in which three alanines are changed to valines, as well as several naturally occurring disease-associated human PrP mutants (such as A117V; Hegde et al., 1998; Hegde et al., 1999; Kim et al., 2001; Stewart and Harris, 2003). Based on analyses of such HD mutants in transgenic mice, increased levels of CtmPrP have been linked to the development of neurodegeneration (Hegde et al., 1998; Hegde et al., 1999; Stewart et al., 2005; Yang et al., 2009; Rane et al., 2010). It is therefore thought that CtmPrP may contribute to at least some genetic human disease caused by such PrP mutations in the central HD. However, numerous PrP mutants lie outside the HD and do not increase CtmPrP production at the ER as assayed in vitro (Stewart and Harris, 2001). Indeed, our own in vitro analysis of two such mutants [PrP(H187R) and PrP(E200K)] by both protease-protection and biotinylation assays confirmed this conclusion (Supplemental Figure S4). How such mutations lead to disease is unknown. Given that CtmPrP is capable of causing disease, we decided to ask whether in vivo, any of the non-HD mutants might somehow affect CtmPrP metabolism.
An initial screen using the biotinylation assay of most of the inherited disease-causing PrP mutants suggested that several (but not all) mutants have somewhat increased biotinylated products (unpublished observations). Among these, PrP(H187R) was especially obvious, and we therefore focused further efforts on this mutant. Our earlier analysis of PrP(H187R) had shown that relative to wild-type PrP, a greater proportion of the mutant is detergent insoluble, and this population is specifically enriched in immaturely glycosylated, EndoH-sensitive, intracellularly localized species (Ashok and Hegde, 2009). Because these properties are in many ways similar to the behavior of CtmPrP (Figure 5), we asked if the mutant-enriched detergent insoluble fraction contained CtmPrP as judged using our biotinylation assay.
When PrPBio and PrP(H187R)Bio cell lysates were fractionated on the basis of detergent solubility and probed for biotinylated species, nearly all of the biotinylated species in both samples was insoluble, and clearly increased for PrP(H187R)Bio (Figure 7A). This difference was consistently seen regardless of how it was visualized: PrP immunoprecipitates probed with Avidin-HRP, and Avidin-pulldowns probed with anti-PrP both showed increased biotinylated products for PrP(H187R)Bio (Figure 7B). This suggested that the total amount of CtmPrP (and cyPrP) was greater for PrP(H187R) than for PrP. Yet, in vitro translocation analysis of PrP(H187R) did not show increased production of either CtmPrP or cyPrP (Supplemental Figure S4). To resolve this discrepancy, we pulse-labeled cells and used Avidin-pulldowns to visualize the cyPrP and CtmPrP being produced by PrPBio and PrP(H187R)Bio. An autoradiograph of the pulldowns showed that both the wild type and mutant generate comparable amounts of biotinylated species after a 30 min pulse labeling (Figure 7C). When the exact same samples were instead blotted with anti-PrP antibodies to visualize total cellular levels of biotinylated product, PrP(H187R)Bio clearly had ∼2–3-fold more than PrPBio. Thus, PrP and PrP(H187R) produce biotinylated species (i.e., CtmPrP and cyPrP) at similar rates, consistent with in vitro translocation analyses; yet, the mutant has higher steady state levels. This strongly suggested that the turnover of the mutant CtmPrP and cyPrP is slower compared with wild-type CtmPrP and cyPrP.
To test this directly, we performed pulse-chase analysis (Figure 7D). When total PrP was analyzed by anti-PrP immunoprecipitation, a subtle difference was observed between PrPBio and PrP(H187R)Bio wherein some bands were slightly overrepresented in the chase sample of the mutant (as has been previously observed; Ashok and Hegde, 2009). Pulldowns with Avidin of these same samples revealed the overrepresented species to be biotinylated. While the biotinylated species were reduced by ∼50% at the 7 h chase time for PrPBio, it was hardly diminished (<20%) for PrP(H187R)Bio. Although the absolute amount of biotinylated species varied somewhat from experiment to experiment, their selective stabilization with the mutant was consistently observed (Supplemental Figure S5). Thus, we conclude that the H187R mutation has the effect of stabilizing the turnover of CtmPrP, and possibly also cyPrP.
