Abstract
RNA polymerase II (RNAPII), the 12-subunit enzyme that synthesizes all mRNAs and several non-coding RNAs in eukaryotes, plays a central role in cell function. Although multiple proteins are known to regulate the activity of RNAPII during transcription, little is known about the machinery that controls the fate of the enzyme before or after transcription. We used systematic protein affinity purification coupled to mass spectrometry (AP-MS) to characterize the high resolution network of protein interactions of RNAPII in the soluble fraction of human cell extracts. Our analysis revealed that many components of this network participate in RNAPII biogenesis. We show here that RNAPII-associated protein 4 (RPAP4/GPN1) shuttles between the nucleus and the cytoplasm and regulates nuclear import of POLR2A/RPB1 and POLR2B/RPB2, the two largest subunits of RNAPII. RPAP4/GPN1 is a member of a newly discovered GTPase family that contains a unique and highly conserved GPN loop motif that we show is essential, in conjunction with its GTP-binding motifs, for nuclear localization of POLR2A/RPB1 in a process that also requires microtubule assembly. A model for RNAPII biogenesis is presented.
Significant effort has been made over the past 4 decades to identify and characterize the factors that regulate the activity of RNA polymerase II (RNAPII),1 the eukaryotic enzyme that synthesizes mRNA and several non-coding RNA. A myriad of protein factors have the ability to regulate the activity of RNAPII during the act of transcription. DNA-binding transcriptional regulators are known to control the activity of the RNAPII transcription machinery in a gene- and cell type-specific manner (1–4), whereas general transcription factors act as RNAPII accessory proteins required for the transcription of all (or most) class II genes (5–8), and co-regulators (co-activators and co-repressors) serve as bridges between DNA-bound factors and the RNAPII machinery (9–12), some affecting the organization and/or chemical modification of the chromatin template of RNAPII (13–15).
Quite surprisingly and despite extensive efforts to analyze the regulatory mechanisms targeting transcription and transcription factors themselves, very little is known about the molecular machinery that regulates the fate of RNAPII before and after transcription. For example, the process of biogenesis of the three nuclear RNAPs (RNAPI, -II, and -III), which comprise both common and specific subunits, has been the subject of only a few reports (16).
We hypothesized that the protein complexes involved in the assembly, folding, and nuclear import of RNAPII are likely to be found in the human cell soluble fraction as opposed to the insoluble fraction that contains chromatin and actively transcribing RNAP molecules. We therefore conducted a survey of the soluble protein complexes that associate with RNAPII using protein affinity purification coupled to mass spectrometry (AP-MS) to identify the factors involved in the biogenesis of RNAPII. Twenty-eight tagged proteins were purified, and their associating partners were identified by MS. High confidence interactions were selected computationally and then used to draw a map of the interactions connecting these complexes. The composition and organization of this network revealed important features about the eukaryotic transcriptional machinery. Most notably, the highly conserved GTPase RNAPII-associated protein 4 (RPAP4)/GPN1 was found to have multiple interactions with the subunits of RNAPs, tubulins, and components of the microtubule assembly machinery, including the chaperonins (chaperonin containing TCP-1 (CCT) complex) and prefoldins (prefoldin-like complex). Our results indicate that both RPAP4/GPN1 activity and microtubule assembly/integrity are required for nuclear localization of the largest RNAPII subunits, POLR2A/RPB1 and POLR2B/RPB2.
EXPERIMENTAL PROCEDURES
Generation of Cell Lines for Expressing TAP-tagged Polypeptides
Selected human polypeptides were cloned into the mammalian expression vector pMZI (17) carrying a TAP tag at its C terminus (18, 19). Stable human embryonic kidney cell lines (EcR-293; derived from HEK293) carrying these constructs were produced as described previously (20, 21).
Expression of TAP-tagged Proteins and Purification of Protein Complexes
Induction for 24–72 h with 3–6 μm ponasterone A (Invitrogen) was used to express the TAP-tagged proteins. Whole cell extracts prepared from induced and non-induced stable EcR-293 cell lines were subjected to purification by the TAP procedure as described previously (20, 21).
Protein Identification by Mass Spectrometry
The TAP eluates were run on SDS gels and stained with silver, and gel slices were excised and digested with trypsin as described previously (20, 21). The resulting tryptic peptides were purified and identified by LC-tandem mass spectrometry (MS/MS) using a microcapillary reversed-phase high pressure liquid chromatography-coupled LTQ-Orbitrap (ThermoElectron) quadrupole ion trap mass spectrometer with a nanospray interface.
The peak list files were generated with extract_msn.exe (version February 15, 2005) using the following parameters: minimum mass set to 600 Da, maximum mass set to 6000 Da, no grouping of MS/MS spectra, precursor charge set to auto, and minimum number of fragment ions set to 10. Protein database searching was performed with Mascot 2.2.04 (Matrix Science) against the human NCBInr protein database (version April 2, 2009). There are 10,427,007 sequences in this database. The mass tolerances for precursor and fragment ions were set to 10 ppm and 0.6 Da, respectively. Trypsin was used as the enzyme allowing for up to two missed cleavages. Carbamidomethyl and oxidation of methionine were allowed as variable modifications.
