By comparing computer modeling predictions with observations, we conclude that strong dynein and weaker myosin-generated forces pull the microtubules inward competing with microtubule plus-ends pushing the microtubule aster outward and that the balance of these forces positions the centrosome at the cell center.
Abstract
The centrosome position in many types of interphase cells is actively maintained in the cell center. Our previous work indicated that the centrosome is kept at the center by pulling force generated by dynein and actin flow produced by myosin contraction and that an unidentified factor that depends on microtubule dynamics destabilizes position of the centrosome. Here, we use modeling to simulate the centrosome positioning based on the idea that the balance of three forces—dyneins pulling along microtubule length, myosin-powered centripetal drag, and microtubules pushing on organelles—is responsible for the centrosome displacement. By comparing numerical predictions with centrosome behavior in wild-type and perturbed interphase cells, we rule out several plausible hypotheses about the nature of the microtubule-based force. We conclude that strong dynein- and weaker myosin-generated forces pull the microtubules inward competing with microtubule plus-ends pushing the microtubule aster outward and that the balance of these forces positions the centrosome at the cell center. The model also predicts that kinesin action could be another outward-pushing force. Simulations demonstrate that the force-balance centering mechanism is robust yet versatile. We use the experimental observations to reverse engineer the characteristic forces and centrosome mobility.
INTRODUCTION
Position and orientation of the nucleus (Burke and Roux, 2009), membrane organelles (Wada and Suetsugu, 2004), and mitotic spindles (Grill et al., 2001) in cells are of crucial importance for their function in health and disease. Similarly, centrosome (CS) localization is essential for neural and epithelial differentiation, cell polarization, spindle positioning, and orientation and control of cell migration (Manneville and Etienne-Manneville, 2006). What are the mechanisms governing these phenomena is a fundamental question of cellular organization. Broadly, three factors—feedback in the reaction–diffusion signaling mechanisms, architectural heterogeneity of the cell, and cytoskeleton network mechanics—can be responsible for the spatial organization of the cell (Mullins, 2010). Here, we investigate the particular question of how the CS finds the cell center, in which the third factor, cytoskeletal mechanics, is crucial.
In many cell types, the CS in interphase is found at the centroid or geometric center of the cell (Dujardin and Vallee, 2002). CS is the focal point of microtubule (MT) aster, so it is not surprising that MTs play a key role in the CS centering, because their length approaches that of the whole cell and also because their rapid growth and shortening dynamics allow them to explore the entire cell space (Wühr et al., 2009). The ability of MTs growing against an obstacle to generate pushing forces by polymerization ratchet mechanism (Dogterom and Yurke, 1997) is at the core of the MT aster centering in vitro (Holy et al., 1997): if the aster's focal point is closer, for example, to the left edge of the experimental chamber (see Figure 1A), then shorter MTs at that side grow against the boundary and buckle. Mechanically, MT filaments are elastic rods, and their buckling forces are inversely proportional to the square of their lengths. Thus, at the left, short MTs buckling against the boundary push the aster to the right with a significant force, whereas at the right fewer MTs reach the boundary because of the periodic shortening, and those that do reach the boundary are long and buckle at a weaker force. The resulting imbalance of the pushing forces drives the MT aster to the central position. This elegant MT pushing mechanism also works in vivo: in the small fission yeast cells, the nucleus can be centered by pushing forces that are generated when growing MTs hit the cell edges (Tran et al., 2001; Tolić-Nørrelykke et al., 2004). The growing MTs also can push against barriers scattered throughout the cytoplasm, such as yolk granules (Bjerknes, 1986; Wühr et al., 2009) in some cells, but the respective mechanical effect was never studied.
Figure 1.
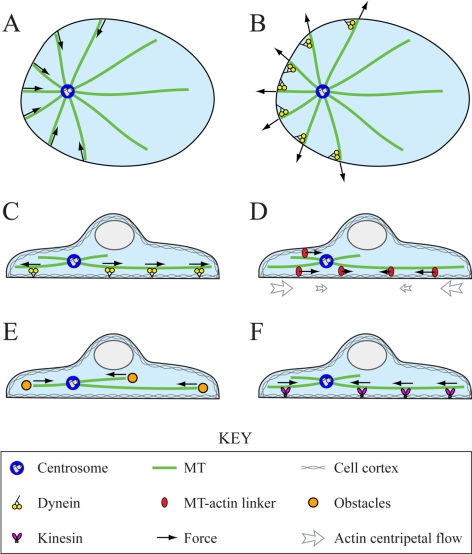
Hypothesized force-generation mechanisms. Top (A and B) and side (C–F) views of the cell. (A) MT pushing against the cell periphery stabilizes the CS centering because if the CS is closer to the left edge, more MTs will reach this edge and push the CS to the right. (B) Dynein pulling from the cell periphery destabilizes the CS centering, if the MT number is the limiting factor. (C) Dynein pulling on the MT sides stabilizes the CS at the center because if the CS is closer to the left edge, more motors will interact with the longer MTs at the right and pull the CS to the right. (D) Actin centripetal flow stabilizes the CS at the center because the dense MTs near the CS shift to the left, from where they are dragged toward the center by the flow. (E) Growing MT plus-ends' pushing against obstacles in the cytoplasm destabilizes the centering, because more MT plus-ends are oriented toward the distal cell edge. (F) Kinesin pushing on the MT sides destabilizes the CS because if the CS is closer to the left edge, more motors will interact with the longer MTs at the right and push the CS to the left.
Typically, however, the forces in the cell are generated not by MTs directly but by the host of molecular motors using the MTs as tracks (Civelekoglu-Scholey and Scholey, 2010). There are many examples of pulling motor forces positioning cell structures (Grill et al., 2001; Pearson and Bloom, 2004). The most well known of them occurs in Caenorhabditis elegans eggs where dyneins associated with the actin cortex at the cell boundary through dynactin, attempt to move toward the MT minus-ends, thereby generating pulling forces on MTs reaching the cell cortex (Grill and Hyman, 2005; see Figure 1B). At first glance, this pulling mechanism should be destabilizing (see Figure 1B): if the aster's focus is closer to the left, more filaments will reach the cortex there, and the force pulling to the left will be stronger decentering the aster. However, if the number of pulling dyneins is limiting, while an abundant number of MTs reach the cortex at all sides of the cell, then this mechanism, in which the motors pull on the MT plus-ends, becomes centering (Grill and Hyman, 2005).
Another possibility is for the dynein motors to be distributed throughout the cytoplasm and attached to structures not easily displaced, e.g., endoplasmic reticulum, yolk, intermediate filaments, or actin (Reinsch and Gönczy, 1998). Then, the longer the MT, the more motors it can engage along its length, leading to a length-dependent pulling force. This servomechanism proposed in Hamaguchi and Hiramoto (1986) (for review, see Dujardin and Vallee, 2002) should stabilize the centering: the aster experiences a net force in the direction of the longest MTs and thus toward the center of the cell (see Figure 1C). The necessary interactions of dyneins with lateral MT surface were observed in fission yeast (Vogel et al., 2009), budding yeast (Adames and Cooper, 2000), and Dictyostelium cells (Koonce and Khodjakov, 2002). For this mechanism to work, the force generators have to be distributed uniformly in the cytoplasm. In many cells, this cannot be the case, because many motors are localized to the dense, yet thin, actin layer of the cell cortex underlying the plasma membrane, whereas the cell interior has vast regions with large fluid fraction of the cytoplasm that the motors are unlikely to fasten to. However, in flat cells, the cortex is close to any point in the interior, and MTs can align along the cortex and thus experience cortical length-dependent forces (O'Connell and Wang, 2000) and get engaged in the servomechanism. Note also that although dynein, anchored to the cortex via dynactin, is the most prominent candidate for forcing MTs (Dujardin and Vallee, 2002), kinesins enmeshed into the actin-rich cortex also can engage MTs at or near their ends and push on them (Brito et al., 2005).