To further validate this conclusion, we asked whether improving signal sequence efficiency, which reduces CtmPrP generation, would at least partially normalize the steady-state levels of detergent-insoluble immature species of PrP(H187R). Indeed, replacement of the PrP signal sequence with that from Prl did reduce the detergent insoluble, immaturely glycosylated species of PrP(H187R), with a corresponding increase in mature glycosylated soluble forms (Figure 7E). Had these mutant-specific aberrant species been generated from fully translocated forms of PrP (e.g., via their misfolding), it is difficult to see how they could have been partially rescued by improving signal sequence efficiency.
Thus, we conclude that among the aberrant species generated by PrP(H187R), CtmPrP is likely to be a contributor, despite the fact that the mutation does not increase CtmPrP production. Rather, it appears that the mutation selectively stabilizes, at least partially, the CtmPrP form to increase its steady state levels and perhaps its trafficking. Given the association between CtmPrP and neurodegeneration, it is attractive to speculate that at least one mechanism contributing to disease caused by these mutants is via increased levels of CtmPrP, which among other things may titrate away key cellular factors important for neuronal function (Chakrabarti and Hegde, 2009).
Although we have not yet done a complete analysis of all mutants or many cell types, we are able to confirm that the selective CtmPrP stabilization is seen in other cell types and for at least one other disease-causing mutation (Supplemental Figure S6). Hence, in HEK293, we again observed increased biotinylated glycosylated PrP(H187R)Bio. In this case, the unglycosylated species was not noticeably increased, perhaps because the cytosolic quality control systems are somewhat different in these cells. In addition to H187R, another disease-causing mutant, PrP(E200K), also showed selective increase in glycosylated biotinylated species (Supplemental Figure S6). Similar results were obtained in HeLa and HT1080 cells (data not shown). Thus, mutants in the PrP C terminus can selectively influence the metabolism of CtmPrP, resulting in its increased levels at steady state. Importantly, not all mutations in the C terminus can be explained by this mechanism. For example, we found that PrP(V210I) is unchanged in the amount of CtmPrP at steady state (data not shown), despite the fact that relative to wild type, a substantial amount of it is immature and detergent insoluble (Ashok and Hegde, 2009). Thus, there appear to be multiple effects of mutations in the C-terminal domain, and the relatively crude parameters of detergent insolubility and immature glycans cannot be used to equate all of the mutants. However, the more precise compartment-restricted biotinylation assay for CtmPrP allowed us to reveal a novel mechanism by which at least some mutants of PrP influence its metabolism.
Conclusions
We have illustrated the ability of site-specific biotinylation to covalently mark the cytosolically exposed population of a protein ordinarily directed to the secretory pathway. In the case of PrP, this method could be combined with an ER-specific modification (glycosylation) to selectively identify and track the low-abundance but biologically important CtmPrP species. This method substantially improves upon other methods to detect CtmPrP, which are either biochemically cumbersome (e.g., topology assays on ER microsomes) or based on a faulty premise (such as lack of signal sequence processing). The method is bio-orthogonal, has a high signal-to-noise, can be readily applied to other systems, and works in formats that allow both steady state analysis and pulse-chase studies. While there are endogenous biotinylated proteins, they do not seem to pose a major source of background in our biochemical assays. Furthermore, it is straightforward to pre-enrich for one's protein of interest (e.g., with immunoprecipitation or ConA in the case of CtmPrP) before detecting biotinylation to markedly improve signal-to-noise. While direct spatial visualization by fluorescence (e.g., using fluorophore-labeled Avidin on fixed cells) is also feasible, this remains to be investigated for the PrP system.
