Abstract
The accelerating pace of technological and analytical development in the fields of genetic and phenotypic profiling has ushered in an era of great promise for multiple sclerosis (MS) research. As we continue to identify modest but meaningful associations to MS susceptibility, disease course, treatment response, and other clinical or paraclinical phenotypes, we must begin to (1) embark on the challenging set of studies that will integrate disparate observations into clinical algorithms, and (2) validate their clinical utility. Genetic data are receiving muchofthe attention today, but they are unlikelytobesufficienttooffer a personalized approach to disease management in MS. Rather, the genetic architecture of the disease, once uncovered, will offer a fixed platform upon which more dynamic molecular profiles can be assembled to deconstruct the structure of the patient population that we label with a diagnosis of MS. The tools and methods to gain insight into the heterogeneity of MS patients are available today; we must now realize their potential in enhancing the care of MS patients.
Keywords: multiple sclerosis, immunotherapy, genetics
Introduction
Multiple sclerosis (MS) research is entering an exciting phase as new technologies enable increasingly more powerful investigations of human samples and reveal modest but meaningful differences between subsets of patients with a diagnosis of MS. Thanks to parallel advances in analytic methodology, these technological breakthroughs are beginning to uncover the structure of the population of MS patients.
The architecture of this chronic neurologic disease is quite complex, as studies suggest the existence of pathophysiologically distinct subsets of patients not only at the onset of MS but also over the course of the disease [Compston and Coles, 2008]. In other words, patient heterogeneity is likely to be dynamic and will require both fixed biomarkers (e.g. DNA polymorphisms) and variable biomarkers (such as RNA, protein and other molecules) to be fully characterized. These biomarkers are also likely to vary depending on the question being pursued. Most efforts to date have investigated the immunologic component of this disease and will therefore form the focus of this review; however, the methodology is the same to explore the neurodegenerative and neurorepair component of MS. In fact, current studies that have unbiased approaches may well reveal useful hints relating to noninflammatory mechanisms in MS.
To date, there are no useful biomarkers for treatment selection in MS, and only MRI offers coarse estimates of recurrence rate early in the disease at the clinically isolated demyelinating syndrome (CIS) stage [Compston and Coles, 2008]. However, methods and platforms are now available to characterize MS with a broad perspective at a new level of detail, and some interesting observations have emerged. It is time to begin to think about the study designs that will validate these observations and transform a correlated biomarker into a useful clinical algorithm. These studies will rigorously test the concept of ‘personalized medicine’, the tailoring of immunotherapy in MS to an individual patient by using information gleaned from their genetic and other tissue-derived material.
The personalized medicine concept
Personalized medicine is not a new concept in the medical arena: over the past century, as the pace of discovery in the biomedical sciences has accelerated, we have been able to develop new treatments and to enhance the targeting of existing treatments to subsets of patients who were most likely to benefit from them based on clinical or paraclinical data. The emphasis on performing ‘evidence-based medicine’ enhanced patient outcomes by recommending diagnostic and treatment algorithms that were most likely to be effective. However, it is true that such treatments were tested in and targeted to patient populations rather than individual patients because our ability to differentiate patient subsets was limited. Enhanced targeting will come from the use of different types of information: (1) markers that are highly correlated to clinical outcome and are therefore informative by themselves, and (2) groups of markers with weak individual correlations to clinical traits that are predictive when considered together. Epidermal growth factor receptor (EGFR) mutations are examples of the first type of marker. The presence of one of a small number of mutations in nonsmall cell lung cancers correlates with better response to treatment with EGFR inhibitors such as gefitinib. Recently, a small phase II trial tested this observation and found longer time to treatment failure to be associated with the presence of EGFR mutations [Yang et al. 2008]. This and many similar efforts illustrate (1) the utility of leveraging genetic and other information to better target treatment and (2) the need for prospective clinical trials to validate any prognostic tool. In this case, analysis of single mutations is informative as their effect on treatment outcome is profound.
The second type of information that is beginning to be used in a clinical setting is illustrated by studies seeking to predict warfarin dosing. In this case, genetic variation in human populations have modest but important effects on warfarin metabolism, and pooling of these different pieces of genetic information, along with important clinical parameters such as age and smoking status, lead to clinical tools that may enhance patient management. One such algorithm is provided at www.warfarindosing.org and is based on a trial that revealed that clinical information could explain 17–22% of the warfarin dose variability and that adding information from three genetic markers enhances the algorithm: with genetic data, it explains 53–54% of the variability in drug response in the screening and validation cohorts [Gage et al. 2008]. The effect of meaningfully combining genetic and clinical data that convey different pieces of information therefore provides a more accurate ability to predict the dose of warfarin that a patient must take, reducing the risk of adverse events that happen commonly as a result of underdosing or overdosing patients.
