Abstract
The human gastrointestinal tract comprises a series of complex and dynamic organs ranging from the stomach to the distal colon, which harbor immense microbial assemblages that are known to be vital for human health. Until recently, most of the details concerning our gut microbiota remained obscure. Over the past several years, however, a number of crucial technological and conceptual innovations have been introduced to shed more light on the composition and functionality of human gut microbiota. Recently developed high throughput approaches, including next-generation sequencing technologies and phylogenetic microarrays targeting ribosomal RNA gene sequences, allow for comprehensive analysis of the diversity and dynamics of the gut microbiota composition. Nevertheless, most of the microbes especially in the human large intestine still remain uncultured, and the in situ functions of distinct groups of the gut microbiota are therefore largely unknown, but pivotal to the understanding of their role in human physiology. Apart from functional and metagenomics approaches, stable isotope probing is a promising tool to link the metabolic activity and diversity of microbial communities, including yet uncultured microbes, in a complex environment. Advancements in current stable isotope probing approaches integrated with the application of high-throughput diagnostic microarray-based phylogenetic profiling and metabolic flux analysis should facilitate the understanding of human microbial ecology and will enable the development of innovative strategies to treat or prevent intestinal diseases of as yet unknown etiology.
Keywords: microbiota, gastrointestinal tract, diagnosis, physiology, stable isotope probing
Introduction
The human body typically harbors 10 times more microbial cells than human cells, which is mainly due to the extremely high density of microorganisms found to be present in the human gastrointestinal (GI) tract [Backhed et al. 2005; Berg, 1996; Savage, 1977]. The vast majority of this microbiota is located particularly at the distal region of the human GI tract, which is the colon [Eckburg et al. 2005; Suau et al. 1999]. From a medical perspective, the importance of this part of the human GI tract to host health was acknowledged even by early observers such as Hippocrates in 400 BC, who stated that ‘death sits in the bowel’ [Kolida et al. 2000]. Most of the current clinical knowledge focuses on the pathogenesis of disease and its appropriate therapy rather than giving us a clear definition of health [Neish, 2009; Arebi et al. 2008; Tannock, 2006]. However, there is growing evidence of the important impact of the colonic microbiota on human gut physiology and health which is strongly affected by a number of microbial activities. These activities include, but are not restricted to, fermentation of dietary compounds that escape digestion in the upper GI tract, processing of mucosal cells shed in the small intestine, and degradation of intestinally secreted mucus [Srikanth and McCormick, 2008; Fava et al. 2006; Noverr and Huffnagle, 2004; Xu and Gordon, 2003]. For understanding the functionality of microbial communities, it is necessary to elucidate the role of individual species within a community. However, it is estimated that approximately 80% of species comprising the human gut microbiota are yet to be cultured [Rajilic-Stojanovic et al. 2007; Egert et al. 2006]. Hence, insights into the function and metabolic potential of these uncultured microbes are lacking. This indicates that culture-independent approaches are crucial to comprehensively study the ecology of the GI tract microbiota. In addition, our knowledge of gut microbiota is in general restricted to the luminal part at the end of the colon — reflected by feces — as other parts of the GI tract can so far only be accessed using invasive procedures. However, minimally invasive experimental techniques that can be used in vivo can now be applied in combination with stable isotope probing (SIP) [Kreuzer-Martin, 2007; Whiteley et al. 2006; Radajewski et al. 2000] to link in situ microbial activity and the diversity of GI tract microbiota. SIP is a powerful tool that can be used in human studies to delineate bacterial food webs that may ultimately influence human wellbeing [Dolnikowski et al. 2005; Kelleher, 2004; Pouteau et al. 2003].
This review presents promising strategies to delve into the functionality of the GI tract microbiota, including fermentation processes in the human colon. Recent scientific advances discussed here could assist in expanding the knowledge of microbial determinants for a healthy gut definition based on key functional properties of gut microbiota. This will also enable the development of direct nutritional strategies for intestinal disease prevention and health promotion.
Overview of human gut microbiota — microbial diversity
The human gut is one of the most densely populated ecosystems, comprising members of the three domains of life on Earth — bacteria, archaea and eucarya [Finegold et al. 1983]. Bacteria dominate this complex ecosystem, where more than 90% of the phylotypes are member of two bacteria divisions: the Bacteroidetes and the Firmicutes [Turroni et al. 2008; Zoetendal et al. 2006; Backhed et al. 2005]. The Gram-positive Firmicutes include numerous different phylogenetic clusters of Clostridia, with clusters IV, IX and XIVa being the most abundant clusters. The predominant genera are Clostridium, Eubacterium, Roseburia and Ruminococcus. Furthermore, the Actinobacteria, including the genera Bifidobacterium and Atopobium, represent important members of the gut microbial community [Turroni et al. 2008; Van Der Waaij et al. 2005; Harmsen et al. 2002; Franks et al. 1998]. In terms of functional diversity, recent metage-nomics-based studies have indicated that the gut microbiome has a coding capacity that vastly exceeds that of the human genome and encodes biochemical pathways that humans have not evolved [Kurokawa et al. 2007; Turnbaugh et al. 2007; Gill et al. 2006; Ley et al. 2006; Backhed et al. 2005].
Recent studies of the gut microbial ecosystem have identified more than 1000 species and possibly over 7000 strains, of which the largest part (~80%) remains uncultured [Zoetendal et al. 2008; Blaut and Clavel, 2007; Rajilic-Stojanovic et al. 2007; Backhed et al. 2005]. However, new approaches for culturing previously uncultured colonic microbes are being developed [Zoetendal et al. 2008; Duncan et al. 2007; Ingham et al. 2007]. In addition to this, new powerful tools for amplification and sequencing of genomic DNA from minute quantities of a sample and barcoded pyrosequencing can be expected to give new insights into the composition of the gut microbiota at high spatiotemporal resolution [Andersson et al. 2008; Marcy et al. 2007].
Almost sterile at birth [Digiulio et al. 2008], the development of the infant gut proceeds to extremely dense colonization, reaching by the age of 2 years a climax mixture of microbes similar to the microbiota found in the adult intestine [Wall et al. 2009]. The composition of infant gut microbiota is determined by several factors that include the mode of delivery, maternal microbiota, diet and environmental hygiene [Palmer et al. 2007; Hallstrom et al. 2004; Fanaro et al. 2003; Favier et al. 2003]. In contrast to the developing infant gut microbiota, each healthy adult's gut appears to have a unique and stable microbiota, as evidenced by molecular fingerprinting, over the time scale of months [Frank and Pace, 2008; Turnbaugh et al. 2007; Zoetendal et al. 1998]. Recent studies have also indicated aberrations in the composition of the human microbiome in obese individuals [Ley et al. 2006], as well as in individuals with a variety of other diseases [Turnbaugh et al. 2009; Zoetendal et al. 2008]. Furthermore, Ley et al. [2006] reported that the composition of the human gut microbiota is responsive to dietary modulation for weight reduction.
Metabolic roles of gut microbiota
An important role of the human gut microbiota is that of a metabolic ‘organ’, which delicately affects our physiology with functions that we have not had to evolve on our own [Turnbaugh et al. 2007; Gill et al. 2006; Backhed et al. 2005]. The ability to process otherwise indigestible components of our diet is one of these vital microbial activities that significantly influences the gut environment and the host, such as providing an energy source and maintaining gut health [Guarner and Malagelada, 2003; Xu and Gordon, 2003; Savage, 1986].