Last but not least, MTs interact with actin gel mechanically through molecular complexes that can simultaneously associate with actin and MT filaments (Huang et al., 1999; Kodama et al., 2003; Weber et al., 2004). Myosin-powered contraction causes ubiquitous centripetal flow of F-actin in cells (Yam et al., 2007; Alexandrova et al., 2008). MTs that are coupled to this flow are dragged and transported to the center (Mikhailov and Gundersen, 1995; Yvon and Wadsworth, 2000; Salmon et al., 2002; Rosenblatt et al., 2004; see Figure 1D). In addition, MTs can be pulled by myosin motors directly on actin cables (Hwang et al., 2003).
Here, we focus on the phenomenon of the CS centering in flat mammalian tissue culture cells in the interphase. Our experimental study (Burakov et al., 2003) revealed that dynein motors' pulling on MTs is responsible for the force stabilizing the CS at the cell center. This force is assisted by a myosin-dependent centering force. The latter is not strong enough to stabilize the symmetric MT aster position by itself due to the third factor that destabilizes the aster and moves the CS to the cell edge. This third factor is associated with the MT turnover dynamics, because using Taxol to stabilize MTs nullifies the respective force. The nature of this MT dynamics-dependent anticentering force, however, remains unknown.
Mathematical and computational modeling was used extensively to complement traditional cell biological and biophysical methods to elucidate mechanistic details of the centering mechanisms in several systems (Holy et al., 1997; Grill et al., 2001; Vogel et al., 2009). Modeling is especially useful because individual MTs and motors are next to impossible to resolve microscopically in many systems and because measuring forces directly is too difficult. Here, we use the reverse engineering approach that has been successfully applied to cytoskeletal mechanics problems (Wollman et al., 2008; Foethke et al., 2009), and we use the observations and measurements reported in Burakov et al. (2003) to answer the following questions: Do dyneins pull on the MT plus-ends or along their length? What is the nature of the anticentering force? How many motors and MTs are involved and what are the characteristic forces in the centering mechanism?
MATERIALS AND METHODS
Modeling
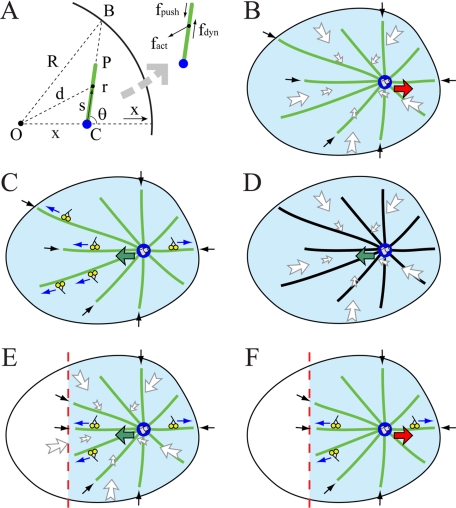
We developed both a continuous deterministic model and a discrete stochastic model in which the flat cell is represented as a disk of ∼20 μm in radius that can be gleaned from the microscopic images. In the continuous model, we place the CS at a distance x from the cell center; from the symmetry considerations, the net force applied to all MTs on the CS is directed along the x-axis toward the cell center (see Figure 2A). We consider an individual MT (Figure 2A) and three forces applied to it: a pushing force f→push acting on its plus-end and directed toward the minus-end, a dynein force f→dyn pulling the MT side and directed toward the plus-end, and an actin-flow-induced drag force f→act pulling the MT toward the cell center. The elementary dynein and pushing forces are constant, while the actin drag force increases from the center to the edges of the cell because actin flow decelerates from the periphery to the center of the cell. We integrate the dynein and actin forces along the length of each MT and then integrate the results over all the MTs to get the total force on the CS as described in Supplemental Material. When integrating, we assume that there are a constant number of motors per MT unit length, that the motor forces are additive, and that the force per motor is independent of the MT movement. The last two assumptions are justified because MTs move much slower than free dyneins glide, so that each dynein motor operates near its stall force. In the continuous model, we assume that the MT aster is radially symmetric about the CS. We use the dynamic instability theory (Dogterom and Leibler, 1993) to find the steady-state continuous distribution of MT plus-ends that is used in the integration. When the nocodazole is applied to the cell locally, we assume that any MT reaching for the edge of the nocodazole-affected field undergoes a catastrophe and that there are no MTs in the wedge shown in Figure 2E. We repeat all the described calculations for the elliptical and one-dimensional (1D) cells. We also consider hypothetical kinesin forces along the MT length and dynein forces from the cell boundary. Respective mathematics is described in Supplemental Material.
Figure 2.
Geometry of the force generation. (A) Three principal forces on a single MT (green line) of length r in the cell with radius R: length-dependent force fdyn by outward-pulling dyneins, flow- and length-dependent force fact generated by the centripetal actin drag, and inward pushing force fpush. CS (blue) is displaced from the cell center by x. O, C, P, B, d, s, and θ are the geometric variables characterizing MT position and orientation used for force calculations in the Supplemental Material. (B) Perturbation (i): When dynein is inhibited and only actin drag and dynamic MT forces are present, centering is destabilized and the CS moves away from the center. (C) Perturbation (ii): When myosin is inhibited, the actin flow stops and only dynein and dynamic MT forces are present; the CS is stabilized at the center. (D) Perturbation (iv): When both dynein is inhibited and MTs are stabilized by Taxol, the actin flow stabilizes the CS at the center. (E) Perturbation (v): After the local application of nocodazole (modeled by eliminating MTs from the wedge of the cell), the CS shifts toward this wedge. (F) Perturbation (vi): After the local application of nocodazole and inhibition of myosin, the CS shifts away from the wedge. Green arrows, the CS is stabilized and moves toward the center; red arrows, the CS is destabilized and moves away from the center; blue arrows, pulling dynein forces; white arrows, actin–myosin drag forces; and black arrows, decentering forces associated with dynamic MTs. Dynamic MTs are shown in green, and Taxol-treated stable MTs are shown in black.
Because the number of MTs estimated from the experiment is on the order of 100 and the known dynamic instability time scale is less than an order of magnitude faster than the characteristic time scale of the CS's movement, the stochastic effects also should be considered. Thus, we developed a discrete stochastic model to verify the results of the continuous model and to make a visual presentation of the model simulations resulting in Supplemental Movies 1–5. In the stochastic model, individual MTs and their dynamic instability are treated explicitly as described in Paul et al. (2009). As described in Supplemental Material, MTs are nucleated at the CS at a constant rate. At each time step, they grow or shorten with fixed speeds. The transitions between the growing and shortening states take place randomly with observed constant catastrophe and rescue rates. At each time step, the force on each MT is calculated numerically according to the formulae from the continuous model, and the forces from all the current MTs are summated to obtain the total force on the CS. The CS is then displaced according to the equation dx→ / dt = μF→ (Civelekoglu-Scholey and Scholey, 2010), where x→ is the CS's coordinate in two dimensions, F→ is the total current force on the CS, and μ is the CS's mobility. Preliminary estimates showed that making the model fully stochastic and introducing random flickering on and off in the forces do not qualitatively change the results.
Experiment
Images displayed in Figures 4 and 5, CS movement rates, MT dynamic instability parameters, and cell dimensions used to develop and calibrate the quantitative model are obtained as described in detail in Burakov et al. (2003).
Figure 4.