A priori, either type of marker may have a role in the field of MS, but none of the candidate markers explored to date has been validated [Miller et al. 2008]. How easy are they to find? Nelson et al. [2009] provide an interesting outline of several reported associations between genetic markers and adverse events following treatment with one of eight different drugs. All of these examples illustrate the fact that a number of different genetic variants (with frequencies ranging from 0.04 to 0.30) have strong effects (genotype risk ratios of 4 to >50) on adverse responses to very different classes of drugs. Further, they present interesting simulations suggesting that genome-wide screening methods have reasonable statistical power to detect such strong associations in as few as 15 cases and 200 control subjects. Thus, analytic methods are at hand to discover genetic associations relating to drug treatment in MS, and sample sizes are not prohibitive if strong effects are sought. Even more modest effects are approachable, since the critical issue in discovering more modest effects is sample size and large numbers of MS patients are treated with several of the currently approved treatments such as glatiramer acetate (GA), interferon beta (IFNß) mitoxanthrone or natalizumab.
Whether strong or more modest predictors of MS treatment response are ultimately discovered, this information on the role of human genetic variation in treatment efficacy will provide an immutable framework upon which measurements of dynamic biomarkers can be overlaid. This approach will lead to a more accurate understanding of the patient subset and disease stage to which an individual belongs. In most MS cases, DNA analysis is unlikely to be sufficient to answer a clinical question, but it will reduce the complexity of challenges such as treatment selection in MS.
Advances in MS genetics
Based on what we know of MS today, it is unlikely that assessing single novel or familial mutations will have a direct impact on patient diagnosis or management. They may have a role in small subsets of patients, but we have yet to find evidence of such patients. For most MS patients, therefore, the warfarin example illustrates the way forward: it is by combining different pieces of information together and collapsing them into a single estimate of risk or outcome that we can have an impact on diagnosis and management in the near term. We are most advanced in our understanding of MS susceptibility for which genetic markers are now being discovered at a rapid pace [De Jager, 2009; Aulchenko et al. 2008; Comabella et al. 2008; Hafler et al. 2008, 2007]. Today, up to 16 risk alleles have been discovered, and, outside of the major histocompatability complex, each has only a modest effect on disease risk, with odds ratios estimated to be between 1.15 and 1.25 [De Jager, 2009; De Jager et al. 2008] We further estimate that 50–100 such risk alleles may exist and that an ongoing, large collaborative experiment involving the International MS Genetics Consortium and the Wellcome Trust will identify most of these risk alleles as it explores the genetic architecture of over 11 000 subjects with MS in 2009. Thus, we will soon have a nearly comprehensive map of genetic susceptibility factors for MS, and it is time to develop clinical tools with which to test the utility of this information in the clinical setting. Already, an early version of such a tool, based on 16 risk alleles known today, can divide subjects into seven categories of genetic risk, with the highest risk category having five times greater risk of MS than the lowest risk category [De Jager et al. unpublished observations]. Thus, these modest pieces of information, when collapsed together can provide information that is potentially meaningful in a clinical setting, and these early results suggest that the impact on diagnosis will only improve as we consider the full spectrum of 50 or more MS risk variants that will be identified in the coming year. Other pieces of clinical information such as age, presence of oligoclonal bands, proportion of CD8lowCD56+CD3- cells in peripheral blood, and results of electrophysiological tests that each are modestly informative will further enhance our initial assessment of a patient as we consider a diagnosis of MS [De Jager et al. 2008; McDonald et al. 2001]. As we gather a comprehensive map of genetic variants involved in triggering an inflammatory demyelinating process, we may identify different pathways that are especially prone to the effects of genetic variation, and different subsets of patients with different combinations of genetic vulnerabilities to MS may emerge.
Interestingly, early assessments of the role of known susceptibility alleles such as those at CD58, IL2RA and IL7R in disease progression suggest that these alleles do not have a significant role in the course of MS once it has started [Baranzini et al. 2009]. Even the HLA DRB1*1501 risk allele, which has an odds ratio of 2.7 for MS susceptibility [Barcellos et al. 2006], has at best only modest evidence of an effect on disease course as assessed in both cross-sectional MRI and clinical data [Okuda et al. 2009; Zivadinov et al. 2007]. This intriguing observation needs validation, but it does support the existence of genetic loci influencing disease severity, as do other studies implicating the DRB1*01 allele and the DRB5 gene [Caillier et al. 2008; Deluca et al. 2007]. Several genome scans for outcomes related to disease course, such as the MS severity score (MSSS) [Roxburgh et al. 2005] or brain volume are ongoing, and their preliminary results are concordant with the one published report in that the associations that are being found appear to be distinct from those found in susceptibility scans [Baranzini et al. 2009]. These observations suggest that the genetic architecture of disease onset and disease course may be quite different: a different set of genes may take over once an inflammatory process directed against the CNS has been initiated. However, we are still in early stages of such evaluations, and definitive assessments await the establishment of large subject cohorts with DNA samples and robust estimates of disease course.