The utility of this method is exemplified by several new insights into CtmPrP biosynthesis and metabolism. Not only were we able to directly detect the CtmPrP population of wild-type PrP, but we also could show that it is a relatively stable membrane protein that samples much of the secretory pathway including the cell surface. The finding that CtmPrP is not rapidly degraded from the ER is especially noteworthy, because it suggests that this species may not be recognized as aberrant by the cellular quality control machinery. Indeed, the elements involved in CtmPrP production (the signal sequence and central hydrophobic domain) are highly conserved, and it is possible that it has a normal function that remains to be elucidated. In this scenario, only under conditions of excess production or stabilization in certain cell types would CtmPrP be detrimental. The ability to set up a system for selectively isolating this population (e.g., by ConA followed by Avidin affinity steps) now paves the way to determining whether its levels vary in vivo in a cell type–specific, aging-dependent, or disease-dependent manner in mice engineered to express PrPBio and BirA. That CtmPrP is degraded in lysosomes may suggest a means by which accumulation of transmissible prions could lead to increased CtmPrP (Hegde et al., 1999). In this view, prion accumulation would lead to lysosomal dysfunction, which would delay CtmPrP degradation. Being able to follow CtmPrP metabolism should permit this and related hypotheses to be tested.
At least in cell culture, the ability to readily detect CtmPrP revealed an unexpected mechanism leading to its increased levels. Here, mutants in the C-terminal domain of PrP act preferentially on the CtmPrP form to stabilize it. This finding would have been very difficult to obtain by other means given the rather subtle effect and the difficult biochemical properties of CtmPrP. Whether this stabilization is due to increased aggregation, altered trafficking, or another reason remains to be determined. However, the increase, while relatively small, is nonetheless likely to be important given that even small increases in CtmPrP seem to be capable of causing neurodegeneration in transgenic mouse models (Hegde et al., 1998; Hegde et al., 1999; Rane et al., 2010).
Supplementary Material
ACKNOWLEDGMENTS
We thank members of the Hegde Lab, particularly Aarthi Ashok, for useful discussions. This work was supported by the Intramural Research Program of the National Institute of Child Health and Human Development at the National Institutes of Health.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-09-0742) on October 27, 2010.
REFERENCES
- Addya S., Anandatheerthavarada H. K., Biswas G., Bhagwat S. V., Mullick J., Avadhani N. G. Targeting of NH2-terminal-processed microsomal protein to mitochondria: a novel pathway for the biogenesis of hepatic mitochondrial P450MT2. J. Cell Biol. 1997;139:589–599. doi: 10.1083/jcb.139.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguzzi A., Baumann F., Bremer J. The prion's elusive reason for being. Annu. Rev. Neurosci. 2008;31:439–477. doi: 10.1146/annurev.neuro.31.060407.125620. [DOI] [PubMed] [Google Scholar]
- Aguzzi A., Calella A. M. Prions: protein aggregation and infectious diseases. Physiol. Rev. 2009;89:1105–1152. doi: 10.1152/physrev.00006.2009. [DOI] [PubMed] [Google Scholar]