A cutoff score of 30 for the first peptide (15 for the additional peptides) for accepting individual MS/MS spectra was established as optimal for the determination of interaction reliability (IR) scores (see supplemental Table S1 for a list of IR scores for individual interactions). IR scores of each interaction between a prey P and a bait B were computed by a predictor using a logistic regression approach as described previously (21). The predictor outputs the probability that an interaction between P and B is correct as a function of a weighted sum of the Mascot score of P, the highest Mascot score of all peptides of P, the number of common interacting partners between P and B, the presence of the interaction in reciprocal purifications, the number of baits that found prey P, and the combinations of these. More precisely, the logistic regression is made of five terms representing the five features described with the addition of the squares of four of these features (the reciprocity feature, which is binary, is not squared) and of 10 terms corresponding to the products of the feature values for a total of 19 terms. To train the predictor, a set of 248 positive and 2403 negative examples was derived from gene ontology (22) with the hypothesis that proteins sharing at least one gene ontology annotation are likely to be interacting and vice versa. Although these sets do not only contain true positive and true negative interactions, they are enriched for a representative subset of correct interaction identifications, which is sufficient for proper training of the predictor. The regression weights of all 19 terms were computed to minimize the cross-entropy error function between the predictions made by the classifier and the labels of all training interactions. To evaluate the accuracy of the predictor, a test set was built from 149 manually identified correct and 54 incorrect interactions based only on strong literature support and not on our interaction data (20). With the IR score threshold used in this study (>0.7337), we estimated a specificity of about 88%, suggesting an overall rate of false positives lower than 12%. The use of this IR score threshold lowered the sensitivity to 72%, suggesting that 28% of the relevant interactions are left out of the graph in Fig. 1B.
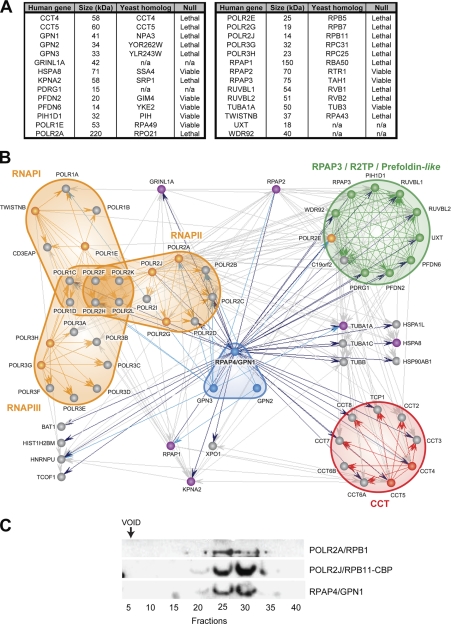
Fig. 1.
High density network of high confidence interactions formed by RNAPII and RPAPs in soluble fraction of human cells. A, table listing the 28 tagged proteins used in this study with their size, homolog in S. cerevisiae, and requirement for yeast growth. B, diagram of the RNAPII-RPAP interaction network. In this diagram, blue arrows indicate proteins that were pulled down by tagged RPAP4/GPN1 (dark blue) and GPN2 and GPN3 (light blue), and gray arrows indicate interactions detected between the various RPAP4/GPN1 interactors in reciprocal TAP tag purifications. Tagged proteins used in affinity purification experiments are colored according to their association with nuclear RNAPs (orange), GPN (blue), CCT (red), RPAP3/R2TP/prefoldin-like (green), or not associated with (or ascribed to) a specific complex (magenta). Official gene symbols are used for convenience in the bioinformatics analysis (POLR2 is used to designate RNAPII subunits instead of RPB). All interactions with an IR score >0.7337 are represented by edges (specificity of 88%; expected rate of false positive <12%). C, RPAP4/GPN1 and RNAPII (POLR2A/RPB1 and tagged POLR2J/RPB11) co-fractionate in gel filtration experiments. n/a, not applicable.
In cases where multiple gene products were identified from the same peptide set, all were unambiguously removed from the data set. In the case of multiple isoforms stemming from a unique gene, the isoform with the best sequence coverage was reported. Proteins identified on the basis of a single peptide are listed in supplemental Tables S4 (human) and S5 (yeast), and the individual spectra are comprehensively presented in supplemental Figs. S1 (human) and S2 (yeast).
Gel Filtration Chromatography
Affinity-purified protein complexes were concentrated by dialysis in buffer F containing 10 mm Hepes, pH 7.9, 100 mm NaCl, 0.1 mm EDTA, 5% glycerol, and 0.5 mm DTT. An aliquot (50 μl) of the concentrated eluate was fractionated on a Superose 6 PC 3.2/30 column (2.4 ml) previously equilibrated in buffer F using the ÄKTA FPLC system (GE Healthcare). The column was run in buffer F at a flow rate of 0.04 ml/min, and 50-μl fractions were collected. Aliquots of each five fractions were pooled, concentrated, and analyzed by Western blot.
siRNA Silencing
RPAP4/GPN1 (ON-TARGETplus SMART pool) and control (siCONTROL non-targeting pool) siRNAs (Dharmacon) were transfected into HeLa cells using Oligofectamine (Invitrogen) at an siRNA final concentration of 100 nm. At various time intervals post-transfection, cells were lysed, and RPAP4/GPN1 expression levels were monitored by Western blotting.