Understanding the role of genetic and other markers in prognosticating disease course is critical as such information may be some of the earliest to find its way in the clinic. Knowing where a patient lies on a gradient of disease severity may not help a physician to select a specific drug, but it could provide a valuable insight in selecting a category of drug from the rapidly expanding armamentarium of MS treatments. Specifically, after consulting with her neurologist, a patient at high risk of malignant disease may choose a more effective drug with potentially more serious adverse events as her first treatment option. Thus, we can potentially use nondrugspecific information on likelihood of disease severity to stratify patients and inform a treatment selection decision between treatment strata ranging from lower efficacy/low adverse event choices to higher efficacy/greater adverse event risk options. The approach to the use of such data is therefore clear and awaits the conclusion of pertinent gene discovery and validation studies in coming years. Such studies will provide us with a foundation with which to develop an effective trial to test such a disease severity-based algorithm. Currently, we do not have validated genetic factors that are associated with disease course in MS, but, given the architecture of disease susceptibility, we suspect that there are likely to be dozens of genes of modest effect involved in this process.
Pharmacogenetics in MS
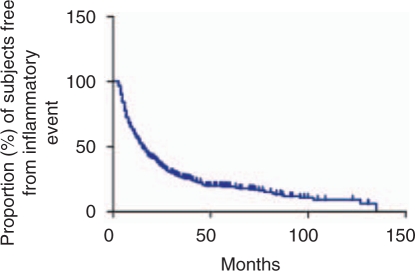
While we have made tremendous progress in understanding the genetic architecture of MS susceptibility, we have yet to apply the same analytic methods to treatment response in a powerful manner. Clinical experience suggests that there is a substantial subset of patients (∼20%), whether treated with GA or IFNß, that have rare or no relapses (Figure 1) and that this subset is larger than the one of patients with benign disease. The main barrier to exploring this observation with genetic methods has been the lack of large independent cohorts with DNA samples that have consistent, detailed phenotypes related to treatment response. Until these are assembled, we will be plagued with many intriguing genetic and other associations with treatment response that are difficult to substantiate, such as the association of HLA DRB1*1501 with response to glatiramer acetate [Fusco et al. 2001]. Nonetheless, we are beginning to see studies that illustrate the direction that we must take. Byun et al. [2008] interrogated 206 subjects with MS at a genome-wide level for clinical response to interferon beta therapy over a 2-year period. The main limitation of this interesting study was the lack of a powerful replication sample, largely because equivalent data matched to DNA samples is not readily available. While we are left without definitive results, the study is nonetheless important because it appears that none of the interrogated sites has overwhelming evidence of association to IFNß response. One caveat is that the Affymetrix 100K SNP array used in this experiment does not provide dense coverage genome-wide, and a common variant with a strong effect on IFNß may therefore exist. Nonetheless, this study suggests that common variants (those assessed by current genotyping arrays) do not have a strong effect on responses to IFNß. Thus, as with susceptibility to MS, effect sizes of common genetic variants (frequency >0.05) are likely to be modest in the context of IFNß treatment. We must therefore design future studies accordingly. We must think about sample sizes in the thousands of subjects to effectively uncover the genetic architecture of treatment response to IFNß. This is true both to identify the common variants using existing genotyping technologies or rare variants with more profound effects that will require one of the emerging high-throughput sequencing platforms to be discovered. Time will tell us whether MS treatments have a similar or different profile from that seen with the susceptibility phenotype; in some cases, genetic variation may directly affect the drug target and could have very strong effects.
Figure 1.
Time to a first clinically or radiologically evident episode of CNS inflammation in treated subjects with MS. The data presented are collected prospectively in a population of patients seen at the Partners MS Center in Boston. Data are included from both 330 subjects starting treatment with interferon beta and 244 subjects starting treatment with glatiramer acetate. There is no significant difference between the treatment-specific curves. This graph illustrates heterogeneity in MS treatment response, with over one-half of patients having a new inflammatory event within 2 years of treatment onset (events are recorded starting week 13 after treatment initiation) and a subset of patients having no new radiographic or clinical events over long periods of treatment.