Microbial performance, growth and metabolism in the human colon depends to a large extent on the supply of substrates that resist digestion in the upper GI tract and endogenous substrates, such as mucin, secreted by the host [Blaut and Clavel, 2007]. The main dietary products, which serve as food for the colonic microbiota, are complex carbohydrates (starches, nonstarch polysaccharides) and proteins [Cummings and Englyst, 1987]. The majority of microorganisms in the human colon ferment carbohydrates and then switch to protein fermentation when these are not available [Ouwehand et al. 2005]. Carbohydrate metabolism is of great importance in the large intestine, as in terms of absolute numbers, the vast majority of culturable microorganisms are saccharolytic [Macfarlane and Macfarlane, 1997]. Numerous different types of carbohydrates reach the colon, where their rates of fermentation are affected by the transit time and vary according to substrate availability, chemical structure and composition [Englyst et al. 1992]. Several studies suggest that dietary carbohydrates are protective against several GI disorders such as colorectal cancer [Guarner, 2005; Topping and Clifton, 2001] but many of these studies have been performed with animal models [Young et al. 2005, Le Leu et al. 2007a, 2007b; Pool-Zobel, 2005; Cassidy et al. 1994]. The underlying mechanism of protection could be associated with the end products of these anaerobic bacterial fermentations, but other metabolic interactions cannot be excluded [Roediger, 1988]. In the human colon, the end products of fermentation are short-chain fatty acids (SCFAs) such as butyrate, acetate, propio-nate, as well as other terminal products such as lactate. SCFAs lead to lowering of the luminal pH, an increase in the bacterial biomass and fecal bulk, and modification of the microbial composition, especially by stimulating the growth of beneficial bacteria including bifidobac-teria and lactobacilli [Le Leu et al. 2005].
Butyrate, one of the major SCFAs, has been the focus of studies aimed at understanding the role of SCFAs in nourishing the colonic epithelium and in the prevention of colon cancer [Hamer et al. 2008; Bauer-Marinovic et al. 2006; Sengupta et al. 2006; Cummings and Bingham, 1987]. Recently, it was observed in healthy individuals that colonic butyrate application resulted in reduced visceral pain perception [Vanhoutvin et al. 2009]. In contrast, colonic protein fermentation is often associated with an increased colon cancer risk as this fermentation results in the production of branched chain fatty acids and potentially toxic metabolites such as amines, ammonia, phenolic compounds and thiols [Bingham et al. 1996; Cummings et al. 1979]. This is also indicated by the fact that colon cancer mostly occurs at the distal end of the colon [Muir et al. 2004; Bufill, 1990]. Therefore, an intake of more slowly fermentable carbohydrates could result in prolongation of the potentially beneficial saccharoly-tic activity, which would lead to an increased production and delivery of SCFA, particularly butyrate, to the distal colon [Wong et al. 2005; Topping and Clifton, 2001; Jacobasch et al. 1999].
Obviously, diet affects colonic nutrition mainly through its effects on gut microbiota. Increasing evidence defines the roots of many colonic diseases and particularly colonic cancer risk to be determined by interactions between the diet and gut microbiota. However, further studies should focus on unraveling the in situ functionality of the gut microbiota and improving the understanding of the impact of the microbiota on host health and wellbeing. This is a difficult task because of the individuality and complexity of a microbial community in a largely inaccessible environment.
In vitro models of the human colon
The human colon is a largely inaccessible part of the GI tract, and a difficult area to study the gut microbiota and microbial activities in vivo. To this end, in vitro modeling represents an elegant way to study the microbial processes, such as carbohydrate and protein fermentation [Macfarlane and Macfarlane, 2007]. In vitro studies are less expensive using pure cultures, defined mixed cultures and stool material as inoculum. Furthermore, in vitro models allow for fast and reproducible experiments under standardized conditions. The strength of in vitro models, however, has also been questioned with respect to several issues. The degree to which the inoculum represents the human colonic microbiota [Drasar, 1988] and the precise mimicking of the colonic conditions [Edwards and Rowland, 1992] are recurring points of discussion. Another limitation of in vitro modeling is that it does not represent the colon as an open system with respect to the absence of excretion of fecal content which inevitably results in changes in bacterial composition and subsequently metabolic activity [Christian et al. 2003]. Similarly, in vitro models lack host cells, thus, their activity and interaction with colonic microbiota cannot be measured. Despite these constraints, in vitro model systems can serve as tools (Table 1) to study microbe-mediated processes occurring in the human colon and to estimate the consequences of these activities on gut health.
Table 1.
In vitro systems used to study human gut microbiota.
| In vitro model | Targeted part of the human Gl-tract | Reference |
| SHIME–model (Simulator of the Human Intestinal Microbial Ecosystem) | Small intestine and colon | Molly et al. [1993] |
| Continuous three-stage system | Proximal and distal colon | Macfarlane et al. [1998] |
| TIM-model (TNO intestinal model) | Stomach, small intestine and colon | Minekus et al. [1995], Minekus et al. [1999] |
| Continuous two-stage system | Proximal and distal colon | Brück et al. [2003] |
| Three-stage culture system | Infant proximal, transverse and distal colon | Cinquin et al. [2006] |
| Human proximal colon system | Proximal colon | Jiménez-Vera et al. [2008] |
Many studies on the fermentation characteristics of relevant dietary carbohydrates have been performed with the use of in vitro models of the gut [Jiménez-Vera et al. 2008; Van De Wiele et al. 2007; Probert et al. 2004; Van Nuenen et al. 2003]. Additionally, in vitro modeling systems are also used to study human intestinal microbes able to colonize mucus and to establish biofilm communities [Macfarlane et al. 2005]. Recently the TNO in vitro model of the large intestine — termed TIM-2 [Minekus et al. 1999] — was used in combination with isotopically labeled substrates to identify colonic populations actively involved in the fermentation of glucose [Egert et al. 2007] and potato starch [Kovatcheva-Datchary et al. 2008]. An important advantage of this in vitro system is the fact that metabolites and water can be constantly removed from the module. In this computer-controlled model, parameters such as transit time and pH are regulated and, for example, age-dependent colon simulations can be achieved. Moreover, peristaltic mixing is simulated and microorganisms reach physiological densities (about 1 × 109-1010ml-1).
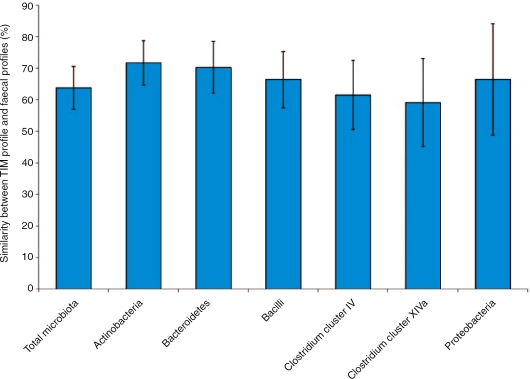
We have recently used high-throughput phylogenetic microarray analysis to compare the microbial community that colonized the TIM-2 model with the fecal community of randomly selected adult volunteers. The data indicated that TIM-2 microbiota is not significantly different from the fecal microbial community of the human volunteers with respect to composition and diversity of the major microbial groups (Figure 1). This is further evidence that the TIM-2 system appears to be representative of the human large intestinal microbiota [Kovatcheva-Datchary et al. 2008].
Figure 1.
Similarity of the total microbiota and major phylogenetic groups between TIM-2 profile and human faecal profiles. Samples were analyzed using the Human Intestinal Tract Chip (HITChip) [Rajilić-Stojanović et al. 2009].
Diagnostic tools to assess microbial diversity of the human gut
Our current knowledge about the microbial composition of the colonic ecosystem in health and disease is still limited. In order to be able to diagnose the presence and abundance of key players of the gut microbiota, a number of culture-independent approaches have been applied.