Antibodies
The following antibodies were used in this study and were obtained from various sources: primary antibody against RPAP4/GPN1 (CIM Antibody Core, Arizona State University, Tempe, AZ), monoclonal antibody against the RNAPII POLR2A/RPB1 subunit (8WG16; Covance), TAP specific anti-calmodulin binding peptide antibody (clone C16T; Upstate), anti-CDK9 antibody (C-20; Santa Cruz Biotechnology), monoclonal anti-β-tubulin antibody (clone TUB 2.1; Sigma), and horseradish peroxidase-conjugated secondary antibody (GE Healthcare).
Proliferation Curves
HeLa cells were seeded into 6-well dishes at a starting density of 10,000 cells/well. Double siRNA treatments were on days 0 and 3. Cells were trypsinized on days 1–7 and counted with a hemocytometer.
Immunofluorescence and Imaging
HeLa cells were grown on Lab-Tek (Nunc). Twenty-four hours post-transfection, cells were fixed with 3.7% formaldehyde in PBS and permeabilized with 0.3% Triton X-100 in PBS. DNA was stained with TO-PRO®-3 (Molecular Probes). For immunofluorescence studies, cells were incubated with the first antibody diluted in 5% donkey serum in PBS for 1 h followed by 1 h of incubation with a 1:200 dilution of Alexa Fluor 488- or Cy3-conjugated secondary antibody. Cells were washed with PBS after each step. Cells were mounted using Prolong Gold and SlowFade Gold antifade reagents (Invitrogen). Images were acquired using an LSM 510 or LSM 710 confocal laser scanning microscope and analyzed using LSM Image Browser, version 3.2.0.70 (Zeiss, Toronto, Canada).
Yeast cell culture, fixation, conversion to spheroplasts, and permeabilization were performed as previously described (23). Slides were blocked and hybridized as described above for mammalian cells. Yeast nuclei were treated with 2.5 μg/ml Hoechst 33342 (Molecular Probes) for 30 min.
Green Fluorescent Protein Fusions
The GFP-RPAP4(Mut) construct was obtained by cloning the full-length RPAP4/GPN1 open reading frame into pGFP2-N1 (PerkinElmer Life Sciences) using the EcoRI and ApaI sites. Transfection of HeLa cells was performed using the Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's protocol.
Cytoplasmic and Nuclear Extracts
Cells grown in a 150-mm dish were washed with PBS and resuspended in 1 ml of ice-cold lysis buffer (50 mm Tris-HCl, pH 8, 5 mm MgCl2, 0.5% Nonidet P-40, 0.5% sodium deoxycholate, and one tablet of Complete Mini EDTA-free (Roche Applied Science)). The lysate was centrifuged for 2 min at 9400 × g. The supernatant, representing the cytoplasmic fraction, was kept on ice. The pellet was resuspended in lysis buffer and submitted to three freeze-thaw cycles in liquid nitrogen. The lysate was centrifuged for 10 min at 18,400 × g, and the supernatant was kept as the nuclear fraction (24). The presence of the protein of interest was analyzed by Western blotting.
Yeast Strains and Growth Media
NPA3, the yeast homolog of RPAP4/GPN1, was cloned in plasmid pRS316 (URA3, ampR, CEN6, ARSH4) and pRS415 (LEU2, ampR, CEN6, ARSH4). Plasmid pRS415-NPA3 was used as a template for all mutagenic polymerase chain reactions (PCRs) using phosphorylated primers (details on the primers used in this study are available upon request). Changes in nucleotide sequences were confirmed by DNA sequencing.
Yeast strain BY4743–22550 (MATa/α his3Δ1/his3Δ1 leu2Δ0/leu2Δ0 met15Δ0/MET15 ura3Δ0/ura3Δ0 lys2Δ0/LYS2 npa3::kanMX/NPA3) obtained from the yeast knock-out collection (Invitrogen) was transformed with pRS316-NPA3 using a standard protocol (25). Transformed cells were sporulated and dissected on yeast peptone-dextrose (YPD) medium. Haploid knocked out strains were selected on yeast complete medium (Yc) lacking uracil containing G418 (200 μg/ml) and counterselected on Yc containing 5-fluoroorotic acid (5FOA) (1 mg/ml). All pRS415-NPA3 plasmids obtained by mutagenesis were transformed into this haploid strain named BCY74 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 lys2Δ0 npa3::kanMX pRS316-NPA3). Yeast strains and plasmids used in this study are listed in supplemental Table S3. The function of each yeast mutant was assessed by plasmid shuffling on 5FOA lacking leucine so that the only copy of the NPA3 gene is mutated. These strains were cultured to an A600 nm of 1.0, serially diluted (1, 1:5, 1:50, and 1:500), and spotted on Yc-Leu−-Ura− and Yc-Leu− containing 1 mg/ml 5FOA. Each plate was incubated for 3–5 days at 30 °C. Yeast mutants that showed a slow growth phenotype on 5FOA were then spotted on Yc-Leu− with or without 20 μg/ml benomyl with the same serial dilutions.