Given that the genetic architecture of IFNß treatment response is likely to be similar to that of MS susceptibility and to require study sizes of thousands of patients to produce validated results, it appears that the formation of a consortium of interested parties would provide the platform with the greatest probability of success for this endeavor. Given the existence of three separate versions of IFNß on the market, and the existence of vast amounts of clinical response data in three separate companies, the ideal situation would be a consortium grouping all three companies as well as academic investigators to explore this question in the most powerful manner possible. While many practical challenges to the formation of such a consortium exist, the emergence of multiple new drugs with enhanced efficacy for MS in the near future may provide the impetus to such a shared approach that could provide tools to direct IFNß to the subset of MS patients that clearly benefit from it in the long term.
Response to IFNß therapy has been the focus of most MS pharmacogenetic studies to date, and we are beginning to develop an idea of the study designs that will be necessary to implement such studies effectively. However, one should not overgeneralize the genetic architecture of a single drug response in MS: the mechanism of action of a given drug will dictate the genetic architecture to its response. It is therefore possible that common variants with large effects exist for other drugs, and each novel drug will have to be investigated appropriately, preferably during its development as the existence of a common variant that has a strong effect on drug response would significantly impact the statistical power of a clinical trial. Finally, we should not focus solely on drug response, as genetic variation may be informative for predicting adverse events related to treatment: Nelson et al. [2009] present several such examples as well as simulation studies suggesting that genome-wide analyses can be used effectively to explore the etiology of severe events even in small numbers of subjects.
RNA and other dynamic markers
Unlike DNA sequence, the level and isoform ratios of expressed RNA are highly dynamic and are therefore likely to contain clues that could help us characterize where in the disease cycle a particular patient is located. Similarly, the study of proteins, lipids and other metabolites will provide different types of information that rapidly reflect ongoing physiological processes. However, robust platforms to effectively pursue the latter classes of biomarker discovery projects are only now beginning to emerge [Ekegren et al. 2008; Steinman, 2008], and we will therefore focus our discussion on RNA for which quantitative methods are relatively robust, are widespread and have well-known limitations. Published efforts in this field have recently been well summarized elsewhere [Miller et al. 2008], and while much has been done in this arena, particularly with IFNß response, we have yet to deploy or even assess a clinical test. The challenge here is to sample the appropriate tissue, cell mixture or cell type. Clearly, the most informative tissue, CNS lesion tissue, is only rarely accessible and is not a practical target for the development of clinical tools in MS. Cerebrospinal fluid would arguably best reflect processes ongoing in the CNS and is an excellent substrate for biomarker discovery. However, blood remains the target of choice, given its accessibility, particularly if repeated or frequent sampling is desired.
The most promising of current observations probably lies in those that relate to the detection of a class I interferon response signature in the peripheral blood of a subset of MS patients [Van Baarsen et al. 2006]. These patients may respond less effectively to exogenous IFNß, either because they have maximized the effects of IFNß following endogenous self-treatment or because the interferon signature is an epiphenomenon in a subset of patients which does not respond to IFNß treatment. What this and other observations now require is a rigorous clinical trial to assess whether capturing and using information on RNA expression in the peripheral blood of untreated patients can guide treatment selection and enhance the efficacy of first line MS treatment. For the foreseeable future, GA and IFNß will remain the mainstay of first-line treatment in most patients, and therefore a pharmacogenomic trial will ideally have two treatment arms and will test whether random or directed assignment to a treatment arm (GA versus IFNß) is more effective.
The principal challenge to the use of RNA and other dynamic biomarkers is their variability, particularly when we consider mixed cell populations such as those sampled in peripheral blood. It is likely that incorporating pertinent genetic markers that have strong effects on RNA expression will reduce the complexity of this question and result in more effective biomarkers. When considered together, these different forms of information therefore show great promise, not only to answer a particular question such as treatment response to a particular drug, but more generally in deconstructing the heterogeneity of MS patients which is typically not apparent at the time of initial clinical assessment.
MS population structure
The class I interferon signature that is seen in a large subset of MS patients is also seen in approximately one-half of subjects with dermatomyositis, systemic lupus erythematosus and rheumatoid arthritis [Van Der Pouw Kraan et al. 2007; Greenberg et al. 2005; Baechler et al. 2003]. This suggests that a subset of subjects within different inflammatory diseases may share pathophysiologic features, regardless of the target organ(s). Thus, the class I interferon signature may distinguish a functionally distinct subset of patients that may not only have a different response to IFNß but also have differences in disease course. An RNA interferon response pattern may therefore discriminate one subset of MS patients, but how many such subsets exist?