The gut microbiota composition is likely to be influenced by complex interactions between host, microbes and the environment. Diet is an important factor, which undoubtedly shapes the gut microbiota, and has been explored in detail using the power of molecular fingerprinting techniques and 16S ribosomal RNA gene sequencing [Abell et al. 2008; Bartosch et al. 2005; Hayashi et al. 2002]. Recently, the influence of dietary factors was studied by Ley et al. [2006], where a high throughput sequencing approach was applied to characterize the fecal microbiota of 12 obese individuals who received either fat-restricted or carbohydrate-restricted low-caloric diets. It was shown that the stool samples of obese subjects were significantly enriched in Firmicutes and depleted in Bacteroidetes, in comparison to samples obtained from lean individuals. In addition, the microbiota of obese individuals became more similar to that of lean subjects over the course of 52 weeks of treatment. In a separate study, fluorescent in situ hybridization (FISH) analysis was applied to investigate the effect of reduced carbohydrate intake on fecal microbiota composition of twenty obese individuals [Duncan et al. 2007]. A progressive decrease was observed in populations related to Roseburia spp., E. rectale and Bifidobacterium spp., as a fraction of total bacterial cells, after decreasing carbohydrate intake. These data showed that dietary carbohydrate supply is an important factor for these microbial groups in order to maintain their populations in the human colon [Duncan et al. 2007].
The two primary human inflammatory bowel diseases (IBD), Crohn's disease (CD) and ulcerative colitis (UC), are usually associated with unstable and disturbed composition of the gut microbiota in comparison with healthy individuals. In the last few years, a number of research groups have focused their activities on determining the gut microbial composition in patients and to define how it is impacted by disease. The composition of the gut microbiota in concordant and discordant identical twins with CD, and healthy
twins, was studied to identify members of the microbiota, which could be linked to CD incidence or development [Dicksved et al. 2008]. Molecular fingerprinting analyses based on T-RFLP of the 16S rRNA sequences revealed a higher microbial diversity in the healthy twins compared to the CD twins. Moreover, the fecal microbiota of the healthy individuals was found to be less variable than those of CD twins. The microbial community profiles of individuals with ileal CD were significantly different from healthy individuals and those with colonic CD. Furthermore, a lower relative abundance of B. uniformis and higher abundance of B. ovatus and B. vulgatus was observed in all patients with ileal involvement in comparison to both healthy twins and twins with colonic disease [Dicksved et al. 2008]. In another study, temperature gradient gel electrophoresis (TGGE) of PCR-ampli-fied 16S rRNA gene fragments was applied to investigate the effect of enteral nutrition therapy on the fecal microbiota in children with CD [Lionetti et al. 2005]. This revealed differences in the microbial composition of healthy subjects and CD patients, but also within the latter, between patients in remission and relapse. Recently, 16S rRNA sequence data were collected from fecal and biopsy samples from CD and UC patients, and compared to those from healthy individuals [Frank et al. 2007]. Significant differences between the gut micro-biota of the two patient groups were detected, including depletion in the level of Firmicutes and Bacteroidetes in comparison with healthy controls. Determining the differences in microbial composition in patients and healthy controls may thus provide novel therapeutic targets. For this purpose, high-throughput, cost-effective methods for microbiota characterization are needed. Recently the application of 454-pyrose-quencing of hypervariable regions of the 16S rRNA gene revealed taxonomic richness of the gut microbiota that exceed any previously reported estimates [Andersson et al. 2008; Dethlefsen et al. 2008]. Pyrosequencing analyses were applied recently in order to study the role of the gut microbiota in the development of obesity. High bacterial diversity and enrichment of H2-producing Prevotellaceae accompanied by a high abundance of H2-utilizing methanogenic Archaea, was found [Zhang et al. 2009].
Phylogenetic microarrays are high-throughput analytical tools, which can be used to measure diversity and abundance of the human gut microbiota. Recently, such a DNA microarray, the Human Intestinal Tract Chip (HITChip), was developed, combining the power of fingerprinting, phylogenetic and quantitative community analysis [Rajilić-Stojanović et al. 2009]. The HITChip targets over 1000 phylotypes of intestinal microbiota, and its application for the analysis of intestinal samples of patients and healthy individuals can provide novel insights into the relationship between the gut microbiota in health or disease [Zoetendal et al. 2008].
Diagnostic tools to assess microbial functionality of the human gut
To understand the complex changes in gut microbiota composition that may predispose towards intestinal disorders or promote human health, techniques that can assay and link metabolic activity to the diversity of intestinal bacteria are needed. Recently explored metagenomics approaches allow the comprehensive study of phylogenetic, physical and functional properties of complex microbial communities, providing a full picture of microbiota dynamics [Handelsman, 2004]. Because metagenomic analyses allow the study of phylogenetic diversity, as well as providing an inventory of potential functions of gut microbiota, it can be used as a tool to link diversity to functionality [Booijink et al. 2007]. Metagenomics screening approaches can be divided into functional and sequence-based driven analyses of collective microbial genomes in complex environments [Gabor et al. 2007]. Sequence-based metagenomic investigations have started to reveal core metabolic functions of the gut microbiota. An early metagenomic study on two healthy adults showed that their fecal microbial metagenomes were enriched with genes involved in energy metabolism, which also include the production of SCFAs as the pivotal energy supply for the intestine [Gill et al. 2006]. Additionally, a recent study where metaproteomics analyses were applied to study distal gut microbiota of a healthy twin pair, indicated that more than 50% of the detected proteins were involved in translation, energy production and carbohydrate metabolism [Verberkmoes et al. 2009]. Comparison of metagenomics [Gill et al. 2006] and metaproteomics data [Verberkmoes et al. 2009] indicated matches in the fucose and butyrate colonic fermentation pathways. Recent large-scale comparative metagenomic analyses demonstrated a clear effect of diet and age on the gut microbiome [Kurokawa et al. 2007].
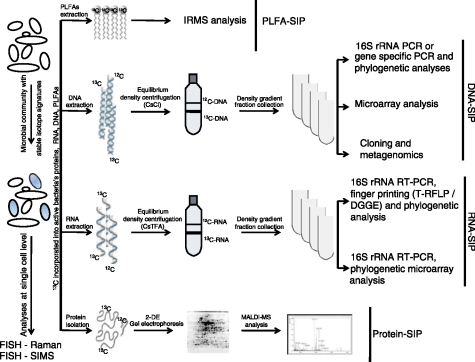
However, an ongoing challenge for microbiologists is to be able to identify which microbes in the human gut carry out a specific metabolic conversion, the products of which may promote intestinal disorders and/or gut health. Recently, isotope probing approaches have been developed, offering great potential to identify microbes that are involved in the metabolism of specific substrates. These molecular tools involve the use of commercially prepared substrates highly enriched in a stable isotope (e.g. 13C) or radioisotope (e.g. 14C), which is added to an environmental sample. Endogenous microbes that metabolize the labeled substrate will incorporate the isotope into components of the microbial cells that provide phylogenetic information [Dumont et al. 2006; Manefield et al. 2006; Lueders et al. 2004; Manefield et al. 2002; Radajewski et al. 2000]. Such components are often referred to as biomarkers, and nucleic acids and fatty acids are most commonly used. FISH-microautoradiography (FISH-MAR) and isotope array technology both use radioactive tracers to study the incorporation of substrate. With FISH-MAR, a direct monitoring of the incorporation of the substrate labeled with a radioactive isotope into single microbial cells is performed [Lee et al. 1999]. In a different way, the analyses with the isotope arrays are performed, requiring the isolation of the labeled biomarker. Ribosomal RNA is hybridized to oligonucleotide arrays to target the 16S rRNA of the bacteria of interest [Adamczyk et al. 2003]. However, since both methods use radioisotopes their application is limited, especially in animal and human subjects, and therefore stable isotopes can offer a safe and convenient alternative for in vivo analysis. SIP methodologies vary in the use of biomarkers, but also the means by which biomarkers are analyzed for isotopic and phylogenetic content (Figure 2). The first application of SIP was in the analysis of phospholipid fatty acids (PLFA) that can be extracted from environmental samples and analyzed by isotope-ratio mass spectro-metry (IRMS) [Boschker et al. 1998]. Microbial populations often have signature PLFA molecules, which allow identification of microbes that have incorporated the 13C-substrate. However, the interpretation of the PLFA patterns of microbes for which there are no cultivated representatives still remain limited, which is the main restriction of the PLFA-SIP [Dumont and Murrell, 2005].