- Anandatheerthavarada H. K., Biswas G., Robin M. A., Avadhani N. G. Mitochondrial targeting and a novel transmembrane arrest of Alzheimer's amyloid precursor protein impairs mitochondrial function in neuronal cells. J. Cell Biol. 2003;161:41–54. doi: 10.1083/jcb.200207030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok A., Hegde R. S. Retrotranslocation of prion proteins from the endoplasmic reticulum by preventing GPI signal transamidation. Mol. Biol. Cell. 2008;19:3463–3476. doi: 10.1091/mbc.E08-01-0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashok A., Hegde R. S. Selective processing and metabolism of disease-causing mutant prion proteins. PLoS Pathog. 2009;5:e1000479. doi: 10.1371/journal.ppat.1000479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckett D., Kovaleva E., Schatz P. J. A minimal peptide substrate in biotin holoenzyme synthetase-catalyzed biotinylation. Protein Sci. 1999;8:921–929. doi: 10.1110/ps.8.4.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boopathi E., Srinivasan S., Fang J. K., Avadhani N. G. Bimodal protein targeting through activation of cryptic mitochondrial targeting signals by an inducible cytosolic endoprotease. Mol. Cell. 2008;32:32–42. doi: 10.1016/j.molcel.2008.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti O., Ashok A., Hegde R. S. Prion protein biosynthesis and its emerging role in neurodegeneration. Trends Biochem. Sci. 2009;34:287–295. doi: 10.1016/j.tibs.2009.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti O., Hegde R. S. Functional depletion of mahogunin by cytosolically exposed prion protein contributes to neurodegeneration. Cell. 2009;137:1136–1147. doi: 10.1016/j.cell.2009.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S., Longhi R., Alcaro S., Ortuso F., Sprocati T., Flora A., Borgese N. N-myristoylation determines dual targeting of mammalian NADH-cytochrome b5 reductase to ER and mitochondrial outer membranes by a mechanism of kinetic partitioning. J. Cell Biol. 2005;168:735–745. doi: 10.1083/jcb.200407082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasari V. R., Anandatheerthavarada H. K., Robin M. A., Boopathi E., Biswas G., Fang J. K., Nebert D. W., Avadhani N. G. Role of protein kinase C-mediated protein phosphorylation in mitochondrial translocation of mouse CYP1A1, which contains a non-canonical targeting signal. J. Biol. Chem. 2006;281:30834–30847. doi: 10.1074/jbc.M510725200. [DOI] [PubMed] [Google Scholar]
- Drisaldi B., Stewart R. S., Adles C., Stewart L. R., Quaglio E., Biasini E., Fioriti L., Chiesa R., Harris D. A. Mutant PrP is delayed in its exit from the endoplasmic reticulum, but neither wild-type nor mutant PrP undergoes retrotranslocation prior to proteasomal degradation. J. Biol. Chem. 2003;278:21732–21743. doi: 10.1074/jbc.M213247200. [DOI] [PubMed] [Google Scholar]
- Fons R. D., Bogert B. A., Hegde R. S. Substrate-specific function of the translocon-associated protein complex during translocation across the ER membrane. J. Cell Biol. 2003;160:529–539. doi: 10.1083/jcb.200210095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goder V., Crottet P., Spiess M. In vivo kinetics of protein targeting to the endoplasmic reticulum determined by site-specific phosphorylation. EMBO J. 2000;19:6704–6712. doi: 10.1093/emboj/19.24.6704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet B., Baruch A., Moon N. S., Poirier M., Sansregret L. L., Erickson A., Bogyo M., Nepveu A. A cathepsin L isoform that is devoid of a signal peptide localizes to the nucleus in S phase and processes the CDP/Cux transcription factor. Mol. Cell. 2004;14:207–219. doi: 10.1016/s1097-2765(04)00209-6. [DOI] [PubMed] [Google Scholar]
- Hegde R. S., Mastrianni J. A., Scott M. R., DeFea K. A., Tremblay P., Torchia M., DeArmond S. J., Prusiner S. B., Lingappa V. R. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- Hegde R. S., Tremblay P., Groth D., DeArmond S. J., Prusiner S. B., Lingappa V. R. Transmissible and genetic prion diseases share a common pathway of neurodegeneration. Nature. 1999;402:822–826. doi: 10.1038/45574. [DOI] [PubMed] [Google Scholar]