The existence of patient subsets has long been suspected on clinical grounds, but it is not yet clear how best to define these subsets, particularly when a patient presents with their first clinical event. Neuropathologic studies have defined different types of lesion patterns in the brains of MS patients [Lucchinetti et al. 2000], and each of these lesion types were initially proposed to be largely found in a different subset of patients. While this is an area of continued investigation with more recent evidence suggesting that one of these plaque patterns may be more generally shared by MS patients [Breij et al. 2008], the original manuscript nonetheless highlighted the issue of pathophysiologic heterogeneity in this disease.
Currently, one clearly distinct subset of patients with a demyelinating disease consists of patients with a diagnosis of neuromyelitis optica (NMO). Integrating data from the anti-aquaporin 4 antibody biomarker with clinical and MRI data looks promising as a diagnostic tool for NMO [Wingerchuk et al. 2006], and this serum biomarker may be informative both in terms of disease course for optic neuritis cases and perhaps in selecting treatments such as rituximab [Jacob et al. 2008; Matiello et al. 2008]. Similarly, a fraction of acute demyelinating encephalomyelitis cases may be associated with anti-MOG antibodies, although these results require further validation [O'Connor et al. 2007]. More broadly, a tremendous amount of work by many different investigators has gone into identifying antibodies against myelin in MS patients, with inconsistent results. This area of active investigation and the role of B cells in MS have recently been reviewed elsewhere [McLaughlin et al. 2008; Racke, 2008], but high-affinity antibodies against myelin antigens have yet to yield a robust biomarker in MS. More recently, low-affinity autoantibody profiles from serum have been investigated, and these data suggest that such antibodies may hold information that can be used to partition subjects with different neuropathologic lesion patterns in biopsy specimens [Quintana et al. 2008]. Overall, these efforts suggest that biomarkers differentiating subtypes of MS patients may exist.
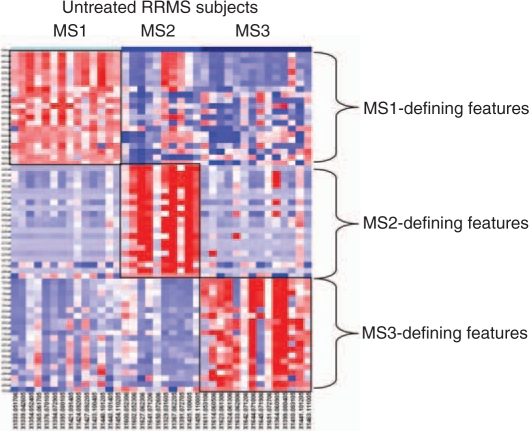
Several different investigations are beginning to explicitly explore the population structure of MS and CIS patients in an unbiased manner to better understand how many subsets of patients may exist. Whether flow cytometric data or RNA expression is used, patient subsets that are defined by the pattern of gene expression in peripheral blood are emerging from these efforts [Corvol et al. 2008; De Jager et al. 2008; Rinaldi et al. 2006]. In these cases, three or four major subsets of subjects are described (Figure 2). These analyses are still in their earliest incarnations, and they clearly are limited by small sample size. Nonetheless, they are conceptually important as they will drive investigators to account for this population structure in future studies, which will result in larger studies that can more effectively assess correlations to particular subgroups. In addition, larger study sizes will refine the resolution of these profiling approaches and will provide better estimates of the true number of MS patient subsets. While there may be large subsets of subjects, it is quite possible that many smaller subsets, such the anti-aquaporin 4 positive cases with an NMO phenotype exist. Notably, the study that used an RNA analysis approach [Corvol et al. 2008] suggests that one of the defined subject subsets is more likely to rapidly progress from the CIS stage to a definite diagnosis of MS. Thus, there may be clinical utility to understanding the architecture of CIS and MS. Even if we do not understand the exact underlying immunologic lesion of each subject subset, these efforts can outline distinct groups of subjects and can be used to characterize correlations to disease course and treatment response within each group.
Figure 2.
A heatmap highlights the difference in expression patterns between the three subsets of untreated subjects with RRMS defined using cytometric data. In this heatmap, each column is an individual subject, with subjects grouped together based on the MS subset (MS1–3) to which they have been assigned by consensus clustering. Each row is a single feature; in this case, each feature is the proportion of a certain cell type present within a sample of peripheral blood mononuclear cells from MS patients. For each MS subset, the 20 features that are most differentiated in that subset were selected for inclusion in this heatmap. Each cell in the heatmap is colored along a gradient with red denoting high relative expression and blue low expression (reproduced from De Jager et al. [2008]).