RESULTS
High Resolution Mapping of Protein Interaction Network for Human RNA Polymerase II
In previous work, we used AP-MS to begin to characterize the network of interactions of the human RNAPII transcription machinery in the soluble fraction of human cell extracts (20, 26, 27). This work identified a number of previously uncharacterized proteins that associate with known transcription factors to regulate their activity prior to their involvement in active transcription on template chromatin. A subset of these factors was found to be tightly connected to RNAPII in the cellular soluble fraction, suggesting a possible role in RNAPII biogenesis and in regulating the activity of this enzyme before its involvement in active transcription (20). To further characterize the network of interactions that connects these RPAPs to RNAPII itself, we proceeded to affinity purify additional components of the network present in human 293 cells (see Fig. 1A for a list of TAP-tagged proteins; note that in Fig. 1, A and B, the official gene symbols are used for all proteins, including the RNAPII subunits (POLR2A–POLR2L)). Overall, 28 members of the RNAPII-RPAP network were used in AP-MS experiments. Demonstrative silver-stained gels are shown in supplemental Fig. S3. Fig. 1B illustrates the high density interaction network mapped using this procedure. As we described above, high confidence interactions were selected by applying a computational algorithm that assigns IR scores to each detected interaction according to the strength of the MS score and the local topology of the network (e.g. conservation of the interaction in reciprocal purifications and number of shared partners). Interactions with an IR score over a threshold that minimizes the rate of false positives were selected (20, 21). Fig. 1B shows all the interactions with IR scores higher than 0.7337. Above this threshold, we estimated a specificity of about 88%, suggesting an overall rate of false positives lower than 12%. Of note, the use of an IR score threshold of 0.7337 lowered the sensitivity to 72%, suggesting that 28% of the relevant interactions are left out of the graph in Fig. 1B. The list of interactions obtained using our experimental procedure with their associated IR scores is provided in supplemental Table S1.
GTPase RPAP4/GPN1 Is a Central Component of RNA Polymerase II Interaction Network
Examination of the diagram in Fig. 1B reveals that a protein termed RPAP4 (official gene symbol, GPN1) occupies a central position in the RNAPII interaction network by connecting RNAPII to: (i) the CCT complex (28, 29); (ii) the RPAP3/R2TP/prefoldin-like complex (30–32), which comprises 11 subunits, including some small molecular weight chaperones called prefoldins; and (iii) a number of additional polypeptides involved in protein assembly and/or folding, including some chaperones. Of note, RPAP4/GPN1 is the only protein in the network connected to the majority of the proteins forming the RNAPII, RPAP3/R2TP/prefoldin-like, and CCT complexes. Affinity purification of tagged NPA3, the yeast homolog of RPAP4/GPN1, confirmed its association with RNAPII and the RPAP3 complex in Saccharomyces cerevisiae and demonstrated the conservation of these interactions in eukaryotes (see supplemental Table S2). To further confirm the association of RPAP4/GPN1 with RNAPII, we performed gel filtration experiments using the material from a POLR2J/RPB11-TAP purification, which contained both RNAPII and RPAP4/GPN1, according to our AP-MS data (see Fig. 1B). As shown in Fig. 1C, RPAP4/GPN1 was found to co-fractionate with the RNAPII subunits POLR2A/RPB1 and POLR2J/RPB11 in these experiments, strengthening the conclusion that RNAPII and RPAP4/GPN1 associate with each other.
RPAP4/GPN1 Is a Conserved GTPase Essential for Growth of Yeast and Human Cells
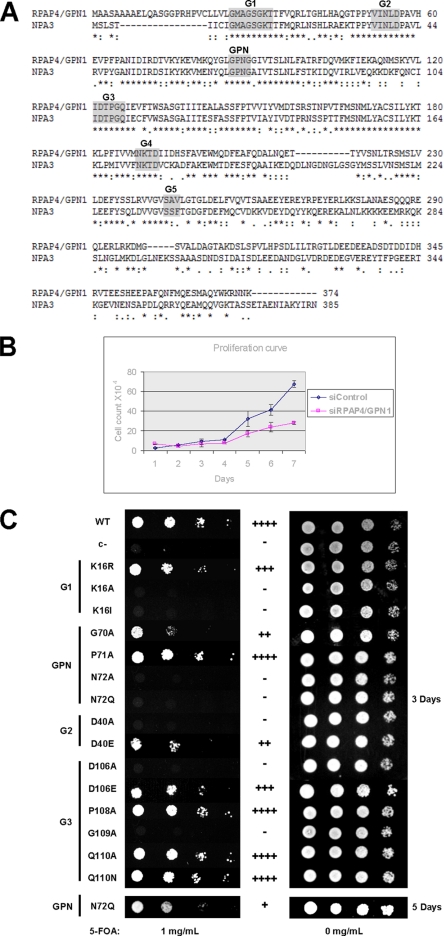
RPAP4/GPN1 (also called XAB1) was described previously as a GTPase that interacts with the DNA repair protein Xeroderma pigmentosum complementation group A (33) as well as a protein that regulates transcription in human cells (34). Although its detailed mechanism of action is not known, RPAP4/GPN1 is part of a newly defined family of GTPases that contains a conserved 3-amino acid (Gly-Pro-Asn or GPN) loop as a unique feature (35). Two other members of this family, GPN2 and GPN3, are also components of the RNAPII interaction network (see Fig. 1B). The GPN loop is located between the G2 and G3 motifs of the RPAP4/GPN1 GTP-binding site elements (Fig. 2A). Our experiments indicated that both RPAP4/GPN1 (Fig. 2B) and yeast NPA3 (Fig. 2C) are essential for cell growth. Amino acid substitutions in the G1, G2, and G3 motifs of the GTP-binding site as well as in the GPN loop impaired the activity of NPA3 (Fig. 2C); some of these were lethal (K16A, K16I, N72A, D40A, D106A, and G109A), and others generated a slow growth phenotype (K16R, G70A, D40E, D106E, and N72Q). Together, these results indicate that both the GTP-binding motif and the GPN loop are essential for NPA3 activity and cell proliferation.