- Howarth M., Takao K., Hayashi Y., Ting A. Y. Targeting quantum dots to surface proteins in living cells with biotin ligase. Proc. Natl. Acad. Sci. USA. 2005;102:7583–7588. doi: 10.1073/pnas.0503125102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S. W., Rane N. S., Kim S. J., Garrison J. L., Taunton J., Hegde R. S. Substrate-specific translocational attenuation during ER stress defines a pre-emptive quality control pathway. Cell. 2006;127:999–1013. doi: 10.1016/j.cell.2006.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungnickel B., Rapoport T. A. A posttargeting signal sequence recognition event in the endoplasmic reticulum membrane. Cell. 1995;82:261–270. doi: 10.1016/0092-8674(95)90313-5. [DOI] [PubMed] [Google Scholar]
- Kim S. J., Hegde R. S. Cotranslational partitioning of nascent prion protein into multiple populations at the translocation channel. Mol. Biol. Cell. 2002;13:3775–3786. doi: 10.1091/mbc.E02-05-0293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S. J., Mitra D., Salerno J. R., Hegde R. S. Signal sequences control gating of the protein translocation channel in a substrate-specific manner. Dev. Cell. 2002;2:207–217. doi: 10.1016/s1534-5807(01)00120-4. [DOI] [PubMed] [Google Scholar]
- Kim S. J., Rahbar R., Hegde R. S. Combinatorial control of prion protein biogenesis by the signal sequence and transmembrane domain. J. Biol. Chem. 2001;276:26132–26140. doi: 10.1074/jbc.M101638200. [DOI] [PubMed] [Google Scholar]
- Krieg U. C., Walter P., Johnson A. E. Photocrosslinking of the signal sequence of nascent preprolactin to the 54-kilodalton polypeptide of the signal recognition particle. Proc. Natl. Acad. Sci. USA. 1986;83:8604–8608. doi: 10.1073/pnas.83.22.8604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulman J. D., Satake M., Harris J. E. A versatile system for site-specific enzymatic biotinylation and regulated expression of proteins in cultured mammalian cells. Protein Expr. Purif. 2007;52:320–328. doi: 10.1016/j.pep.2006.09.011. [DOI] [PubMed] [Google Scholar]
- Kurzchalia T. V., Wiedmann M., Girshovich A. S., Bochkareva E. S., Bielka H., Rapoport T. A. The signal sequence of nascent preprolactin interacts with the 54K polypeptide of the signal recognition particle. Nature. 1986;320:634–636. doi: 10.1038/320634a0. [DOI] [PubMed] [Google Scholar]
- Land T., Rouault T. A. Targeting of a human iron-sulfur cluster assembly enzyme, nifs, to different subcellular compartments is regulated through alternative AUG utilization. Mol. Cell. 1998;2:807–815. doi: 10.1016/s1097-2765(00)80295-6. [DOI] [PubMed] [Google Scholar]
- Levine C. G., Mitra D., Sharma A., Smith C. L., Hegde R. S. The efficiency of protein compartmentalization into the secretory pathway. Mol. Biol. Cell. 2005;16:279–291. doi: 10.1091/mbc.E04-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz H., Windl O., Kretzschmar H. A. Cellular phenotyping of secretory and nuclear prion proteins associated with inherited prion diseases. J. Biol. Chem. 2002;277:8508–8516. doi: 10.1074/jbc.M110197200. [DOI] [PubMed] [Google Scholar]
- Lu N. Z., Cidlowski J. A. Translational regulatory mechanisms generate N-terminal glucocorticoid receptor isoforms with unique transcriptional target genes. Mol. Cell. 2005;18:331–342. doi: 10.1016/j.molcel.2005.03.025. [DOI] [PubMed] [Google Scholar]
- Lu N. Z., Collins J. B., Grissom S. F., Cidlowski J. A. Selective regulation of bone cell apoptosis by translational isoforms of the glucocorticoid receptor. Mol. Cell. Biol. 2007;27:7143–7160. doi: 10.1128/MCB.00253-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma J., Lindquist S. Conversion of PrP to a self-perpetuating PrPSc-like conformation in the cytosol. Science. 2002;298:1785–1788. doi: 10.1126/science.1073619. [DOI] [PubMed] [Google Scholar]