Conclusion
Personalized medicine has often been presented as being driven purely by genetic information, but this relatively simple concept is not likely to be sufficient for most questions in the field of MS. In some cases, such as when polymorphisms affect the amino acid sequence of a target molecule, drug response could be profoundly affected in the subset of patients with an allele that disrupts a drug interaction site. However, the preliminary evidence produced by our research community to date suggests that the approach of integrating genetic data with clinical and other biomarker data (including imaging data) is likely to be the more fruitful one in most cases. In general, assessing dynamic biomarkers in the context of a fixed architecture defined by variation in DNA sequence will provide a conceptual framework that reduces the complexity of discovering meaningful associations that are either treatment-specific or nonspecific and associated with disease severity. Either type of information is useful in informing treatment selection and enhancing the likelihood of treatment efficacy.
Current reports in this arena are exciting, but they highlight the fact that we are woefully underpowered in most studies and suffer from the absence of a replication sample set of equal or greater size than the discovery sample. The way forward is clear: it has been laid out by the International MS Genetics Consortium and its successes in identifying susceptibility alleles for MS. Consortia of investigators are the vehicles that will allow the accumulation and harmonization of treatment responses and disease course across MS centers and will provide the nexus of resources, expertise and samples needed to first discover and then test clinical algorithms. Some of these collaborations are emerging, particularly in Europe, and will hopefully include industry partners that have much experience and potentially many samples to provide to this type of endeavor.
A nontrivial consideration in these endeavors is the cost involved for these various tests once they are implemented in the clinical arena, and this important question should be assessed as part of the clinical trials validating any new algorithm. However, in general, the unit cost of genetic and many biomarker tests is small, particularly when considering the annual cost of MS treatment, and the rational targeting of treatment is therefore likely to be cost-effective. One interesting idea in the evaluation of genetic variation is that of a single genotyping array that could yield information for a patient's entire disease cycle. Specifically, one of the genome-wide genotyping arrays may not appear to be attractive as a clinical test in the context of a single question. However, because DNA sequence variation is fixed, this type of array may be attractive if one takes a longer view: it will provide information for the individual patient as she progresses through the course of her illness, including diagnosis, prognosis on disease course, and successive iterations of treatment selection. Genotyping a single array at the onset of the disease will obviate the need for repeated bouts of targeted genotyping during the course of MS and will also be helpful in managing other common diseases that a patient may face with increasing age, such as cardiovascular disease and dementing illnesses. It may thus be cost-effective in the long run, even if most of the genotyping data is never used clinically.
Overall, the next 5–10 years will see the development and testing of algorithms that may implement the promise of personalized medicine in the field of MS. The challenge ahead is not to rapidly implement interesting tests into the clinic; rather, it is for our community to work together to establish the framework of investigators that will validate robust clinical tools with which to deconstruct the heterogeneity of the MS patient population. All too often in medicine, we have seen the utility of promising approaches to disease management being oversold, which leads to a loss of interest in their application. The potential of a personalized approach to MS disease management is clear, and we should move together to realize those aspects of this approach that provide meaningful benefits to our patients.
Conflict of interest statement
The author has received compensation for speaking engagements from Biogen IDEC. He has served on advisory boards for Biogen IDEC and TEVA Neuro-Science. He receives research support from Source MDx, Inc.