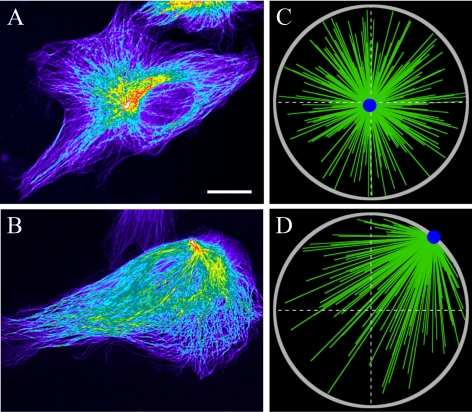
Centering in control cells and loss of stability in dynein-inhibited cells. (A) Centered CS in the control cell. (B) CS shifted to the cell edge in the dynein-inhibited cell. Hot-cold colors illustrate high–low tubulin density, respectively. Note that in B, the destabilized CS is at the cell edge closest to the centroid of the cell. Bar, 10 μm. (C) Snapshot of stochastic simulations from Supplemental Movie 1, corresponding to the situation in (A). (D) Snapshot of stochastic simulations from Supplemental Movie 2, corresponding to the situation in B. Gray circle, cell periphery, green lines, MTs; and blue dot, CS.
Figure 5.
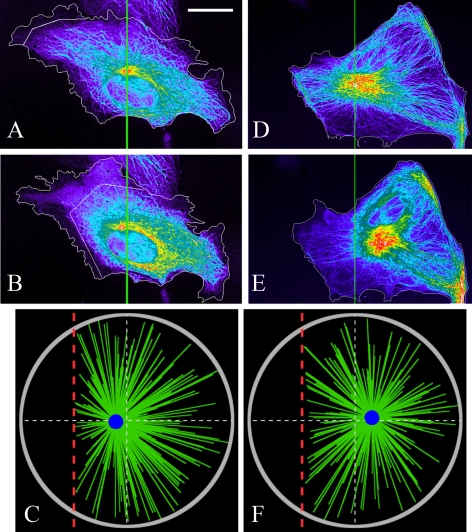
Effects of the local nocodazole application on the CS's positioning. Observed CS positions before (A) and after (B) the local nocodazole-application at the left side of the cell. The zigzag line shows the boundary of the nocodazole-affected region. The centrosome shifts slightly toward the nocodazole source, which is in agreement with the model result (C)—the snapshot of the stochastic simulations from Supplemental Movie 3 corresponding to the situation in A and B. (D and E) Observed CS positions before (D) and after (E) the local nocodazole application at the left side of the myosin-inhibited cell. The centrosome shifts slightly away from the nocodazole source, which is in agreement with the model result (F)—the snapshot of the stochastic simulations from Supplemental Movie 4 corresponding to the situation in D and E. Bar, 10 μm (A, B, D, and E), and the colors are the same as those in Figure 4. Red dashed lines indicate the boundary of the nocodazole-affected region in the simulations.
RESULTS
Qualitative Analysis Suggests That MT Pushing on Obstacles throughout the Cortex Destabilizes the CS Positioning
Our model is based on the following observations (Burakov et al., 2003). 1) When dynein is inhibited, centering is destabilized and the CS moves away from the center (Figure 2B, red arrow). 2) Inhibition of myosin stops the centripetal actin flow but does not affect the centering (Figure 2C, green arrow illustrates the CS centering). 3) When both dynein and myosin are inhibited, the CS moves away from the center. 4) When dynein is inhibited, and in addition the cell is treated with Taxol inhibiting MT dynamics, the CS stays at the center (Figure 2D, green arrow illustrates the CS centering). 5) When nocodazole is applied locally to the cell edge disrupting MTs there, the CS shifts away from the center toward the nocodazole source (Figure 2E, green arrow). 6) When nocodazole is applied locally, and in addition myosin is inhibited in the cell, the CS shifts away from the nocodazole source (Figure 2F, red arrow). 7) Finally, when nocodazole is applied locally, together with myosin inhibition and dynein weakening, the CS oscillates near the cell center.
These results indicate that three mechanisms participate in CS positioning: one mechanism is dynein dependent; another mechanism is myosin powered; and the third mechanism relies on the MT dynamics, meaning that either growing or shortening MT plus-ends are involved. Result 1 indicates that dynein's action is to stabilize the CS at the center, probably by pulling astral MTs along their length, because pulling only from the cell boundary would destabilize the centering. In Supplemental Material, we provide calculations showing that the centering effect of a limited dynein number pulling from the cell boundary is less likely. The reason is that when nocodazole is applied locally, dynein from the unaffected part of the cell boundary reorients the pulling force so effectively that the CS is likely to be shifted away from the nocodazole source contradicting experimental result 5. Result 4 that deals with the situation, in which the dynein- and MT dynamics-related forces are suppressed and only the myosin-generated force acts (Figure 2D), indicates that the myosin-powered mechanism is also to stabilize the CS at the center. Probably, the interaction is through transient associations between the MTs and the centripetally flowing F-actin, which causes effective inward drag force on the MTs. Result 3 associates the destabilizing mechanism with the MT dynamics. Results 1 and 2 also hint that the dynein-depended centering is stronger than the destabilizing mechanism, whereas the myosin-dependent centering is weaker.
Plausible positioning mechanisms based on the MT dynamics could result from the interactions between MT plus-ends and cell boundary or obstacles, or kinesin motors that are anchored throughout the cortex. Lateral interactions along the sides of MTs are less likely to contribute because such interactions do not require MT dynamic instability. However, under Kinesin Pushing along the MT Lengths Can Generate the Decentering Force, we discuss a possibility that the off-centering force originates from kinesin motors' pushing along the MT length (Figure 1F). If some motors pull on the MT plus-ends throughout the cytoplasm (or shortening MTs pull on cortex structures that remain attached to the MTs; Grishchuk et al., 2005), they would stabilize the aster at the center because more growing plus-ends would be located between the CS and the distal side of the cell. Polymerizing MT plus-ends pushing on the cell boundary would lead to centering (Figure 1A), as discussed in the Introduction. One additional possibility is that when the CS is closer to one side of the cell, the MTs reorient such that they push the cell boundary only at the distal side, causing destabilization (Supplemental Figure S7). But in this case, the myosin-powered drag is also destabilizing (Supplemental Figure S7), which contradicts experimental result 4. If some motors pull the growing MT plus-ends from the cell boundary, they would destabilize the aster (Figure 1B). But this would contradict experimental result 7: MTs remaining on the opposite side of the nocodazole-application region would directly pull the CS toward that side, because myosin activity is inhibited. This leaves us with the only plausible mechanism: the growing MT plus-ends push on structures that are associated with the cortex, which lines up both the ventral and dorsal surfaces of the cell (Figure 1E). The following mathematical model and simulations demonstrate that this hypothesis agrees with all the observations.
Force Balance at the Centrosome Has Centering Effect If the Dynein Pulling Is Strong
We computed three major forces acting on the CS as functions of the distance from the center of a disk-like cell (Figure 3A). Confirming the qualitative analysis, Figure 3A (dotted) shows that when the CS shifts to the right from the center, then positive net force from MT pushing will move the CS further to the right, whereas negative net forces from dynein pulling and actin drag (solid and dashed, respectively) will return the CS to the center. Note that for each MT, dynein is pulling in the outward direction, whereas myosin is pulling inward. So, the elementary forces applied to individual MTs are in different directions and have different signs. However, after integrating the elementary forces over all the MTs, both (dynein and myosin) net forces act inward and therefore are stabilizing. Magnitudes of all three (dynein, myosin, and pushing) forces increase in a roughly linear manner as the CS shifts away from the center. This prediction agrees with the observation that the CS accelerates away from the center when dynein is inhibited (Burakov et al., 2003). The calculation suggests that the sum of the three forces will have the centering effect if the net dynein-force is strong enough (density of dynein motors exceeds a threshold). Similarly, in absence of dynein, if the net force from myosin is less than that from MTs' pushing, the CS will be destabilized and will move to the cell edge.
Figure 3.