- Ma J., Wollmann R., Lindquist S. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science. 2002;298:1781–1785. doi: 10.1126/science.1073725. [DOI] [PubMed] [Google Scholar]
- Matthews G. D., Gur N., Koopman W. J., Pines O., Vardimon L. Weak mitochondrial targeting sequence determines tissue-specific subcellular localization of glutamine synthetase in liver and brain cells. J. Cell Sci. 2010;123:351–359. doi: 10.1242/jcs.060749. [DOI] [PubMed] [Google Scholar]
- Mize G. J., Harris J. E., Takayama T. K., Kulman J. D. Regulated expression of active biotinylated G-protein coupled receptors in mammalian cells. Protein Expr. Purif. 2008;57:280–289. doi: 10.1016/j.pep.2007.09.018. [DOI] [PubMed] [Google Scholar]
- Naamati A., Regev-Rudzki N., Galperin S., Lill R., Pines O. Dual targeting of Nfs1 and discovery of its novel processing enzyme, Icp55. J. Biol. Chem. 2009;284:30200–30208. doi: 10.1074/jbc.M109.034694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negro A., Ballarin C., Bertoli A., Massimino M. L., Sorgato M. C. The metabolism and imaging in live cells of the bovine prion protein in its native form or carrying single amino acid substitutions. Mol. Cell Neurosci. 2001;17:521–538. doi: 10.1006/mcne.2000.0953. [DOI] [PubMed] [Google Scholar]
- Pop C., Salvesen G. S. Human caspases: activation, specificity, and regulation. J. Biol. Chem. 2009;284:21777–21781. doi: 10.1074/jbc.R800084200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prusiner S. B. Prions. Proc. Natl. Acad. Sci. USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane N. S., Chakrabarti O., Feigenbaum L., Hegde R. S. Signal sequence insufficiency contributes to neurodegeneration caused by transmembrane prion protein. J. Cell Biol. 2010;188:515–526. doi: 10.1083/jcb.200911115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane N. S., Kang S. W., Chakrabarti O., Feigenbaum L., Hegde R. S. Reduced translocation of nascent prion protein during ER stress contributes to neurodegeneration. Dev. Cell. 2008;15:359–370. doi: 10.1016/j.devcel.2008.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rane N. S., Yonkovich J. L., Hegde R. S. Protection from cytosolic prion protein toxicity by modulation of protein translocation. EMBO J. 2004;23:4550–4559. doi: 10.1038/sj.emboj.7600462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev-Rudzki N., Karniely S., Ben-Haim N. N., Pines O. Yeast aconitase in two locations and two metabolic pathways: seeing small amounts is believing. Mol. Biol. Cell. 2005;16:4163–4171. doi: 10.1091/mbc.E04-11-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regev-Rudzki N., Pines O. Eclipsed distribution: a phenomenon of dual targeting of protein and its significance. Bioessays. 2007;29:772–782. doi: 10.1002/bies.20609. [DOI] [PubMed] [Google Scholar]
- Robin M. A., Anandatheerthavarada H. K., Biswas G., Sepuri N. B., Gordon D. M., Pain D., Avadhani N. G. Bimodal targeting of microsomal CYP2E1 to mitochondria through activation of an N-terminal chimeric signal by cAMP-mediated phosphorylation. J. Biol. Chem. 2002;277:40583–40593. doi: 10.1074/jbc.M203292200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannerud R., Annaert W. Trafficking, a key player in regulated intramembrane proteolysis. Semin. Cell Dev. Biol. 2009;20:183–190. doi: 10.1016/j.semcdb.2008.11.004. [DOI] [PubMed] [Google Scholar]
- Sass E., Blachinsky E., Karniely S., Pines O. Mitochondrial and cytosolic isoforms of yeast fumarase are derivatives of a single translation product and have identical amino termini. J. Biol. Chem. 2001;276:46111–46117. doi: 10.1074/jbc.M106061200. [DOI] [PubMed] [Google Scholar]