Figure 2.
Summary of stable isotope probing (SIP) approaches, suitable diagnostic tools, to assess gut microbiota functionality and link it to phylogeny.
Nucleic acids (NA) have a higher phylogenetic resolution than PLFA-SIP and enable identification of active but as yet uncultured populations at the species level. NA-based SIP works on the principle of separation of isotopically labeled DNA or RNA from unlabeled NA. The isolated labeled DNA/RNA represents the microbial populations that incorporated the isotope into the biomarker through metabolic sequestration. NA-based SIP experiments have been applied to a large number of environmental studies focused on the identification of bacteria that carry out specific degradative functions. Different culture independent techniques were used to monitor the 13C-DNA/RNA. DNA-based SIP studies have described the application of polymerase chain reaction (PCR) analyses, targeting functional or taxonomic marker genes [Morris et al. 2002; Radajewski et al. 2002; Radajewski et al. 2000]. As a culture-independent microbial taxo-nomic marker, the 16S rRNA and the encoding gene have been applied most frequently. Subsequent fingerprinting (e.g. denaturing gradient gel electrophoresis [DGGE] or T-RFLP), 16S rRNA clone library construction and/or microarray analyses, are further used to reveal the microbial populations involved in the degradation of the particular substrate [Cupples et al. 2007; Neufeld et al. 2007; Dumont et al. 2006]. DNA-SIP enables analysis of isotopically labeled functional genes [Dumont and Murrell, 2005], which provides a further functional view of the active microbiota. Moreover, DNA-SIP in combination with metagenomics can provide a broad insight into the genetic potential of microorganisms that are attributed to the in situ use of specific substrates [Egert et al. 2006]. An important limitation of DNA-SIP is the requirement for DNA synthesis and cell division in order to obtain sufficient incorporation of the label into the DNA for gradient separation. Conversely, RNA occurs in greater cellular copy numbers, has a higher turnover rate than DNA and is produced independent of cellular replication. For this reason, RNA will be labeled more rapidly than DNA making it a highly responsive biomar-ker in SIP analyses [Manefield et al. 2002]. Due to its greater buoyant density, isopycnic centrifu-gation of RNA is performed in cesium trifluoroa-cetate (CsTFA) rather than CsCl. Additionally, based on the reduced loading capacity of CsTFA, lower RNA loading amounts (250–500ngml_1) [Egert et al. 2007; Whiteley et al. 2006; Lueders et al. 2004] are required for a successful separation in comparison with the DNA-SIP, where 5?gml_1 of DNA is an optimal concentration Jensen et al. 2008; Lueders et al. 2004]. To analyze the fractionated RNA, qualitative analyses such as reverse transcriptase PCR-based fingerprinting methods (DGGE T-RFLP) and subsequent cloning and sequencing, or phylogenetic microarray analysis, are applied, which enable phylogenetic identification of the active microbial population [Kovatcheva-Datchary et al. 2008; Egert et al. 2007; Lueders et al. 2004]. Additionally, quantitative evaluation of the isopycnic RNA gradients can be performed using reverse transcriptase quantitative PCR (RT-qPCR), which leads to high precision and better resolution for recovery of the labeled nucleic acids [Lueders et al. 2004]. The success of NA-SIP depends mostly on the sufficient degree of labeling required for the separation of labeled and unlabeled nucleic acids by buoyant density centrifugation. To this end, extended incubation times are often required. However, increasing the time of nucleic acid enrichment has to be balanced in order to avoid changes that can occur in the bacterial community after addition of the substrate of interest. An example could be the effect of secondary degradation of the substrates (cross-feeding), which can affect bacterial diversity and metabolic activity [Belenguer et al. 2006]. However, such cross-feeding effects can also be instrumental in identifying food chains in the human intestinal systems. Recent studies with 16S rRNA-based SIP performed using in vitro conditions in the human intestine showed that a high concentration of labeled tracer is necessary to have good separation between the labeled and unlabeled fractions of the nucleic acids [Egert et al. 2007].
Protein-based stable isotope probing (protein-SIP) is a novel approach that analyzes specific metabolic activity of a single bacterial species within a community, which incorporate the labeled substrate using proteins as a biomarker [Jehmlich et al. 2008]. The most important advantage of the protein analysis is its direct connection to physiological function, as proteins are known to catalyze the biochemical reaction. Thus, proteins are a source of phylogenetic and functional information, making them ideal bio-markers for monitoring community structure and function.
Recently, new elegant tools have been developed that combine single-cell technologies with stable isotope analysis of microbial communities to monitor stable-isotope uptake at the single-cell level. These include technologies such as Raman microspectroscopy [Huang et al. 2004] and nano-secondary ion mass spectrometry (nano-SIMS) [Kuypers and Jørgensen, 2007]. Raman spectroscopy analyses enable the detection of clear shifts in key regions (phenylalanine, proteins and nucleic acids) of bacterial Raman spectral profiles, which allow detection of the incorporation of the stable isotope into an individual cell. Furthermore, the Raman approach can be combined with FISH (Raman-FISH), which facilitates the understanding of the link between individual bacterial cells and their metabolic functions [Huang et al. 2007]. The nano-SIMS technology analyses both stable- and radioactive-isotope content at single cell resolution, which exceeds the capacity of a Raman microscope [Kuypers and Jørgensen, 2007]. Combination of FISH-nanoSIMS allows the phylogenetic and isotopic analysis of a sample in a single scan. Nevertheless, this technology is far from becoming commonplace and affordable, mostly because of the high cost of the infrastructure required for nanoSIMS analysis.
Furthermore, SIP techniques are also suitable for obtaining qualitative and quantitative information about metabolic fluxes in the colon. Isotopically labeled compounds enable the selective study of that part of the microbial or host metabolism that involves the isotopic tracer. NMR and gas- or liquid-chromatography can be used to measure the labeled compounds and further identify active metabolic pathways [Egert et al. 2006; Bacher et al. 1998]. In a very recent study, we reported the application of RNA based-SIP in combination with liquid-chromatography (LC-MS). The molecular data indicated Ruminococcus bromii as the primary degrader of starch in an in vitro model of the human colon, as it was found to predominate in the labelled fractions. Furthermore, the integration of molecular and metabolite data suggested metabolic cross-feeding in the system, where populations related to R. bromii are the primary starch degrader, while those related to Prevotella spp., Bifidobacterium adolescentis and Eubacterium rectale might be further involved in the trophic chain.
Future perspectives
Identification of the prime functions of human gut microbiota in maintaining human health requires a better understanding of its diversity and functionality, which can also facilitate its manipulation. Most intestinal microbes have not been cultured and the in situ functions of distinct groups of gut microbiota are largely unknown but pivotal to the understanding of their role in health and disease. Technological advances in culture-independent microbiology have revolutionized gastroenterological microbiology. Recently introduced metagenomics approaches have become extremely useful in addressing knowledge gaps on gut microbiota composition and have started to reveal core metabolic functions of the gut microbiota. However, it is not known which members of the gut micro-biota are involved in specific metabolic activities in situ. An important function of the gut micro-biota is related to fermentation of nondigestible dietary residue, metabolites of which are considered to be essential for intestinal health. New developments in stable isotope-based approaches can be used in identifying the key players of gut microbiota, the functions of which may have a direct impact on human wellbeing. Furthermore, extending the in vitro models to human feeding trials, in which relevant dietary oligo- and polymeric carbohydrates are delivered to the human colon, will allow the exploration of the real power of these molecular approaches.