Calculated distance dependence of the forces on the CS. Calculated forces on the CS. All distances x are normalized by the cell radius R. (A) Normalized net forces on the CS as functions of the normalized distance from the CS to the center (CS shifts to the right side of the center). Solid, dashed, and dotted curves correspond to the dynein, myosin, and pushing forces, respectively. The dynein force is in the unit of aL (average dynein force per MT; a is the dynein force per unit length, and L is the dynamic instability length). The myosin force is in the unit of bL2 (average actin drag force per MT; b is the drag force per unit area). The pushing force is in the unit of fpush (average pushing force per MT). (B) The total net force on the CS in units of fpush in the case when aL ≈ 3fpush and bL2 = 8fpush. Solid line, control cell; dashed line, dynein-inhibited cell; dotted line, myosin-inhibited cell; and dot-dashed line, cell with both dynein and myosin inhibited. (C) In the case when aL ≈ 3fpush and bL2 = 8fpush, the total force on the CS calculated in the nocodazole-affected cell (the nocodazole-affected wedge extends half-way to the center) is shown for the control cell (solid line) and myosin-inhibited cell (dashed line). The inset zooms-in to the region near the cell center to illustrate the signs of the forces there. Black dots show the predicted equilibrium CS positions. Green arrows are centering, inward (negative) forces; red arrows are decentering, outward (positive) forces.
We found the balance of CS in a cell to be determined by three characteristic forces: fpush, the average pushing force per MT; aL, the average dynein force per MT, and bL2, the average myosin-driven force per MT. Here, L is the length scale for MT dynamics instability (see Supplemental Material), a is the dynein force per unit length of MT, and b is the characteristic actin drag-force per unit area. Note that parameter L predicted by the model (see Supplemental Material) is on the order of 60 μm, whereas the cell radius is ∼20 μm. This means that most MTs reach the cell boundary, which is observed, and most individual MTs are ∼20 μm in length. The parameter scan (see below) suggests that when the relations aL ≈ 3fpush and bL2 ≈ 8 fpush are satisfied between these three main force scales, the total force on the CS becomes negative on the right side of the cell (Figure 3B, solid) and thus stabilizes the CS at the center against mechanical fluctuations. When myosin is inhibited, the sum of the net forces from dynein and MT-pushing remains negative (Figure 3B, dotted), so the centering persists. However, when either dynein alone (Figure 3B, dashed) or both dynein and myosin (Figure 3B, dot-dashed) are inhibited, the force becomes positive and the centered position of the CS is destabilized. Stochastic simulations depicted in Supplemental Movies 1 and 2 confirm these predictions. Simulations also demonstrate that away from the center the destabilized CS in the dynein-inhibited cell moves at a speed of the order of 0.1 μm/min, in agreement with the data reported in Burakov et al. (2003). Snapshots from Supplemental Movies 1 and 2 mimicking the respective experimental images are shown in Figure 4.
The Centrosome Undergoes a Small Shift from the Center If the MTs Are Spatially Perturbed
We modeled the local nocodazole application reported in Burakov et al. (2003) by calculating changes in the three major forces after deletion of MTs from the wedge at the cell side (Figures 2, E and F, and 5) and calculating respective changes in the three major forces. Supplemental Figure S3 illustrates how the forces change: after the MT density diminishes at the left, dynein pulls the CS to the right, so the dynein force becomes more positive in attempt to shift the CS to the right (Supplemental Figure S3A). However, two other forces have the opposite effect: more MTs at the right are dragged by the actin flow in the left direction (Supplemental Figure S3B). The pushing from the plus-ends of these dominating MTs also moves the CS to the left (Supplemental Figure S3C). Thus, despite the fact that dynein is stronger than either myosin or MT pushing separately, now that both the myosin-powered flow and the MT pushing oppose the dynein force, the net effect is shifting the CS to the left toward the nocodazole source. This is confirmed by Figure 3C (solid), which shows the net force on the CS in the presence of the nocodazole effect with parameter values satisfying aL ≈ 3fpush and bL2 ≈ 8fpush. This force–distance relation illustrates that the stable equilibrium position of the CS is on the left side of the cell center, close to the nocodazole source (Figure 3C, solid). However, because in this situation two weak forces negate a strong force, the net force is weak, and the CS's shift from the center is predicted to be small, in agreement with the experimental observations (Figure 5, A and B). This prediction is further confirmed by the stochastic simulations (Figure 5C and Supplemental Movie 3). When the nocodazole is applied to a myosin-inhibited cell, the model predicts that the dynein's pulling from the right overwhelms the MTs' pushing at the right. Therefore, with aL ≈ 3fpush and bL2 ≈ 8fpush, the application of nocodazole to a myosin-inhibited cell (Figure 3C, dashed) will shift the CS away from the nocodazole source to the right, in agreement with the experiment (Figure 5, D and E). The stochastic simulations (Figure 5F and Supplemental Movie 4) further support this result.
The Experimental Constraints Allow to Estimate the Forces and Centrosome Mobility in the Centering Mechanism
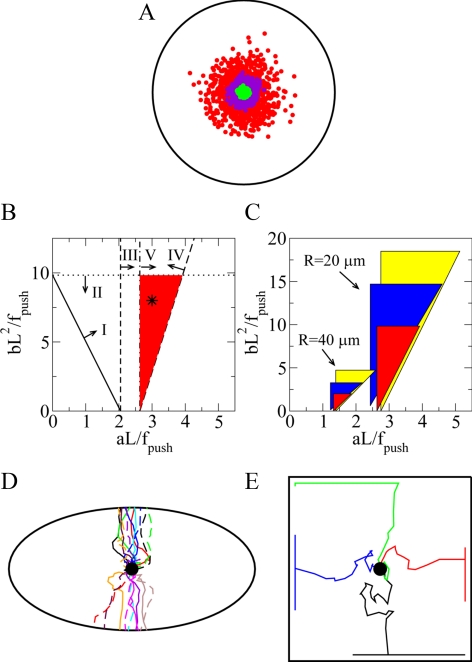
All relevant forces, as well as the effective CS drag coefficient, scale with the number of MTs, so the continuous model results are invariant when the MT number changes, adding to the model robustness. However, the stochastic effects of random imbalances that arise from the MT dynamic instability would increase the fluctuation of CS's position when the number of MT decreases. To test the impact of this effect, we used stochastic simulations (Supplemental Movie 1) and observed that when the average MT number is between 30 and 300, the CS stays very close to the center, but with 3 MTs, the CS wandered relatively far from the center (Figure 6A). This allows us to roughly estimate the necessary number of MTs to be ∼100. Although accurate experimental count is not possible, this number agrees with our rough image analysis.
Figure 6.
Model calibration and predictions. (A) Stochastic simulations illustrate fluctuation of CS position in the control cell with the number of MTs being N ∼ 300 (green dots), N ∼ 30 (purple dots), and N ∼ 3 (red dots). The cell periphery is shown in black. For each case, 1000 simulated CS positions at a 10-min time interval are shown. (B) Parameter values for the disk-like cell with radius R = 20 μm are shown. Lines and associated arrows indicate domains of the parameter values that support the observed CS behavior in control cell (I), dynein-inhibited cell (II), myosin-inhibited cell (III), cell with the local application of nocodazole (IV), and myosin-inhibited cell with the local application of nocodazole (V). The intersection of these domains shown in red is the region of parameters for which the model explains all experimental observations. This region is around the values determined by the relations aL ≈ 3f0 and bL2 = 8f0 shown with the star. (C) Parameter region dependence on the cell shape and size: red regions correspond to circular cells with radius R; blue regions to the elliptical cells with the same areas as those of the disk-like cells and an aspect ratio of 2; and yellow regions to the 1D cells with a half-length R. (D) Trajectories of the CS in a dynein-inhibited ellipsoidal cell from 20 individual stochastic simulations are marked with different colors and line styles. For each trajectory, the CS is initially positioned at the cell center (black dot). (E) Trajectories of the CS in a dynein-inhibited square cell from four individual stochastic simulations are marked with different colors. For each trajectory, the CS is initially positioned at the cell center (black dot). In two simulations, the CS ended at the edge of the cell, and in other two simulations in the corners.