- Shaffer K. L., Sharma A., Snapp E. L., Hegde R. S. Regulation of protein compartmentalization expands the diversity of protein function. Dev. Cell. 2005;9:545–554. doi: 10.1016/j.devcel.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Schatz P. J. Use of peptide libraries to map the substrate specificity of a peptide-modifying enzyme: a 13 residue consensus peptide specifies biotinylation in Escherichia coli. Biotechnology. 1993;11:1138–1143. doi: 10.1038/nbt1093-1138. [DOI] [PubMed] [Google Scholar]
- Sharma A., Mariappan M., Appathurai S., Hegde R. S. In vitro dissection of protein translocation into the mammalian endoplasmic reticulum. Methods Mol. Biol. 2010;619:339–363. doi: 10.1007/978-1-60327-412-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein I., Peleg Y., Even-Ram S., Pines O. The single translation product of the FUM1 gene (fumarase) is processed in mitochondria before being distributed between the cytosol and mitochondria in Saccharomyces cerevisiae. Mol. Cell. Biol. 1994;14:4770–4778. doi: 10.1128/mcb.14.7.4770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R. S., Drisaldi B., Harris D. A. A transmembrane form of the prion protein contains an uncleaved signal peptide and is retained in the endoplasmic Reticulum. Mol. Biol. Cell. 2001;12:881–889. doi: 10.1091/mbc.12.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart R. S., Harris D. A. Most pathogenic mutations do not alter the membrane topology of the prion protein. J. Biol. Chem. 2001;276:2212–2220. doi: 10.1074/jbc.M006763200. [DOI] [PubMed] [Google Scholar]
- Stewart R. S., Harris D. A. Mutational analysis of topological determinants in prion protein (PrP) and measurement of transmembrane and cytosolic PrP during prion infection. J. Biol. Chem. 2003;278:45960–45968. doi: 10.1074/jbc.M307833200. [DOI] [PubMed] [Google Scholar]
- Stewart R. S., Harris D. A. A transmembrane form of the prion protein is localized in the Golgi apparatus of neurons. J. Biol. Chem. 2005;280:15855–15864. doi: 10.1074/jbc.M412298200. [DOI] [PubMed] [Google Scholar]
- Stewart R. S., Piccardo P., Ghetti B., Harris D. A. Neurodegenerative illness in transgenic mice expressing a transmembrane form of the prion protein. J. Neurosci. 2005;25:3469–3477. doi: 10.1523/JNEUROSCI.0105-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong W. H., Jameson G. N., Huynh B. H., Rouault T. A. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. USA. 2003;100:9762–9767. doi: 10.1073/pnas.1732541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westergard L., Christensen H. M., Harris D. A. The cellular prion protein (PrP(C)): its physiological function and role in disease. Biochim Biophys Acta. 2007;1772:629–644. doi: 10.1016/j.bbadis.2007.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Cook J., Rassbach B., Lemus A., DeArmond S. J., Mastrianni J. A. A new transgenic mouse model of Gerstmann-Straussler-Scheinker syndrome caused by the A117V mutation of PRNP. J. Neurosci. 2009;29:10072–10080. doi: 10.1523/JNEUROSCI.2542-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yogev O., Singer E., Shaulian E., Goldberg M., Fox T. D., Pines O. Fumarase: a mitochondrial metabolic enzyme and a cytosolic/nuclear component of the DNA damage response. PLoS Biol. 2010;8:e1000328. doi: 10.1371/journal.pbio.1000328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudt M. R., Cidlowski J. A. The glucocorticoid receptor: coding a diversity of proteins and responses through a single gene. Mol. Endocrinol. 2002;16:1719–1726. doi: 10.1210/me.2002-0106. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.