Conflict of interest statement None declared.
Contributor Information
Petia Kovatcheva-Datchary, TI Food and Nutrition, Wageningen, The Netherlands; and Laboratory of Microbiology, Wageningen University, Wageningen, The Netherlands.
Erwin G. Zoetendal, TI Food and Nutrition, Wageningen, The Netherlands; and Laboratory of Microbiology, Wageningen University, The Netherlands erwin.zoetendal@wur.nl
Koen Venema, TI Food and Nutrition, Wageningen, The Netherlands; and TNO Quality of Life, Zeist, The Netherlands.
Willem M. de Vos, TI Food and Nutrition, Wageningen, The Netherlands; Laboratory of Microbiology, Wageningen University, Wageningen, The Netherlands; and Department of Basic Veterinary Medicine, Helsinki University, Helsinki, Finland
Hauke Smidt, TI Food and Nutrition, Wageningen, The Netherlands; and Laboratory of Microbiology, Wageningen University, Wageningen, The Netherlands.
References
- Abell G.C., Cooke CM., Bennett C.N., Conlon M.A., Mcorist A.L. (2008) Phylotypes related to Ruminococcus bromii are abundant in the large bowel of humans and increase in response to a diet high in resistant starch. FEMS Microbiol Ecol 66:505–515 [DOI] [PubMed] [Google Scholar]
- Adamczyk J., Hesselsoe M., Iversen N., Horn M., Lehner A., Nielsen P.H.et al. (2003) The isotope array, a new tool that employs substrate-mediated labeling of rRNA for determination of microbial community structure and function. Appl Environ Microbiol 69:6875–6887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson A.F., Lindberg M., Jakobsson H., Bäckhed F., Nyrén P., Engstrand L. (2008) Comparative analysis of human gut microbiota by barcoded pyrosequencing. PLoS ONE 3(7):e2836, 1–8, doi: 10.1371/journal.pone.0002836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arebi N., Gurmany S., Bullas D., Hobson A., Stagg A., Kamm M. (2008) Review article: The psy-choneuroimmunology of irritable bowel syndrome - an exploration of interactions between psychological, neurological and immunological observations. Aliment Pharmacol Ther 28:830–840 [DOI] [PubMed] [Google Scholar]
- Bacher A., Rieder C, Eichinger D., Arigoni D., Fuchs G., Eisenreich W. (1998) Elucidation of novel biosynthetic pathways and metabolite flux patterns by retrobiosynthetic NMR analysis. FEMS Microbiol Rev 22:567–598 [Google Scholar]
- Backhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. (2005) Host-bacterial mutualism in the human intestine. Science 307:1915–1920 [DOI] [PubMed] [Google Scholar]
- Bartosch S., Woodmansey E.J., Paterson J.C., McMurdo M.E., Macfarlane G.T. (2005) Microbiological effects of consuming a synbiotic containing Bifidobacterium bifidum, Bifidobacterium lactis, and oligofructose in elderly persons, determined by real-time polymerase chain reaction and counting of viable bacteria. Clin Infect Dis 40:28–37 [DOI] [PubMed] [Google Scholar]
- Bauer-Marinovic M., Florian S., Muller-Schmehl K., Glatt H., Jacobasch G. (2006) Dietary resistant starch type 3 prevents tumor induction by 1,2-dimethylhydrazine and alters proliferation, apoptosis and dedifferentiation in rat colon. Carcinogenesis 27:1849–1859 [DOI] [PubMed] [Google Scholar]
- Belenguer A., Duncan S.H., Calder A.G., Holtrop G., Louis P., Lobley G.E.et al. (2006) Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72:3593–3599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg R.D. (1996) The indigenous gastrointestinal microflora. Trends Microbiol 4:430–435 [DOI] [PubMed] [Google Scholar]
- Bingham S.A., Pignatelli B., Pollock J.R., Ellul A., Malaveille C, Gross G.et al. (1996) Does increased endogenous formation of N-nitroso compounds in the human colon explain the association between red meat and colon cancer? Carcinogenesis 17:515–523 [DOI] [PubMed] [Google Scholar]
- Blaut M., Clavel T. (2007) Metabolic diversity of the intestinal microbiota: Implications for health and disease. J Nutr 137:751S–755S [DOI] [PubMed] [Google Scholar]
- Booijink C.C., Zoetendal E.G., Kleerebezem M., De Vos W.M. (2007) Microbial communities in the human small intestine: Coupling diversity to metage-nomics. Future Microbiol 2:285–295 [DOI] [PubMed] [Google Scholar]
- Boschker H. T. S., Nold S.C., Wellsbury P., Bos D., De Graaf W., Pel R.et al. (1998) Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801–805 [Google Scholar]
- Brück W.M., Graverholt G., Gibson G.R. (2003) A two-stage continuous culture system to study the effect of supplemental a-lactalbumin and glycomacro-peptide on mixed cultures of human gut bacteria challenged with enteropathogenic Escherichia coli and Salmonella serotype Typhimurium. J Appl Microbiol 95:44–53 [DOI] [PubMed] [Google Scholar]
- Bufill J.A. (1990) Colorectal cancer: Evidence for distinct genetic categories based on proximal or distal tumor location. Ann Intern Med 113:779–788 [DOI] [PubMed] [Google Scholar]
- Cassidy A., Bingham S.A., Cummings J.H. (1994) Starch intake and colorectal cancer risk: An international comparison. Br J Cancer 69:937–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christian M.T., Edwards C.A., Preston T., Johnston L., Varley R., Weaver L.T. (2003) Starch fermentation by faecal bacteria of infants, toddlers and adults: Importance for energy salvage. Eur J Clin Nutr 57:1486–1491 [DOI] [PubMed] [Google Scholar]
- Cinquin C, Le Blay G., Fliss I., Lacroix C. (2006) New three-stage in vitro model for infant colonic fermentation with immobilized fecal micro-biota. FEMS Microbiol Ecol 57:324–336 [DOI] [PubMed] [Google Scholar]
- Cummings J.H., Bingham S.A. (1987) Dietary fibre, fermentation and large bowel cancer. Cancer Surv 6:601–621 [PubMed] [Google Scholar]
- Cummings J.H., Englyst H.N. (1987) Fermentation in the human large intestine and the available substrates. Am J Clin Nutr 45:1243–1255 [DOI] [PubMed] [Google Scholar]
- Cummings J.H., Hill M.J., Jivraj T., Houston H., Branch W.J., Jenkins D.J. (1979) The effect of meat protein and dietary fiber on colonic function and metabolism. I. Changes in bowel habit, bile acid excretion, and calcium absorption. Am J Clin Nutr 32:2086–2093 [DOI] [PubMed] [Google Scholar]
- Cupples A.M., Shaffer E.A., Chee-Sanford J.C., Sims G.K. (2007) DNA buoyant density shifts during 15N-DNA stable isotope probing. Microbiol Res 162:328–334 [DOI] [PubMed] [Google Scholar]
- Dethlefsen L., Huse S., Sogin M.L., Relman D.A. (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6(11):2383–2400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dicksved J., Halfvarson J., Rosenquist M., Jarnerot G., Tysk C, Apajalahti J.et al. (2008) Molecular analysis of the gut microbiota of identical twins with Crohn's disease. ISME J 2:716–727 [DOI] [PubMed] [Google Scholar]
- Digiulio D.B., Romero R., Amogan H.P., Kusanovic J.P., Bik E.M., Gotsch F.et al. (2008) Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: A molecular and culture-based investigation. PLoS ONE 3(8):e3056, 1–10, doi: 10.1371/journal.pone.0003056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolnikowski G.G., Marsh J.B., Das S.K., Welty F.K. (2005) Stable isotopes in obesity research. Mass Spectrom Rev 24:311–327 [DOI] [PubMed] [Google Scholar]