In Supplemental Material, we report the calculations that allowed us to use five of six experimental observations discussed above to put stringent constraints on the model parameters (two other observations are explained without such constraints). The model parameter space is simple and two dimensional (Figure 6B). The system behavior is fully determined by two dimensionless ratios: the characteristic dynein force aL divided by the average pushing force fpush, and the characteristic actin–myosin force bL2 divided by fpush. In this parameter space, there is a relatively narrow triangular region of parameters around the values of aL ≈ 3fpush and bL2 ≈ 8fpush (Figure 6B, star), with which the model explains all the experimental observations.
The stringent constraints on the model parameters allow us to estimate the order of magnitude of the characteristic forces on the MTs. When a growing MT runs into an obstacle, it could either undergo catastrophe (Janson et al., 2003) or continue growing by bypassing the obstacle. We assume that these two events occur with comparable possibilities and that the collision-induced catastrophe is a nontrivial part of the total catastrophe events. Then, we estimate that an MT will run into an obstacle about every 30 s, which is roughly the observed characteristic time interval between two catastrophe events (Burakov et al., 2003). We assume that the force is generated for ∼3 s before the MT starts shortening or bypasses the obstacle and continues growing. Because a stalled MT develops a force of ∼6 pN (Dogterom and Yurke, 1997), and it takes a few seconds for the stalled MT to start shortening (Janson et al., 2003), the average force on an MT tip would be fpush ∼ 6 pN × 3 s/30 s = 0.6 pN.
This pushing mechanism is limited by MTs' buckling force, which is the maximal compression force that an MT can sustain. Because the buckling force is inversely proportional to the MT length, it could be very small for long MTs. Indeed, the buckling force for an MT in an aqueous medium can be estimated as ∼10B/l2, where B ∼ 20 pN × μm2 is the MT flexural rigidity and l is its length. So, only MTs shorter than √10B/fpush ∼ 15 μm could push effectively. However, the MTs are embedded in an actin elastic gel. This significantly increases the compressive force that the MTs can sustain (Brangwynne et al., 2006): for a long MT embedded into the elastic gel, the buckling length λ ∼ 2π (B/Y)1/4, where Y is the Young modulus of the actin meshwork, is independent of the pushing force and MT length. This length is λ ∼ 2.5 μm for characteristic cell cortex elasticity Y ∼ 103 pN/μm2. Even for 2 orders of magnitude weaker actin gel that could be above the narrow cortex layer, with Y ∼ 10 pN/μm2, the buckling length remains small, λ ∼ 6 μm. Respective buckling force ∼10B/λ2 ∼ 6 pN is well above the characteristic pushing force which therefore will be transduced without weakening to the CS.
Taking into account the estimate of aL ≈ 3fpush and L ∼ 60 μm, we conclude that a ∼ 0.03 pN/μm. Because a single dynein motor can develop a force of ∼1 pN (Mallik et al., 2004), there should be an average of ∼1 pulling dynein motor per 30 μm along an MT, or roughly one working motor per MT. This allows estimating the necessary density of dynein motors in the cortex. We assume that there are ∼100 MTs in the cell, each being ∼20 μm long. Then, there are ∼100 × 20 μm/30 μm ≈ 70 dynein motors pulling on the MT aster. We further assume that all the dynein motors within 50 nm from any MT can associate with that MT (Oiwa and Sakakibara, 2005) and that only half of the motors are pulling. Then, there should be ∼140 motors localized in an area of 100 × 20 μm × 0.05 μm ≈ 100 μm2. Thus, the necessary dynein density is ∼1.5/μm2. Because the area of the cell is π × (20 μm)2 ≈ 1200 μm2, ∼2000 dynein molecules in total have to be in the cortex. We are not aware of any direct experimental measurement of this number; however, existence of hundreds of foci that are likely to contain a few dynein molecules each was reported in Kobayashi and Murayama (2009), which agrees with our prediction.
In Supplemental Material, we estimate the mobility of the MT aster with the CS at the center to be μ ∼ 0.03 μm/(pN × min), which corresponds to a friction constant of ζ = 1/μ ∼ 30 pΝ × min/μm. Considering that, when the CS is significantly off-center, a force of 50fpush ∼ 30 pN is applied to the aster, we predict that the CS would shift at speed 50 μfpush ∼ 1 μm/min, which is the observed moving speed of the CS (Burakov et al., 2003). Note that this is also the characteristic observed speed of the centripetal flow in the flat cells (Alexandrova et al., 2008). We propose that the drag on the MT aster does not originate from the viscous resistance that is negligibly small but instead is from the protein friction (Bormuth et al., 2009) – transient attachments between the MTs and the actin filaments in the cortex. The effective friction constant for each attachment can be estimated as κτ (Bormuth et al., 2009), where κ ∼ 10 pN/μm is the effective spring coefficient of deformed actin filament (Mogilner and Oster, 1996), and τ ∼ 1 s is the characteristic time before such filament detaches from an MT (Howard, 2001). So, the effective friction constant for each attachment is ∼10 pN × s/μm. To account for the total friction constant of 30 pΝ × min/μm, we estimate that ∼200 such attachments, or approximately two attachments per MT, exist in the cell. This number is also a model prediction, because no relevant data have been reported.
Centering Mechanism in Cells of Different Shapes and Sizes
Because most of the cells are not perfectly round, we investigated how the centering works in elongated cells (Figure 6D). Our simulations confirmed that all model predictions for the round cells remain valid in the ellipsoidal cells. We also noticed an interesting phenomenon: when dynein is inhibited, the destabilized CS invariably moved to the closest edge of the cell (Figure 6D and Supplemental Movie 5), which is a serendipitous test of the model; when we reexamined the respective images obtained for our previous study (Burakov et al., 2003), we saw that this was exactly the case (Figure 4B). The explanation stems from the fact that the CS position in this situation is determined only by the force balance between the myosin–actin and pushing forces. The analysis in Supplemental Material shows that the magnitude of the myosin-powered force is very sensitive to the distances in the cell, because the speed of the actin centripetal flow is proportional to the distance from the cell center. Therefore, in the elliptical cell, the myosin–actin force is weaker along the short axis than along the long axis of the cell. On the other hand, the MT pushing force is less affected by the cell geometry, because most of the MT plus-ends are distributed near the CS. Thus, the orientation-insensitive outward pushing overcomes the inward drag from actin flow more easily along the short axis of the cell. Note that very elongated cell is close to a 1D system, for which we have calculated all the forces analytically (see Supplemental Material), which further strengthens the model's predictive power. We also observed that in the elongated and 1D cells, greater ranges of model parameters could explain all experimental observations (Figure 6C) due to subtle distance and angle dependencies of the three principal forces discussed in Supplemental Material. For cells of greater sizes, the parameter region that explains all experimental observations becomes smaller (Figure 6C). The simple reason is that in a large cell, very few MTs could reach the cell boundary, so the dynamic MT probing would work less efficiently.