References
- International MS Genetics Consortium. (2008) Refining genetic associations in multiple sclerosis. Lancet Neurol 7:567–569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko Y.S., Hoppenbrouwers I.A., Ramagopalan S.V., Broer L., Jafari N., Hillert J.et al. (2008) Genetic variation in the KIF1b locus influences susceptibility to multiple sclerosis. Nat Genet 40:1402–1403 [DOI] [PubMed] [Google Scholar]
- Baechler E.C., Batliwalla F.M., Karypis G., Gaffney P.M., Ortmann W.A., Espe K.J.et al. (2003) Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA 100:2610–2615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baranzini S.E., Wang J., Gibson R.A., Galwey N., Naegelin Y., Barkhof F.et al. (2009) Genome-wide association analysis of susceptibility and clinical phenotype in multiple sclerosis. Hum Mol Genet 18:767–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos L.F., Sawcer S., Ramsay P.P., Baranzini S.E., Thomson G., Briggs F.et al. (2006) Heterogeneity at the HLA-DRB1 locus and risk for multiple sclerosis. Hum Mol Genet 15:2813–2824 [DOI] [PubMed] [Google Scholar]
- Breij E.C., Brink B.P., Veerhuis R., Van Den Berg C., Vloet R., Yan R.et al. (2008) Homogeneity of active demyelinating lesions in established multiple sclerosis. Ann Neurol 63:16–25 [DOI] [PubMed] [Google Scholar]
- Byun E., Caillier S.J., Montalban X., Villoslada P., Fernandez O., Brassat D.et al. (2008) Genome-wide pharmacogenomic analysis of the response to interferon beta therapy in multiple sclerosis. Arch Neurol 65:337–344 [DOI] [PubMed] [Google Scholar]
- Caillier S.J., Briggs F., Cree B.A., Baranzini S.E., Fernandez-Vina M., Ramsay P.P.et al. (2008) Uncoupling the roles of HLA-DRB1 and HLA-DRB5 genes in multiple sclerosis. J Immunol 181:5473–5480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comabella M., Craig D.W., Camina-Tato M., Morcillo C., Lopez C, Navarro A.et al. (2008) Identification of a novel risk locus for multiple sclerosis at 13q31.3 by a pooled genome-wide scan of 500,000 single nucleotide polymorphisms. PLoS ONE 3:e3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compston A., Coles A. (2008) Multiple sclerosis. Lancet 372:1502–1517 [DOI] [PubMed] [Google Scholar]
- Corvol J.C., Pelletier D., Henry R.G., Caillier S.J., Wang J., Pappas D.et al. (2008) Abrogation of T cell quiescence characterizes patients at high risk for multiple sclerosis after the initial neurological event. Proc Natl Acad Sci USA 105:11839–11844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager P.L., Jia X., Wang J., de Bakker P. I.W, Ottoboni L., Aggarwal NT.et al. (2009) Analysis of genome scans and replication identify CD6. ICSBP1, and TNFRSF1a as novel multiple sclerosis susceptibility loci. Nature Genetics.(in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jager P.L., Rossin E., Pyne S., Tamayo P., Ottoboni L., Viglietta Vet al. (2008) Cytometric profiling in multiple sclerosis uncovers patient population structure and a reduction of CD8low cells. Brain 131:1701–1711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deluca G.C., Ramagopalan S.V., Herrera B.M., Dyment D.A., Lincoln M.R., Montpetit A.et al. (2007) An extremes of outcome strategy provides evidence that multiple sclerosis severity is determined by alleles at the HLA-DRB1 locus. Proc Natl Acad Sci USA 104:20896–20901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekegren T, Hanrieder J., Bergquist J. (2008) Clinical perspectives of high-resolution mass spectrometry-based proteomics in neuroscience: exemplified in amyotrophic lateral sclerosis biomarker discovery research. J Mass Spectrom 43:559–571 [DOI] [PubMed] [Google Scholar]
- Fusco C., Andreone V., Coppola G., Luongo V., Guerini F., Pace E.et al. (2001) Hla-Drb1*1501 and response to copolymer-1 therapy in relapsing-remitting multiple sclerosis. Neurology 57:1976–1979 [DOI] [PubMed] [Google Scholar]
- Gage B.F., Eby C, Johnson J.A., Deych E., Rieder M.J., Ridker P.M.et al. (2008) Use of pharmacogenetic and clinical factors to predict the therapeutic dose of warfarin. Clin Pharmacol Ther 84:326–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg S.A., Pinkus J.L., Pinkus G.S., Burleson T., Sanoudou D., Tawil R.et al. (2005) Interferon-alpha/beta-mediated innate immune mechanisms in dermatomyositis. Ann Neurol 57:664–678 [DOI] [PubMed] [Google Scholar]
- Hafler D.A., Compston A., Sawcer S., Lander E.S., Daly M.J., De Jager P.L.et al. (2007) Risk alleles for multiple sclerosis identified by a genomewide study. N Engl J Med 357:851–862 [DOI] [PubMed] [Google Scholar]