- Drasar B.S. (1988) The bacterial flora of the intestine. In: Rowland I. The Role of the Gut Flora in Toxicity and Cancer, Academic Press: London, pp. 23–38 [Google Scholar]
- Dumont M.G., Murrell J.C. (2005) Stable isotope probing - linking microbial identity to function. Nat Rev Microbiol 3:499–504 [DOI] [PubMed] [Google Scholar]
- Dumont M.G., Neufeld J.D., Murrell J.C. (2006) Isotopes as tools for microbial ecologists. Curr Opin Biotech 17:57–58 [Google Scholar]
- Dumont M.G., Radajewski S.M., Miguez C.B., Mcdonald I.R., Murrell J.C. (2006) Identification of a complete methane monooxygenase operon from soil by combining stable isotope probing and meta-genomic analysis. Environ Microbiol 8:1240–1250 [DOI] [PubMed] [Google Scholar]
- Duncan S.H., Belenguer A., Holtrop G., Johnstone A.M., Flint H.J., Lobley G.E. (2007) Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl Environ Microbiol 73:1073–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan S.H., Louis P., Flint H.J. (2007) Cultivable bacterial diversity from the human colon. Lett Appl Microbiol 44:343–350 [DOI] [PubMed] [Google Scholar]
- Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M.et al. (2005) Diversity of the human intestinal microbial flora. Science 308:1635–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards C.A., Rowland I.R. (1992) Bacterial fermentation in the colon and its measurment. In: Schweizer T, Edwards C. Dietary Fibre - a Component of Food, Springer-Verlag: London, pp. 119–136 [Google Scholar]
- Egert M., De Graaf A.A., Maathuis A., De Waard P., Plugge C.M., Smidt H.et al. (2007) Identification of glucose-fermenting bacteria present in an in vitro model of the human intestine by RNA-stable isotope probing. FEMS Microbiol Ecol 60:126–135 [DOI] [PubMed] [Google Scholar]
- Egert M., De Graaf A.A., Smidt H., De Vos W.M., Venema K. (2006) Beyond diversity: functional microbiomics of the human colon. Trends Microbiol 14:86–91 [DOI] [PubMed] [Google Scholar]
- Englyst H.N., Kingman S.M., Cummings J.H. (1992) Classification and measurement of nutritionally important starch fractions. Eur J Clin Nutr 46(Suppl. 2):S33–50 [PubMed] [Google Scholar]
- Fanaro S., Chierici R., Guerrini P., Vigi V. (2003) Intestinal microflora in early infancy: composition and development. Acta Paediatr Suppl 91:48–55 [DOI] [PubMed] [Google Scholar]
- Fava F., Lovegrove J.A., Gitau R., Jackson K.G., Tuohy K.M. (2006) The gut microbiota and lipid metabolism: implications for human health and coronary heart disease. Curr Med Chem 13:3005–3021 [DOI] [PubMed] [Google Scholar]
- Favier CF., De Vos W.M., Akkermans A.D. (2003) Development of bacterial and bifidobacterial communities in feces of newborn babies. Anaerobe 9:219–229 [DOI] [PubMed] [Google Scholar]
- Finegold S.M., Sutter V.L., Mathiesen G.E. (1983) Microflora composition and development. In: Hentges D. J. Human Intestinal Microflora in Health and Disease, Academic Press: New York, pp. 3–119 [Google Scholar]
- Frank D.N., Amand A. L. S., Feldman R.A., Boedeker E.C., Harpaz N., Pace N.R. (2007) Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. PNAS USA 104:13780–13785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D.N., Pace N.R. (2008) Gastrointestinal microbiology enters the metagenomics era. Curr Opin Gastroen 24:4–10 [DOI] [PubMed] [Google Scholar]
- Franks A.H., Harmsen HJ., Raangs G.C., Jansen G.J., Schut F., Welling G.W. (1998) Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl Environ Microbiol 64:3336–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabor E., Liebeton K., Niehaus F., Eck J., Lorenz P. (2007) Updating the metagenomics toolbox. Biotechnol J 2:201–206 [DOI] [PubMed] [Google Scholar]
- Gill S.R., Pop M., Deboy R.T., Eckburg P.B., Turnbaugh P.J., Samuel B.S.et al. (2006) Metagenomic analysis of the human distal gut micro-biome. Science 312:1355–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarner F. (2005) The intestinal flora in inflammatory bowel disease: normal or abnormal? Curr Opin Gastroenterol 21:414–18 [PubMed] [Google Scholar]
- Guarner F., Malagelada J.R. (2003) Gut flora in health and disease. Lancet 361:512–519 [DOI] [PubMed] [Google Scholar]
- Hallstrom M., Eerola E., Vuento R., Janas M., Tammela O. (2004) Effects of mode of delivery and necrotising enterocolitis on the intestinal microflora in preterm infants. Eur J Clin Microbiol Infect Dis 23:463–470 [DOI] [PubMed] [Google Scholar]
- Hamer H.M., Jonkers D., Venema K., Vanhoutvin S., Troost F.J., Brummer R.J. (2008) Review article: The role of butyrate on colonic function. Aliment Pharmacol Ther 27:104–119 [DOI] [PubMed] [Google Scholar]
- Handelsman J. (2004) Metagenomics: Application of genomics to uncultured microorganisms. Microbiol Mol Biol Rev 68:669–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmsen H.J., Raangs G.C., He T, Degener J.E., Welling G.W. (2002) Extensive set of 16S rRNA-based probes for detection of bacteria in human feces. Appl Environ Microbiol 68:2982–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Sakamoto M., Benno Y. (2002) Fecal microbial diversity in a strict vegetarian as determined by molecular analysis and cultivation. Microbiol Immunol 46:819–831 [DOI] [PubMed] [Google Scholar]
- Huang W.E., Griffiths R.I., Thompson I.P., Bailey M.J., Whiteley A.S. (2004) Raman microscopic analysis of single microbial cells. Anal Chem 76:4452–4458 [DOI] [PubMed] [Google Scholar]
- Huang W.E., Stoecker K., Griffiths R., Newbold L., Daims H., Whiteley A.S.et al. (2007) Raman-fish: Combining stable-isotope Raman spectroscopy and fluorescence in situ hybridization for the single cell analysis of identity and function. Environ Microbiol 9:1878–1889 [DOI] [PubMed] [Google Scholar]
- Ingham C.J., Sprenkels A., Bomer J., Molenaar D., Van Den Berg A., Van Hylckama Vlieg J.E.et al. (2007) The micro-petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. PNAS USA 104:18217–18222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobasch G., Schmiedl D., Kruschewski M., Schmehl K. (1999) Dietary resistant starch and chronic inflammatory bowel diseases. Int J Colorectal Dis 14:201–211 [DOI] [PubMed] [Google Scholar]
- Jehmlich N., Schmidt F., Von Bergen M., Richnow H.H., Vogt C. (2008) Protein-based stable isotope probing (Protein-SIP) reveals active species within anoxic mixed cultures. ISME J 2:1122–1133 [DOI] [PubMed] [Google Scholar]
- Jensen S., Neufeld J.D., Birkeland N.-K., Hovland M., Murrell J.C. (2008) Methane assimilation and trophic interactions with marine Methylomicrobium in deep-water coral reef sediment off the coast of Norway. FEMS Microbiol Ecol 66:320–330 [DOI] [PubMed] [Google Scholar]
- Jiménez-Vera R., Monroy O., Corona-Cruz A., Garćia-Garibay M. (2008) Construction of a model of the human proximal colon. World J Microb Biot 24:2767–2774 [Google Scholar]