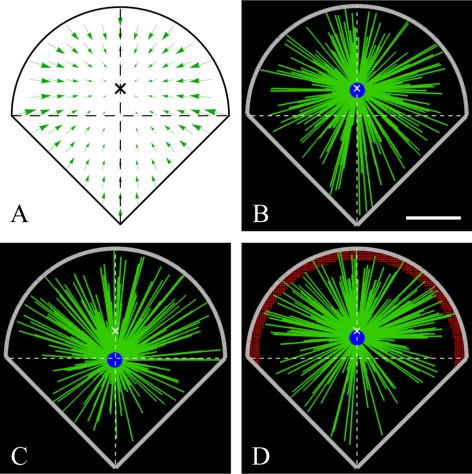
Cells plated on microfabricated substrates can be forced into particular geometries (Théry et al., 2006). To examine the model-predicted behavior on such cells, we simulated the stochastic force-balance model on the square- (Figure 6E) and fan-shaped (Figure 7) domains mimicking the cells shapes reported in Théry et al. (2006). The pattern of the actin centripetal flow in such cells was not observed, so for the square cell we assumed a radially symmetric inward flow, whereas for the fan-shaped cell that resembles motile epithelial cells we chose the flow pattern (Figure 7A) with the convergence point closer to the round edge (and almost at the cell centroid) characteristic for the motile cells. The simulations for the square cell predict, not surprisingly, the stable CS centering (Figure 6E) in agreement with the observations (Théry et al., 2006). Nontrivially, when we switched off dynein force in the simulations, the CS moved to the middle of the cell edge and then either drifted along the edge or went to one of the four cell corners and stayed there (Figure 6E). In the virtual fan-shaped cell, the CS stabilized at the convergence point of the flow (Figure 7B). We investigated what happens if the MT distribution becomes asymmetric, with twice the MTs oriented to the round edge (corresponding to the observed situation in the motile cell where the round edge would be leading). The CS was shifted to the rear (Figure 7C, in agreement with the observations in Théry et al., 2006) due to the dominant effect of the actin retrograde flow near the round edge. The simulations showed that with extra dyneins accumulated at the leading edge of the cell (Figure 7D), the CS shifted toward this edge, similar to what is observed in motile epithelial cells (Dujardin et al., 2003).
Figure 7.
Model predictions for the fan-shaped cell. (A) Hypothesized centripetal actin flow field (green arrows) with flow center (cross) near the centroid. (B–D) Snapshots of the simulations with isotropic MT nucleation (B), anisotropic MT nucleation with density of MTs growing toward the round edge being twice the density of MTs growing toward the corner (C), and anisotropic MT nucleation as described in B, but with additional motors at the cell “leading edge” (marked with red dots). Green lines, MTs. Blue circle, CS. Thick gray lines, cell periphery. White crosses, flow center. Bar, 10 μm.
Kinesin Pushing along the MT Lengths Can Generate the Decentering Force
A distinct possibility for the nature of the decentering force is the pushing action of plus-end–directed kinesin motors along the MT lengths (Figure 1F). It is easy to see from comparison between Figure 1, C and F, that the kinesin pushing is opposite to the dynein pulling; other than this, the kinesin and dynein forces would scale similarly with the sizes and distances. Thus, kinesin pushing along the MT lengths can generate the decentering force. In the Supplemental Material, we demonstrate that as far as the total kinesin force in control cells is less than the total dynein force, the kinesin-based mechanism is consistent with all experimental results. However, this mechanism is subject to two requirements. First, kinesin motors should be anchored to stationary structures in the cell, which is possible: binding of kinesin to intermediate filaments have been described previously (Helfand et al., 2004); in addition, conventional kinesin interacts with myosin V, which in turn interacts with actin filaments (Huang et al., 1999). Second, experimental result 4 indicates that the decentering force is switched off if the MTs are stabilized by Taxol. However, kinesin can push the stabilized MTs. Due to this caveat, we favor the hypothesis that it is MT end pushing, rather than kinesin action, that is responsible for the decentering factor. One possibility, however, is that the Taxol-treated MT is mechanically rigid enough so that the kinesin force cannot move the aster off center. Future inhibition of kinesin experiment will be able to resolve this issue.
DISCUSSION
The fundamental questions of whether it is the pushing or the pulling force that positions the nucleus and organelles in cells and what is the origin of this force have been answered in the past decade with a combination of experimental and modeling research (Kimura and Onami, 2005). Here, we used modeling to address this question for the CS centering in the interphase cell (Burakov et al., 2003). The most important result of our study is that in addition to a strong dynein pulling and a weak myosin-powered actin drag, there is an anticentering pushing force that is generated by the growing MTs throughout the cell. One possible origin for such force is the polymerization ratchet force exerted by the MT plus-ends on obstacles or organelles that are scattered throughout the cell and anchored to the cytoskeletal scaffold (Bjerknes, 1986). Another possibility is that the MTs interact along their lengths with kinesin motors that are anchored to the actin network (Brito et al., 2005). Dynamic MT pushing or pulling on the cell periphery or plus-ends pulling on structures throughout the cell are incompatible with the experiment.
Furthermore, our modeling results argue for the dynein servomechanism—dyneins are anchored to the cortex across the cell and pull on MTs along their lengths—and are inconsistent with the case that dyneins mainly pull on the MT plus-ends from the cell boundary. This conclusion is supported by recent experimental data (Brodsky et al., 2007). By calibrating the model with multiple experimental measurements, we constrain the model parameters to an extent that we are able to predict the order of magnitude of characteristic forces. Namely, we predict that ∼100 dynamic MTs are responsible for average pushing force of ∼1 pN per filament. This anticentering force is overwhelmed by a dynein-generated pN-range pulling force on each 30-μm length of MT and is assisted by a drag force that is caused by 1-2 molecular links between each MT and the centripetally flowing actin network. We also estimate the necessary dynein density to be 1-2 motors per square micron. Finally, we suggest that the viscous-like drag on the shifting CS originates from the dynamic breakage of MT–actin links. We then estimate the CS–MT-aster mobility to be a few hundredths of μm/(min × pN). We also find that a force of the order of 100 pN is needed to push the aster at a characteristic speed of a few microns per minute, in agreement with Reinsh and Gönczy (1998).
We predict that the centering mechanism is robust: all that is needed for the CS to find the cell center is for total dynein force to be greater than a modest threshold of ∼1 motor pulling per MT. The experiments and simulations of the nocodazole application demonstrate that significant perturbations of the MT dynamics lead to relatively small shifts of the CS. The reason is the opposing action of dynein and myosin-powered flow on individual MTs: whereas dynein pulls an MT outward from the center, the actin flow pulls it inward, so altering the MT distribution leads to changes in the opposing forces that partially cancel each other. Additional indication for the robustness of the centering mechanism is that the CS is predicted to be positioned close to the cell center in square and fan-shaped cells (Figures 6E and 7) in a way insensitive to MTs' anisotropy and system perturbations. The scan of the model parameter space shows that the mechanism becomes even more robust in the elongated cells (Figure 6D)—and in practice, all cells are elongated to some extent. Finally, increasing cell size makes the centering mechanism less robust (Figure 6C), because fewer MTs reach the cell boundaries, in agreement with discussion in Wühr et al. (2009). However, a proportional increase of the MT length would restore the centering effectiveness.
The centering mechanism is not only robust but also versatile: the dynein pulling alone can overpower the destabilizing MT pushing to stabilize the CS's centering, so the myosin-powered actin drag seems redundant for the centering. However, the CS's equilibrium point is between the convergence points of the actin centripetal flow and dyneins' pulling field. Therefore, introducing asymmetry and heterogeneity to the two centering forces (by manipulating the dynein and myosin distributions) could shift the CS to a desired position (Figure 7, B and C). For example, concentrated dyneins at the leading edge of the motile cell could shift the CS toward the front (Figure 7D; Dujardin et al., 2003) against the rearward myosin force (Grabham et al., 2007).
Here, we did not discuss the interactions between the CS and nucleus that are linked intimately to the CS (Robinson et al., 1999) and other elements of cytoskeleton (Starr, 2007). We showed above that the centering is nucleus independent, because the experiments in cell cytoplasts resulted in a similar CS behavior (Burakov et al., 2003). Besides, positions of the nucleus and the CS are established by separate regulatory pathways (Gomes et al., 2005). Nevertheless, mechanical effect of the CS–nucleus interaction is an important future challenge.