Fig. 2.
RPAP4/GPN1 is a conserved protein containing GPN loop element and GTP-binding motifs that are essential for cell growth. A, linear representation of human RPAP4/GPN1 and its yeast homolog NPA3. The domains forming the GTP-binding site (G1–G5; shaded) and the GPN loop are shown (* identical residue; : conserved substitution;. semi-conserved substitution). B, siRNA silencing of RPAP4/GPN1 inhibits human cell growth. Proliferation curves of HeLa cells following double treatment with either an siRNA directed against RPAP4/GPN1 (squares) or control siRNA (diamonds) over a 7-day period are shown. Data are presented as means ± Standard Deviation. C, the GTP-binding motif and the GPN loop are essential for NPA3 function in yeast. Amino acid substitutions in the G1, G2, G3, or GPN loop domain of NPA3 are either lethal or produce slow growth phenotypes in yeast (c- = negative control). Growth phenotypes are presented as equivalent to wild-type (WT) (++++), slightly slower (+++), slower (++), very slow (+) and no growth (-).
Silencing of RPAP4/GPN1 Results in Cytoplasmic Accumulation of POLR2A/RPB1, the Largest Subunit of RNAPII
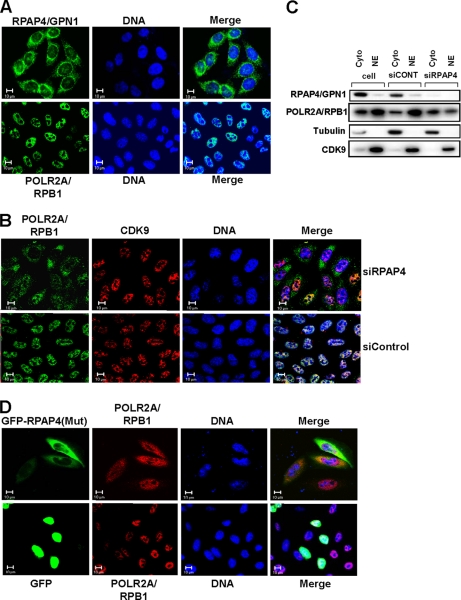
To start elucidating the function of the RPAPs and of their association with RNAPII, we performed siRNA silencing experiments and monitored the effect of RPAP depletions on various parameters, including RNAPII assembly and intracellular localization. Our rationale for suspecting a role for the RPAPs in the biogenesis of RNAPII was based on the observation that RPAP1, -2, -3, and -4 are found mainly in the cytoplasm of yeast (36) and human cells (see Fig. 3A, upper panel, for data on the localization of RPAP4/GPN1). The cytoplasmic localization of the RPAPs contrasts with that of RNAPII, which is primarily nuclear (Fig. 3A, lower panel) as monitored using an antibody directed against its largest subunit, POLR2A/RPB1. The current report addresses the function of RPAP4/GPN1 in RNAPII biogenesis, whereas the role of the other RPAPs will be described elsewhere.2
Fig. 3.
Silencing of RPAP4/GPN1 or overexpression of GFP-RPAP4 fusion protein with amino acid substitutions in GPN loop result in accumulation of POLR2A/RPB1 in cytoplasm. A, localization of RPAP4/GPN1 (upper row) and POLR2A/RPB1 (lower row) in HeLa cells was determined by immunofluorescence. B, localization of POLR2A/RPB1 following treatment of HeLa cells with either an siRNA targeting RPAP4/GPN1 or a control siRNA. The localization of CDK9 is shown as a control. C, cell fractionation showing the effect of RPAP4/GPN1 silencing on the cytoplasmic/nuclear ratio of POLR2A/RPB1. Tubulin and CDK9 are used as cytoplasmic and nuclear localization controls, respectively. D, overexpression of a GFP-RPAP4/GPN1 fusion protein carrying amino acid substitutions in the GPN loop (GPN to AAA), but not GFP alone, leads to the accumulation of POLR2A/RPB1 in the cytoplasm of treated cells. In each case, the location of the DNA is shown (TO-PRO-3 iodide). NE, nuclear extract; siCONT, siCONTROL.
Silencing of RPAP4/GPN1 resulted in the accumulation of RNAPII in the cytoplasm of treated cells with the enzyme found in both the cytoplasm and the nucleus after siRNA treatment (Fig. 3B, upper panel). RPAP4/GPN1 silencing did not affect the localization of CDK9, which was used as a nuclear control in Fig. 3B. Treatment with a control siRNA did not affect nuclear localization of RNAPII (Fig. 3B, lower panel). These results were confirmed by biochemical fractionation experiments that showed a shift in the cytoplasmic/nuclear ratio of POLR2A/RBP1 following RPAP4/GPN1 depletion (Fig. 3C).