- Jacob A., Weinshenker B.G., Violich I., Mclinskey N., Krupp L., Fox R.J.et al. (2008) Treatment of neuromyelitis optica with rituximab: retrospective analysis of 25 patients. Arch Neurol 65:1443–1448 [DOI] [PubMed] [Google Scholar]
- Lucchinetti C, Bruck W., Parisi J., Scheithauer B., Rodriguez M., Lassmann H. (2000) Heterogeneity of multiple sclerosis lesions: implications for the pathogenesis of demyelination. Ann Neurol 47:707–717 [DOI] [PubMed] [Google Scholar]
- Matiello M., Lennon V.A., Jacob A., Pittock S.J., Lucchinetti CF., Wingerchuk D.M.et al. (2008) NMO-IgG predicts the outcome of recurrent optic neuritis. Neurology 70:2197–2200 [DOI] [PubMed] [Google Scholar]
- McDonald W.I., Compston A., Edan G., Goodkin D., Hartung H.P., Lublin F.D.et al. (2001) Recommended diagnostic criteria for multiple sclerosis: guidelines from the International Panel on the Diagnosis of Multiple Sclerosis. Ann Neurol 50:121–127 [DOI] [PubMed] [Google Scholar]
- McLaughlin K.A., Wucherpfennig K.W. (2008) B cells and autoantibodies in the pathogenesis of multiple sclerosis and related inflammatory demyelinating diseases. Adv Immunol 98:121–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller A., Avidan N., Tzunz-Henig N., Glass-Marmor L., Lejbkowicz I., Pinter R.Y.et al. (2008) Translation towards personalized medicine in multiple sclerosis. J Neurol Sci 274:68–75 [DOI] [PubMed] [Google Scholar]
- Nelson M.R., Bacanu S.A., Mosteller M., Li L., Bowman C.E., Roses A.D.et al. (2009) Genome-wide approaches to identify pharmacogenetic contributions to adverse drug reactions. Pharmacogenomics J 9:23–33 [DOI] [PubMed] [Google Scholar]
- O'Connor K.C., McLaughlin K.A., De Jager P.L., Chitnis T., Bettelli E., Xu C.et al. (2007) Self-antigen tetramers discriminate between myelin autoantibodies to native or denatured protein. Nat Med 13:211–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuda D.T., Srinivasan R., Oksenberg J.R., Goodin D.S., Baranzini S.E., Beheshtian A.et al. (2009) Genotype-phenotype correlations in multiple sclerosis: HLA genes influence disease severity inferred by 1HMR spectroscopy and MRI measures. Brain 132:250–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana F.J., Farez M.F., Viglietta V., Iglesias A.H., Merbl Y., Izquierdo G.et al. (2008) Antigen microarrays identify unique serum autoantibody signatures in clinical and pathologic subtypes of multiple sclerosis. Proc Natl Acad Sci USA 105:18889–18894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racke M.K. (2008) The role of B Cells in multiple sclerosis: rationale for B-cell-targeted therapies. Curr Opin Neurol 21(Suppl. 1):S9–S18 [DOI] [PubMed] [Google Scholar]
- Rinaldi L., Gallo P., Calabrese M., Ranzato F., Luise D., Colavito D.et al. (2006) Longitudinal analysis of immune cell phenotypes in early stage multiple sclerosis: distinctive patterns characterize MRI-active patients. Brain 129:1993–2007 [DOI] [PubMed] [Google Scholar]
- Roxburgh R.H., Seaman S.R., Masterman T, Hensiek A.E., Sawcer S.J., Vukusic S.et al. (2005) Multiple sclerosis severity score: using disability and disease duration to rate disease severity. Neurology 64:1144–1151 [DOI] [PubMed] [Google Scholar]
- Steinman L. (2008) New targets for treatment of multiple sclerosis. J Neurol Sci 274:1–4 [DOI] [PubMed] [Google Scholar]
- Van Baarsen L.G., Van Der Pouw Kraan T.C., Kragt J.J., Baggen J.M., Rustenburg F., Hooper T.et al. (2006) A subtype of multiple sclerosis defined by an activated immune defense program. Genes Immun 7:522–531 [DOI] [PubMed] [Google Scholar]
- Van Der Pouw Kraan T.C., Wijbrandts C.A., Van Baarsen L.G., Voskuyl A.E., Rustenburg F., Baggen J.M.et al. (2007) Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis 66:1008–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingerchuk D.M., Lennon VA., Pittock S.J., Lucchinetti C.F., Weinshenker B.G. (2006) Revised diagnostic criteria for neuromyelitis optica. Neurology 66:1485–1489 [DOI] [PubMed] [Google Scholar]
- Yang C.H., Yu C.J., Shih J.Y, Chang Y.C., Hu F.C., Tsai M.C.et al. (2008) Specific EGFR mutations predict treatment outcome of stage IIIb/IV patients with chemotherapy-naive non-small-cell lung cancer receiving first-line gefitinib monotherapy. J Clin Oncol 26:2745–2753 [DOI] [PubMed] [Google Scholar]
- Zivadinov R., Uxa L., Bratina A., Bosco A., Srinivasaraghavan B., Minagar A.et al. (2007) HLA-DRB1*1501, -DQB1*0301, -DQB1*0302, -DQB1*0602, and -DQB1*0603 alleles are associated with more severe disease outcome on mri in patients with multiple sclerosis. Int Rev Neurobiol 79:521–535 [DOI] [PubMed] [Google Scholar]