- Kelleher J.K. (2004) Probing metabolic pathways with isotopic tracers: Insights from mammalian metabolic physiology. Metab Eng 6:1–5 [DOI] [PubMed] [Google Scholar]
- Kolida S., Tuohy K., Gibson G.R. (2000) The human gut flora in nutrition and approaches for its dietary modulation. Nutr Bull 25:223–231 [Google Scholar]
- Kovatcheva-Datchary P., Egert M., Maathuis A., Rajilic-Stojanovic M., De Graaf A.A., Smidt H.et al. (2008) Linking phylogenetic identities of bacteria to starch fermentation in an in vitro model of the large intestine by RNA-based stable isotope probing. Environ Microbiol 11:914–926 [DOI] [PubMed] [Google Scholar]
- Kreuzer-Martin H.W. (2007) Stable isotope probing: Linking functional activity to specific members of microbial communities. Soil Sci Soc Am J 71:611–619 [Google Scholar]
- Kurokawa K., Itoh T., Kuwahara T., Oshima K., Toh H., Toyoda A.et al. (2007) Comparative meta-genomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res 14:169–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuypers M. M. M., Jørgensen B.B. (2007) The future of single-cell environmental microbiology. Environ Microbiol 9:6–7 [DOI] [PubMed] [Google Scholar]
- Le Leu R.K., Brown I.L., Hu Y., Bird A.R., Jackson M., Esterman A.et al. (2005) A synbiotic combination of resistant starch and Bifidobacterium lactis facilitates apoptotic deletion of carcinogen-damaged cells in rat colon. J Nutr 135:996–1001 [DOI] [PubMed] [Google Scholar]
- Le Leu R.K., Brown I.L., Hu Y., Esterman A., Young G.P. (2007a) Suppression of azoxymethane-induced colon cancer development in rats by dietary resistant starch. Cancer Biol Ther 6:1621–1626 [DOI] [PubMed] [Google Scholar]
- Le Leu R.K., Brown I.L., Hu Y, Morita T., Esterman A., Young G.P. (2007b) Effect of dietary resistant starch and protein on colonic fermentation and intestinal tumourigenesis in rats. Carcinogenesis 28:240–245 [DOI] [PubMed] [Google Scholar]
- Lee N., Nielsen P.H., Andreasen K.H., Juretschko S., Nielsen J.L., Schleifer K.H.et al. (1999) Combination of fluorescent in situ hybridization and microautoradiography - a new tool for structure-function analyses in microbial ecology. Appl Environ Microbiol 65:1289–1297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley R.E., Turnbaugh P.J., Klein S., Gordon J.I. (2006) Microbial ecology: Human gut microbes associated with obesity. Nature 444:1022–1023 [DOI] [PubMed] [Google Scholar]
- Lionetti P., Callegari M.L., Ferrari S., Cavicchi M.C., Pozzi E., De Martino M.et al. (2005) Enteral nutrition and microflora in pediatric Crohn's disease. JPEN J Parenter Enteral Nutr 29:S173–175, discussion S175-178, S184-178 [DOI] [PubMed] [Google Scholar]
- Lueders T., Manefield M., Friedrich M.W. (2004) Enhanced sensitivity of DNA- and rRNA-based stable isotope probing by fractionation and quantitative analysis of isopycnic centrifugation gradients. Environ Microbiol 6:73–78 [DOI] [PubMed] [Google Scholar]
- Lueders T., Wagner B., Claus P., Friedrich M.W. (2004) Stable isotope probing of rRNA and DNA reveals a dynamic methylotroph community and trophic interactions with fungi and protozoa in oxic rice field soil. Environ Microbiol 6:60–72 [DOI] [PubMed] [Google Scholar]
- Macfarlane G.T., Macfarlane S. (1997) Human colonic microbiota: Ecology, physiology and metabolic potential of intestinal bacteria. Scand J Gastroenterol Suppl 222:3–9 [DOI] [PubMed] [Google Scholar]
- Macfarlane G.T., Macfarlane S. (2007) Models for intestinal fermentation: Association between food components, delivery systems, bioavailability and functional interactions in the gut. Curr Opin Biotechnol 18:156–162 [DOI] [PubMed] [Google Scholar]
- Macfarlane G.T., Macfarlane S., Gibson G.R. (1998) Validation of a three-stage compound continuous culture system for investigating the effect of retention time on the ecology and metabolism of bacteria in the human colon. Microb Ecol 35:180–187 [DOI] [PubMed] [Google Scholar]
- Macfarlane S., Woodmansey E.J., Macfarlane G.T. (2005) Colonization of mucin by human intestinal bacteria and establishment of biofilm communities in a two-stage continuous culture system. Appl Environ Microbiol 71:7483–7492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manefield M., Griffiths R., Bailey M.J., Whiteley A.S. (2006) Stable isotope probing: A critique of its role in linking phylogeny and function. In: Nannipieri P., Smalla K. Nucleic Acids and Proteins in Soil, Vol. 8, Springer Berlin: Heidelberg, pp. 205–216 [Google Scholar]
- Manefield M., Whiteley A.S., Griffiths R.I., Bailey M.J. (2002) RNA stable isotope probing, a novel means of linking microbial community function to phylogeny. Appl Environ Microbiol 68:5367–5373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy Y, Ishoey T, Lasken R.S., Stockwell TB., Walenz B.P., Halpern A.L.et al. (2007) Nanoliter reactors improve multiple displacement amplification of genomes from single cells. PLoS Genet 3:1702–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minekus M., Marteau P., Havenaar R., Huis In't Veld J. H. J. (1995) A multicompartmental dynamic computer-controlled model simulating the stomach and small intestine. ATLA - Alternatives to Laboratory Animals 23:197–209 [Google Scholar]
- Minekus M., Smeets-Peeters M., Bernalier A., Marol-Bonnin S., Havenaar R., Marteau P.et al. (1999) A computer-controlled system to simulate conditions of the large intestine with peristaltic mixing, water absorption and absorption of fermentation products. Appl Microbiol Biotechnol 53:108–114 [DOI] [PubMed] [Google Scholar]
- Molly K., Vande Woestyne M., Verstraete W. (1993) Development of a 5-step multi-chamber reactor as a simulation of the human intestinal microbial ecosystem. Appl Microbiol Biotechnol 39:254–258 [DOI] [PubMed] [Google Scholar]
- Morris S.A., Radajewski S., Willison T.W, Murrell J.C. (2002) Identification of the functionally active methanotroph population in a peat soil microcosm by stable-isotope probing. Appl Environ Microbiol 68:1446–1453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muir J.G., Yeow E.G., Keogh J., Pizzey C, Bird A.R., Sharpe K.et al. (2004) Combining wheat bran with resistant starch has more beneficial effects on fecal indexes than does wheat bran alone. Am J Clin Nutr 79:1020–1028 [DOI] [PubMed] [Google Scholar]
- Neish A.S. (2009) Microbes in gastrointestinal health and disease. Gastroenterology 136:65–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeld J.D., Schafer H., Cox M.J., Boden R., McDonald I.R., Murrell J.C. (2007) Stable-isotope probing implicates Methylophaga spp and novel gammaproteobacteria in marine methanol and methylamine metabolism. ISME J 1:480–491 [DOI] [PubMed] [Google Scholar]
- Noverr M.C., Huffnagle G.B. (2004) Does the microbiota regulate immune responses outside the gut? Trends Microbiol 12:562–568 [DOI] [PubMed] [Google Scholar]