The model we proposed is minimal and does not consider factors such as orientation-dependent forces (Tsou et al., 2003), MT length regulation (Tolić-Nørrelykke, 2010), force–velocity properties, and force-driven detachment of dynein (Vogel et al., 2009), and MT bending (Bicek et al., 2009). There are also other positioning processes working in the cell—forceless centering mechanism (Malikov et al., 2005) and cell adhesions determining CS stabilization (Théry et al., 2006), to name but a few. Potentially all these factors are not negligible; future investigations will be needed to see whether they change our model predictions. To further test our force-balance model of centering, suggestions for future experiments include 1) using nanotechnology to build local barriers in the cytoplasm, which would perturb the pushing force and shift the CS in a predictable way; 2) using UV light to locally cancel global nocodazole effect (Hamaguchi and Hiramoto, 1986) to dissect three forces locally; and 3) using laser ablation of MTs to segregate pulling and pushing forces and test the length dependence of the pulling force from dynein (Vogel et al., 2009).
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Eric Cytrynbaum for help at early stage of this work, Drs. Iva Tolic-Nørrelykke and Ivan Maly for fruitful discussions, and anonymous reviewers for suggestions. This work was supported by National Institute of Health grants GM-068952 (to A.M.) and GM-62290 (to V.R.).
Abbreviations used:
- CS
centrosome
- MT
microtubule.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-07-0627) on October 27, 2010.
REFERENCES
- Adames N. R., Cooper J. A. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J. Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexandrova A. Y., Arnold K., Schaub S., Vasiliev J. M., Meister J.-J., Bershadsky A. D., Verkhovsky A. B. Comparative dynamics of retrograde actin flow and focal adhesions: formation of nascent adhesions triggers transition from fast to slow flow. PLoS ONE. 2008;3:e3234. doi: 10.1371/journal.pone.0003234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicek A. D., Tüzel E., Demtchouk A., Uppalapati M., Hancock W. O., Kroll D. M., Odde D. J. Anterograde microtubule transport drives microtubule bending in LLC-PK1 epithelial cells. Mol. Biol. Cell. 2009;20:2943–2953. doi: 10.1091/mbc.E08-09-0909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerknes M. Physical theory of the orientation of astral mitotic spindles. Science. 1986;234:1413–1416. doi: 10.1126/science.3787253. [DOI] [PubMed] [Google Scholar]
- Bormuth V., Varga V., Howard J., Schäffer E. Protein friction limits diffusive and directed movements of kinesin motors on microtubules. Science. 2009;325:870–873. doi: 10.1126/science.1174923. [DOI] [PubMed] [Google Scholar]
- Brangwynne C. P., MacKintosh F. C., Kumar S., Geisse N. A., Talbot J., Mahadevan L., Parker K. K., Ingber D. E., Weitz D. A. Microtubules can bear enhanced compressive loads in living cells because of lateral reinforcement. J. Cell Biol. 2006;173:733–741. doi: 10.1083/jcb.200601060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brito D. A., Strauss J., Magidson V., Tikhonenko I., Khodjakov A., Koonce M. P. Pushing forces drive the comet-like motility of microtubule arrays in Dictyostelium. Mol. Biol. Cell. 2005;16:3334–3340. doi: 10.1091/mbc.E05-01-0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky I. B., Burakov A. V., Nadezhdina E. S. Microtubules' interaction with cell cortex is required for their radial organization, but not for centrosome positioning. Cell Motil. Cytoskeleton. 2007;64:407–417. doi: 10.1002/cm.20192. [DOI] [PubMed] [Google Scholar]
- Burakov A., Nadezhdina E., Slepchenko B., Rodionov V. Centrosome positioning in interphase cells. J. Cell Biol. 2003;162:963–969. doi: 10.1083/jcb.200305082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burke B., Roux K. J. Nuclei take a position: managing nuclear location. Dev. Cell. 2009;17:587–597. doi: 10.1016/j.devcel.2009.10.018. [DOI] [PubMed] [Google Scholar]
- Civelekoglu-Scholey G., Scholey J. M. Mitotic force generators and chromosome segregation. Cell. Mol. Life Sci. 2010;67:2231–2250. doi: 10.1007/s00018-010-0326-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dogterom M., Leibler S. Physical aspects of the growth and regulation of microtubule structures. Phys. Rev. Lett. 1993;70:1347–1350. doi: 10.1103/PhysRevLett.70.1347. [DOI] [PubMed] [Google Scholar]
- Dogterom M., Yurke B. Measurement of the force-velocity relation for growing microtubules. Science. 1997;278:856–860. doi: 10.1126/science.278.5339.856. [DOI] [PubMed] [Google Scholar]
- Dujardin D. L., Vallee R. B. Dynein at the cortex. Curr. Opin. Cell Biol. 2002;14:44–49. doi: 10.1016/s0955-0674(01)00292-7. [DOI] [PubMed] [Google Scholar]
- Dujardin D. L., Barnhart L. E., Stehman S. A., Gomes E. R., Gundersen G. G., Vallee R. B. A role for cytoplasmic dynein and LIS1 in directed cell movement. J. Cell Biol. 2003;163:1205–1211. doi: 10.1083/jcb.200310097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foethke D., Makushok T., Brunner D., Nédélec F. Force- and length-dependent catastrophe activities explain interphase microtubule organization in fission yeasts. Mol. Syst. Biol. 2009;5:1–6. doi: 10.1038/msb.2008.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes E. R., Jani S., Gundersen G. G. Nuclear movement regulated by Cdc42, MRCK, myosin, and actin flow establishes MTOC polarization in migrating cells. Cell. 2005;121:451–463. doi: 10.1016/j.cell.2005.02.022. [DOI] [PubMed] [Google Scholar]
- Grabham P. W., Seale G. E., Bennecib M., Goldberg D. J., Vallee R. B. Cytoplasmic dynein and LIS1 are required for microtubule advance during growth cone remodeling and fast axonal outgrowth. J. Neurosci. 2007;27:5823–5834. doi: 10.1523/JNEUROSCI.1135-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill S. W., Gönczy P., Stelzer E.H.K., Hyman A. A. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- Grill S. W., Hyman A. A. Spindle positioning by cortical pulling forces. Dev. Cell. 2005;8:461–465. doi: 10.1016/j.devcel.2005.03.014. [DOI] [PubMed] [Google Scholar]
- Grishchuk E. L., Molodtsov M. I., Ataullakhanov F. I., McIntosh J. R. Force production by disassembling microtubules. Nature. 2005;438:384–388. doi: 10.1038/nature04132. [DOI] [PubMed] [Google Scholar]
- Hamaguchi M. S., Hiramoto Y. Analysis of the role of astral rays in pronuclear migration in sand dollar eggs by the colcemid-UV method. Dev. Growth Differ. 1986;28:143–156. doi: 10.1111/j.1440-169X.1986.00143.x. [DOI] [PubMed] [Google Scholar]
- Helfand B. T., Chang L., Goldman R. D. Intermediate filaments are dynamic and motile elements of cellular architecture. J. Cell Sci. 2004;15:133–141. doi: 10.1242/jcs.00936. [DOI] [PubMed] [Google Scholar]
- Holy T. E., Dogterom M., Yurke B., Leibler S. Assembly and positioning of microtubule asters in microfabricated chambers. Proc. Nat. Acad. Sci. USA. 1997;94:6228–6231. doi: 10.1073/pnas.94.12.6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard J. Mechanics of Motor Proteins and the Cytoskeleton. Sunderland, MA: Sinauer Associates; 2001. [Google Scholar]
- Huang J. D., Brady S. T., Richards B. W., Stenoien D., Resau J. H., Copeland N. G., Jenkins N. A. Direct interaction of microtubule- and actin-based transport motors. Nature. 1999;397:267–270. doi: 10.1038/16722. [DOI] [PubMed] [Google Scholar]