Overexpression of GFP-RPAP4/GPN1 Fusion Protein with Mutation in GPN Loop Also Causes Accumulation of POLR2A/RPB1 in Cytoplasm
To confirm the involvement of RPAP4/GPN1 in the nuclear import of POLR2A/RPB1, we overexpressed in HeLa cells (seeking a dominant negative effect) a fusion protein consisting of an RPAP4/GPN1 variant with a mutated GPN loop motif fused to a green fluorescent protein (GFP) using a CMV promoter-driven expression vector. Expression of this GFP-RPAP4(GPN → AAA) fusion protein led to the accumulation of POLR2A/RPB1 in the cytoplasm of transfected cells (Fig. 3D, upper panel). As expected, the GFP-RPAP4(GPN → AAA) fusion protein was localized in the cytoplasm of expressing cells, whereas the GFP control was found diffused throughout the cell (Fig. 3D, lower panel). The results of these overexpression experiments suggest that interfering with the function of RPAP4/GPN1 induces an accumulation of POLR2A/RPB1 in the cell cytoplasm.
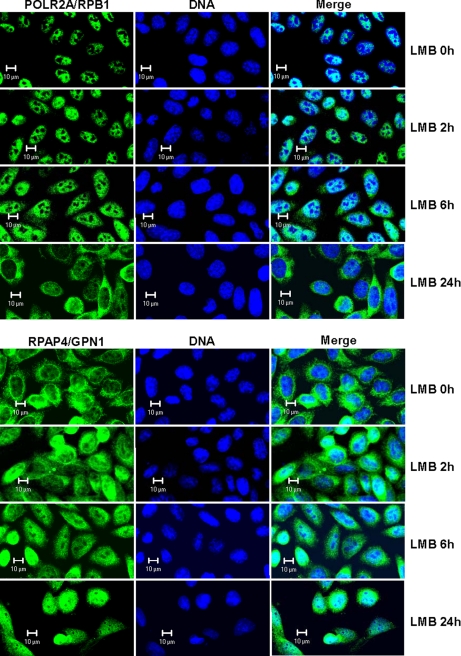
XPO1/CRM1 Inhibitor Leptomycin B Induces Sequestration of RPAP4/GPN1 in Nucleus and Accumulation of POLR2A/RPB1 in Cytoplasm
Proteins with a leucine-rich nuclear export signal (NES) are actively translocated to the cytoplasm via the action of XPO1/CRM1 (37, 38). Because RPAP4/GPN1 was reported to have an NES (34), we treated cultured cells with the XPO1/CRM1 inhibitor leptomycin B (LMB) and monitored the effect on the localization of both RPAP4/GPN1 and POLR2A/RPB1. As expected, RPAP4/GPN1 was sequestered in the nucleus as a result of LMB treatment (Fig. 4, lower panel); the effect was observed after 2 h of treatment and increased up to 24 h. Strikingly, however, LMB treatment led to sequestration of POLR2A/RPB1 in the cytoplasm with time, and this was slightly delayed compared with RPAP4/GPN1 nuclear sequestration (Fig. 4, upper panel). More precisely, POLR2A/RPB1 was not observed in the cytoplasm after 2 h of LMB treatment, only a small amount was seen at 6 h, and it was clearly seen and at a maximum level at 24 h. The same treatment did not affect localization of the transcription elongation factor CDK9, which was used as a control (data not shown). This finding further supports the notion that RPAP4/GPN1 nucleocytoplasmic shuttling through the XPO1/CRM1-NES pathway participates in the nuclear import of POLR2A/RPB1.
Fig. 4.
Inhibition of CRM1-dependent nuclear export results in retention of RPAP4/GPN1 in nucleus followed by accumulation of POLR2A/RPB1 in cytoplasm of treated cells. Cells grown in the absence (0 h) or presence (2, 6, and 24 h) of 10 ng/ml LMB were analyzed by immunofluorescence using antibodies directed against RPAP4/GPN1 and the POLR2A/RPB1 subunit of RNAPII. In each case, the location of the DNA is shown (TO-PRO-3 iodide).
Yeast NPA3 Mutants with Slow Growth Phenotype Accumulate POLR2A/RPB1 in Cytoplasm
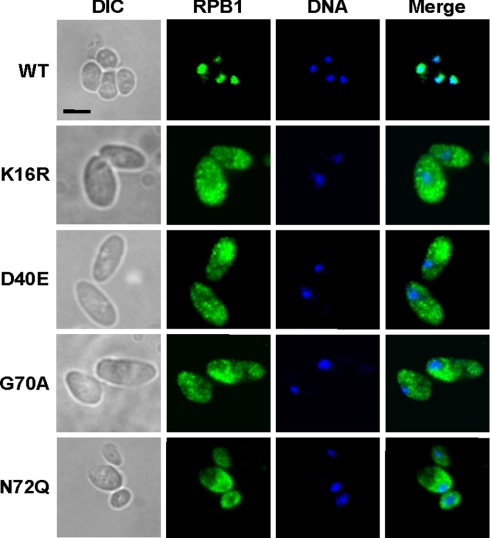
To further define the mechanism by which RPAP4/GPN1 regulates POLR2A/RPB1 nuclear import, we took advantage of our NPA3 mutant alleles that are not lethal but that confer a slow growth phenotype (see Fig. 2C). The subcellular localization of POLR2A/RPB1 was monitored in yeast strain K16R mutated in the G1 domain, strain D40E mutated in the G2 domain, and in strains G70A and N72Q mutated in the GPN loop element. Fig. 5 shows that these slow growing strains accumulated POLR2A/RPB1 in the cytoplasm. These results indicate that the integrity of the GTP-binding domain is important for the POLR2A/RPB1 nuclear import function of NPA3.