- Ouwehand A.C., Derrien M., De Vos W., Tiihonen K., Rautonen N. (2005) Prebiotics and other microbial substrates for gut functionality. Curr Opin Biotechnol 16:212–217 [DOI] [PubMed] [Google Scholar]
- Palmer C., Bik E.M., Digiulio D.B., Relman D.A., Brown P.O. (2007) Development of the human infant intestinal microbiota. PLoS Biol 5:e177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool-Zobel B.L. (2005) Inulin-type fructans and reduction in colon cancer risk: Review of experimental and human data. Br J Nutr 93(Suppl. 1):S73–90 [DOI] [PubMed] [Google Scholar]
- Pouteau E., Nguyen P., Ballevre O., Krempf M. (2003) Production rates and metabolism of short-chain fatty acids in the colon and whole body using stable isotopes. Proc Nutr Soc 62:87–93 [DOI] [PubMed] [Google Scholar]
- Probert H.M., Apajalahti J.H., Rautonen N., Stowell J., Gibson G.R. (2004) Polydextrose, lactitol, and fructo-oligosaccharide fermentation by colonic bacteria in a three-stage continuous culture system. Appl Environ Microbiol 70:4505–4511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radajewski S., Ineson P., Parekh N.R., Murrell J.C. (2000) Stable-isotope probing as a tool in microbial ecology. Nature 403:646–649 [DOI] [PubMed] [Google Scholar]
- Radajewski S., Webster G., Reay D.S., Morris S.A., Ineson P., Nedwell D.B.et al. (2002) Identification of active methylotroph populations in an acidic forest soil by stable-isotope probing. Microbiology 148:2331–2342 [DOI] [PubMed] [Google Scholar]
- Rajilić-Stojanović M., Heilig H. G. H. J., Molenaar D., Kajander K., Surakka A., Smidt H.et al. (2009) Development and application of the human intestinal tract chip (HITChip), a phylogenetic microarray: Analysis of universally conserved phylotypes in the abundant microbiota of young and elderly adults. Environ Microbiol, doi: 10.1111/j.1462-2920.2009.01900.x [DOI] [PMC free article] [PubMed]
- Rajilic-Stojanovic M., Smidt H., De Vos W.M. (2007) Diversity of the human gastrointestinal tract microbiota revisited. Environ Microbiol 9:2125–2136 [DOI] [PubMed] [Google Scholar]
- Roediger W. E. W. (1988) Bacterial short-chain fatty acids and mucosal diseases of the colon. Brit J Surg 75:346–348 [DOI] [PubMed] [Google Scholar]
- Savage D.C. (1977) Microbial ecology of the gastrointestinal tract. Ann Rev Microbiol 31:107–133 [DOI] [PubMed] [Google Scholar]
- Savage D.C. (1986) Gastrointestinal microflora in mammalian nutrition. Ann Rev Nutr 6:155–178 [DOI] [PubMed] [Google Scholar]
- Sengupta S., Muir J.G., Gibson P.R. (2006) Does butyrate protect from colorectal cancer? J Gastroenterol Hepatol 21:209–218 [DOI] [PubMed] [Google Scholar]
- Srikanth C.V., McCormick B.A. (2008) Interactions of the intestinal epithelium with the pathogen and the indigenous microbiota: A three-way crosstalk. Interdisciplin Perspect Infect Dis 14 pages. doi: 10.1155/2008/626827 [DOI] [PMC free article] [PubMed]
- Suau A., Bonnet R., Sutren M., Godon J.J., Gibson G.R., Collins M.D.et al. (1999) Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol 65:4799–4807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannock G.W. (2006) Hygeia versus asclepius. Future Microbiol 1:345–349 [DOI] [PubMed] [Google Scholar]
- Topping D.L., Clifton P.M. (2001) Short-chain fatty acids and human colonic function: Roles of resistant starch and nonstarch polysaccharides. Physiol Rev 81:1031–1064 [DOI] [PubMed] [Google Scholar]
- Turnbaugh P.J., Hamady M., Yatsunenko T., Cantarel B.L., Duncan A., Ley R.E.et al. (2009) A core gut microbiome in obese and lean twins. Nature 457:480–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh P.J., Ley R.E., Hamady M., Fraser-Liggett CM., Knight R., Gordon J.I. (2007) The human microbiome project. Nature 449:804–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turroni F., Ribbera A., Foroni E., Van Sinderen D., Ventura M. (2008) Human gut microbiota and bifidobacteria: From composition to functionality. Antonie van Leeuw 94:35–50 [DOI] [PubMed] [Google Scholar]
- Van De Wiele T, Boon N., Possemiers S., Jacobs H., Verstraete W. (2007) Inulin-type fructans of longer degree of polymerization exert more pronounced in vitro prebiotic effects. J Appl Microbiol 102:452–460 [DOI] [PubMed] [Google Scholar]
- Van Der Waaij L.A., Harmsen H.J., Madjipour M., Kroese F.G., Zwiers M., Van Dullemen H.M.et al. (2005) Bacterial population analysis of human colon and terminal ileum biopsies with 16S rRNA-based fluorescent probes: Commensal bacteria live in suspension and have no direct contact with epithelial cells. Inflamm Bowel Dis 11:865–871 [DOI] [PubMed] [Google Scholar]
- Van Nuenen M. H. M. C., Meyer P.D., Venema K. (2003) The effect of various inulins and Clostridium difficile on the metabolic activity of the human colonic microbiota in vitro. Microb Ecol Health Dis 15:137–144 [Google Scholar]
- Vanhoutvin S., Troost F.J., Lindsey P.J., Kilkens T. O. C., Hamer H.M., Jonkers D.et al. (2009) The effects of butyrate enemas on visceral perception in healthy volunteers. Neurogastroent Motil (in press) [DOI] [PubMed]
- Verberkmoes N.C., Russell A.L., Shah M., Godzik A., Rosenquist M., Halfvarson J.et al. (2009) Shotgun metaproteomics of the human distal gut microbiota. ISME J 3:179–189 [DOI] [PubMed] [Google Scholar]
- Wall R., Ross R.P., Ryan C.A., Hussey S., Murphy B., Fitzgerald G.F.et al. (2009) Role of gut microbiota in early infant development. Clin Med: Pediatrics 3:45–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteley A.S., Manefield M., Lueders T. (2006) Unlocking the ‘microbial black box' using RNA-based stable isotope probing technologies. Curr Opin Biotechnol 17:67–71 [DOI] [PubMed] [Google Scholar]
- Wong C.S., Sengupta S., Tjandra J.J., Gibson P.R. (2005) The influence of specific luminal factors on the colonic epithelium: High-dose butyrate and physical changes suppress early carcinogenic events in rats. Dis Colon Rectum 48:549–559 [DOI] [PubMed] [Google Scholar]
- Xu J., Gordon J.I. (2003) Honor thy symbionts. PNAS USA 100:10452–10459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young G.P., Hu Y., Le Leu R.K., Nyskohus L. (2005) Dietary fibre and colorectal cancer: A model for environment-gene interactions. Mol Nutr Food Res 49:571–584 [DOI] [PubMed] [Google Scholar]
- Zhang H., Dibaise J.K., Zuccolo A., Kudrna D., Braidotti M., Yu Y.et al. (2009) Human gut micro-biota in obesity and after gastric bypass. PNAS USA 106:2365–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal E.G., Akkermans A.D., De Vos WM. (1998) Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl Environ Microbiol 64:3854–3859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoetendal E.G., Rajilic-Stojanovic M., De Vos WM. (2008) High-throughput diversity and functionality analysis of the gastrointestinal tract micro-biota. Gut 57:1605–1615 [DOI] [PubMed] [Google Scholar]
- Zoetendal E.G., Vaughan E.E., De Vos WM. (2006) A microbial world within us. Mol Microbiol 59:1639–1650 [DOI] [PubMed] [Google Scholar]