- Hwang E., Kusch J., Barral Y., Huffaker T. C. Spindle orientation in Saccharomyces cerevisiae depends on the transport of microtubule ends along polarized actin cables. J. Cell Biol. 2003;161:483–488. doi: 10.1083/jcb.200302030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janson M. E., de Dood M. E., Dogterom M. Dynamic instability of microtubules is regulated by force. J. Cell Biol. 2003;161:1029–1034. doi: 10.1083/jcb.200301147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Onami S. Computer simulations and image processing reveal length-dependent pulling force as the primary mechanism for C. elegans male pronuclear migration. Dev. Cell. 2005;8:765–775. doi: 10.1016/j.devcel.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Kobayashi T., Murayama T. Cell cycle-dependent microtubule-based dynamic transport of cytoplasmic dynein in mammalian cells. PLoS One. 2009;4:e7827. doi: 10.1371/journal.pone.0007827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodama A., Karakesisoglou I., Wong E., Vaezi A., Fuchs E. ACF 7, an essential integrator of microtubule dynamics. Cell. 2003;115:343–354. doi: 10.1016/s0092-8674(03)00813-4. [DOI] [PubMed] [Google Scholar]
- Koonce M. P., Khodjakov A. Dynamic microtubules in Dictyostelium. J. Muscle Res. Cell M. 2002;23:613–619. doi: 10.1023/a:1024446821701. [DOI] [PubMed] [Google Scholar]
- Malikov V., Cytrynbaum E. N., Kashina A., Mogilner A., Rodionov V. Centering of a radial microtubule array by translocation along microtubules spontaneously nucleated in the cytoplasm. Nat. Cell Biol. 2005;7:1213–1218. doi: 10.1038/ncb1332. [DOI] [PubMed] [Google Scholar]
- Mallik R., Carter B. C., Lex S. A., King S. J., Gross S. P. Cytoplasmic dynein functions as a gear in response to load. Nature. 2004;427:649–652. doi: 10.1038/nature02293. [DOI] [PubMed] [Google Scholar]
- Manneville J. B., Etienne-Manneville S. Positioning centrosomes and spindle poles: looking at the periphery to find the centre. Biol. Cell. 2006;98:557–565. doi: 10.1042/BC20060017. [DOI] [PubMed] [Google Scholar]
- Mikhailov A. V., Gundersen G. G. Centripetal transport of microtubules in motile cells. Cell Motil. Cytoskeleton. 1995;32:173–186. doi: 10.1002/cm.970320303. [DOI] [PubMed] [Google Scholar]
- Mogilner A., Oster G. The physics of lamellipodial protrusion. Eur. Biophys. J. 1996;25:47–53. [Google Scholar]
- Mullins R. D. Cytoskeletal mechanisms for breaking cellular symmetry. Cold Spring Harb. Perspect. Biol. 2010;2:1–16. doi: 10.1101/cshperspect.a003392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connell C. B., Wang Y.-L. Mammalian spindle orientation and position respond to changes in cell shape in a dynein-dependent fashion. Mol. Biol. Cell. 2000;11:1765–1774. doi: 10.1091/mbc.11.5.1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oiwa K., Sakakibara H. Recent progress in dynein structure and mechanism. Curr. Opin. Cell Biol. 2005;17:98–103. doi: 10.1016/j.ceb.2004.12.006. [DOI] [PubMed] [Google Scholar]
- Paul R., Wollman R., Silkworth W. T., Nardi I. K., Cimini D., Mogilner A. Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proc. Nat. Acad. Sci. USA. 2009;106:15708–15713. doi: 10.1073/pnas.0908261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Bloom K. Dynamic microtubules lead the way for spindle positioning. Nat. Rev. Mol. Cell Biol. 2004;5:481–492. doi: 10.1038/nrm1402. [DOI] [PubMed] [Google Scholar]
- Reinsch S., Gönczy P. Mechanisms of nuclear positioning. J. Cell Sci. 1998;111:2283–2295. doi: 10.1242/jcs.111.16.2283. [DOI] [PubMed] [Google Scholar]
- Robinson J. T., Wojcik E. J., Sanders M. A., McGrail M., Hays T. S. Cytoplasmic dynein is required for the nuclear attachment and migration of centrosomes during mitosis in Drosophila. J. Cell Biol. 1999;146:597–608. doi: 10.1083/jcb.146.3.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt J., Cramer L. P., Baum B., McGee K. M. Myosin II-dependent cortical movement is required for centrosome separation and positioning during mitotic spindle assembly. Cell. 2004;117:361–372. doi: 10.1016/s0092-8674(04)00341-1. [DOI] [PubMed] [Google Scholar]
- Salmon W. C., Adams M. C., Waterman-Storer C. M. Dual-wavelength fluorescent speckle microscopy reveals coupling of microtubule and actin movements in migrating cells. J. Cell Biol. 2002;158:31–37. doi: 10.1083/jcb.200203022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starr D. A. Communication between the cytoskeleton and the nuclear envelope to position the nucleus. Mol. Biosyst. 2007;3:583–589. doi: 10.1039/b703878j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Théry M., Racine V., Piel M., Pépin A., Dimitrov A., Chen Y., Sibarita J.-B., Bornens M. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc. Nat. Acad. Sci. USA. 2006;103:19771–19776. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolić-Nørrelykke I. M., Sacconi L., Thon G., Pavone F. S. Positioning and elongation of the fission yeast spindle by microtubule-based pushing. Curr. Biol. 2004;14:1181–1186. doi: 10.1016/j.cub.2004.06.029. [DOI] [PubMed] [Google Scholar]
- Tolić-Nørrelykke I. M. Force and length regulation in the microtubule cytoskeleton: lessons from fission yeast. Curr. Opin. Cell Biol. 2010;22:21–28. doi: 10.1016/j.ceb.2009.12.011. [DOI] [PubMed] [Google Scholar]
- Tran P., Marsh L., Doye V., Inoué S., Chang F. A mechanism for nuclear positioning in fission yeast based on microtubule pushing. J. Cell Biol. 2001;153:397–412. doi: 10.1083/jcb.153.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou M.F.B., Ku W., Hayashi A., Rose L. S. PAR-dependent and geometry-dependent mechanisms of spindle positioning. J. Cell Biol. 2003;160:845–855. doi: 10.1083/jcb.200209079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel S. K., Pavin N., Maghelli N., Jülicher F., Tolić-Nørrelykke I. M. Self-organization of dynein motors generates meiotic nuclear oscillations. PLoS Biol. 2009;7:e1000087. doi: 10.1371/journal.pbio.1000087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada M., Suetsugu N. Plant organelle positioning. Curr. Opin. Cell Biol. 2004;7:626–631. doi: 10.1016/j.pbi.2004.09.005. [DOI] [PubMed] [Google Scholar]
- Weber K. L., Sokac A. M., Berg J. S., Cheney R. E., Bement W. M. A microtubule-binding myosin required for nuclear anchoring and spindle assembly. Nature. 2004;431:325–329. doi: 10.1038/nature02834. [DOI] [PubMed] [Google Scholar]
- Wollman R., Civelekoglu-Scholey G., Scholey J. M., Mogilner A. Reverse engineering of force integration during mitosis in the Drosophila embryo. Mol. Syst. Biol. 2008;4:1–13. doi: 10.1038/msb.2008.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wühr M., Dumont S., Groen A. C., Needleman D. J., Mitchison T. J. How does a millimeter-sized cell find its center? Cell Cycle. 2009;8:1115–1121. doi: 10.4161/cc.8.8.8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam P. T., Wilson C. A., Ji L., Hebert B., Barnhart E. L., Dye N. A., Wiseman P. W., Danuser G., Theriot J. A. Actin-myosin network reorganization breaks symmetry at the cell rear to spontaneously initiate polarized cell motility. J. Cell Biol. 2007;178:1207–1221. doi: 10.1083/jcb.200706012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yvon A.M.C., Wadsworth P. Region-specific microtubule transport in motile cells. J. Cell Biol. 2000;151:1003–1012. doi: 10.1083/jcb.151.5.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.