Fig. 5.
NPA3 mutant yeast strains with slow growth phenotype accumulate POLR2A/RPB1 in cytoplasm. Yeast strains expressing wild-type NPA3 (WT) or NPA3 variants with amino acid substitutions in the G1 (K16R), G2 (D40E), and GPN loop (G70A and N72Q) and showing slow growth phenotypes (see Fig. 2C) were analyzed by immunofluorescence with an anti-POLR2A/RPB1 antibody. In each case, the location of the DNA is shown (Hoechst staining). Scale bar, 5 μm. DIC, differential interference contrast.
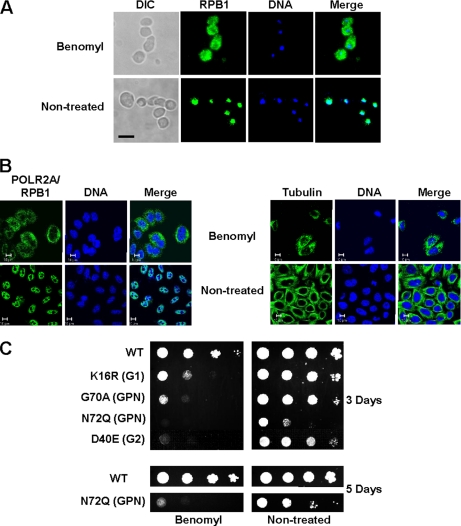
Benomyl, an Inhibitor of Microtubule Assembly, Exacerbates the Growth Phenotype of NPA3 Mutants
Our functional studies indicated that RPAP4/GPN1 is involved in regulating, directly or indirectly, nuclear import of the largest RNAPII subunit, POLR2A/RPB1. At this point, our results do not define the mechanism by which nuclear targeting of POLR2A/RPB1 occurs. However, it is noteworthy that RPAP4/GPN1 specifically connects RNAPII subunits to the CCT complex. The CCT complex is responsible for polymerization of tubulins into microtubules, a process that has been shown to require the action of prefoldins as cofactors, as well as the assembly/folding of many other protein complexes (28, 29). Recently, Dekker et al. (39) reported the association of CCT with RNAPII subunits and provided evidence that RNAPII is not a substrate for CCT, arguing against a direct role for the CCT complex in the assembly of RNAPII. A role for the CCT complex remains possible, however, in the nuclear import of RNAPII subunits in conjunction with RPAP4/GPN1. For example, a mechanism by which CCT couples polymerization of tubulins with POLR2A/RPB1 transport is very attractive and has been reported for other nuclear proteins (38). Our results indicate that treatment of yeast (Fig. 6A) and human (Fig. 6B) cells with concentrations of benomyl that promote depolarization of microtubules (see Fig. 6B, right panel, which shows the effect of high benomyl concentrations on microtubule integrity) led to the accumulation of POLR2A/RPB1 in the cell cytoplasm. Interestingly, yeast strains expressing NPA3 variants with amino acid substitutions conferring a slow growth phenotype (see Fig. 2B) were hypersensitive to benomyl (40); their growth was completely inhibited by sublethal concentrations of benomyl that did not affect microtubule assembly (Fig. 6C). These results indicate that microtubule assembly/integrity and RPAP4/GPN1 function may be coupled.
Fig. 6.
RPAP4/GPN1 function in POLR2A/RPB1 nuclear import is coupled to microtubule assembly. A, treatment of yeast cells (WT strain) with benomyl (60 μg/ml) leads to accumulation of POLR2A/RPB1 in the cytoplasm. B, treatment of HeLa cells with benomyl at concentrations (30 μg/ml) that produce microtubule dismantling (right panel) leads to accumulation of POLR2A/RPB1 in the cytoplasm (left panel). C, NPA3 mutant yeast strains with a slow growth phenotype are hypersensitive to the microtubule assembly inhibitor benomyl. Yeast strains expressing NPA3 variants with amino acid substitutions in the G1 (K16R), G2 (D40E), and GPN loop (G70A and N72Q) and showing slow growth phenotypes (see Fig. 2B) were grown in the absence or the presence of sublethal concentrations of benomyl (20 μg/ml). DIC, differential interference contrast.
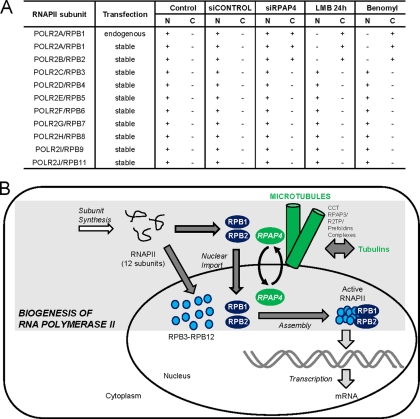
Silencing of RPAP4/GPN1 Also Interferes with Nuclear Import of POLR2B/RPB2, the Second Largest Subunit of RNAPII, but Not the Smaller Enzyme Subunits
To address the effect of RPAP4/GPN1 silencing on nuclear import of the other RNAPII subunits, we performed siRNA silencing experiments in cell lines independently expressing a FLAG-tagged version of 10 RNAPII subunits (POLR2A/RPB1–9 and POLR2J/RPB11). As expected, FLAG-POLR2A/RPB1 accumulated in the cytoplasm upon RPAP4 silencing (Fig. 7A). To our surprise, however, the data indicate that POLR2B/RPB2, the second largest RNAPII subunit, also accumulated in the cytoplasm upon RPAP4/GPN1 silencing (Fig. 7A and supplemental Fig. S4). This effect was not observed for the other RNAPII subunits. Notably, FLAG-POLR2B/RPB2 also accumulated in the cytoplasm upon leptomycin B and benomyl treatment. These results suggest that RNAPII subunits are imported in the nucleus through at least two distinct mechanisms, one involving RPAP4/GPN1 and microtubule assembly for its two largest subunits and a second in which RPAP4/GPN1 and microtubule assembly are dispensable.
Fig. 7.
Nuclear import of POLR2A/RPB1 and POLR2B/RPB2 subunits of RNAPII requires RPAP4/GPN1 and microtubule assembly. A, table showing that RPAP4/GPN1 silencing and LMB and benomyl treatments result in the accumulation of POLR2A/RPB1 and POLR2B/RPB2 in the cytoplasm (C) of treated cells, whereas all the other tested RNAPII subunits retained a nuclear (N) localization upon treatment. B, model for the role of RPAP4/GPN1 during RNAPII biogenesis. Biogenesis of RNAPII requires the synthesis of its 12 subunits (POLR2A/RPB1–POLR2K/RPB12) in the cytoplasm. POLR2A/RPB1 and POLR2B/RPB2, the two largest RNAPII subunits, are imported to the nucleus through a mechanism that requires RPAP4/GPN1 shuttling and microtubule assembly in a process previously shown to involve CCT/chaperonin and prefoldin complexes. Nuclear import of other RNAPII subunits proceeds through alternative mechanisms. This finding implies that the assembly of the 12-subunit active RNAPII occurs in the nuclear space following subunit import.
DISCUSSION
Biogenesis of nuclear RNAPs is an important process as these three molecular machines decode the information contained in eukaryotic genomes by synthesizing RNA molecules. RNAPII synthesizes all mRNAs and some small nuclear RNAs. Although many factors that regulate RNAPII activity during transcription have been identified and characterized, little is known about the machinery that controls its biogenesis, a process that delivers active enzymes to sites of transcription in the nucleus. To start elucidating the molecular mechanisms of RNAPII biogenesis, we reasoned that a survey of protein complexes that associate with RNAPII in the cellular soluble fraction, which is likely to be enriched in factors that control pre- or post-transcriptional stages of the life cycle of the enzyme, would reveal the identity of such factors. We elected to use AP-MS to perform a systematic survey of these complexes and to map the interaction networks of RNAPII present in the cellular soluble fraction. As shown in Fig. 1, this network links RNAPII to a number of previously characterized proteins with known or predicted functions in protein complex assembly, including the CCT chaperonin complex and the prefoldins. The network also revealed connections with previously uncharacterized (or poorly characterized) proteins (see below for details). Because the data set used to draw this protein interaction map has been filtered to minimize the rate of false positives (estimated below 12%), we propose that the network represented in Fig. 1B comprises many factors that regulate the fate of RNAPII before and/or after its involvement in active transcription on genomic DNA. We also estimate that some additional components of this regulatory network are absent in Fig. 1B because our computational approach was tuned to minimize false positives to the detriment of false negatives. Many of the factors that were not included in Fig. 1 (to ensure a minimal number of false positives) are included in supplemental Table S1.
Our AP-MS analysis of the RNAPII transcription machinery reveals a network of protein complexes that control the biogenesis of this essential enzyme. This conclusion is supported by two main findings. First, as mentioned above, the set of proteins that is connected to RNAPII with a high degree of confidence is significantly enriched in proteins previously shown to function in protein complex assembly and in proteins with sequence features found in factors involved in protein complex assembly (the armadillo domain of RPAP1 and the tetratricopeptide repeat domains of RPAP3 are examples) (41–44). Second, our functional analysis of components of this network indicates that they play a role in the nuclear import and/or assembly of RNAPII before its involvement in transcription per se (see below and data not shown). Such enrichment in specific chaperones is unique to this part of the network and not obtained using other tagged proteins (a total of 77 tagged proteins have now been used in our experiments), indicating that it cannot merely be the result of improper folding of the expressed tagged proteins.
The first component of this network to draw our attention because of its central location at the interface of RNAPII and other protein complexes known to participate in protein complex formation, assembly, and/or folding was the GTPase RPAP4/GPN1. Modifying the steady-state level of RPAP4/GPN1 or introducing amino acid substitutions into the RPAP4/GPN1 GTP-binding or GPN loop motifs dramatically affected the cellular localization of RNAPII and promoted its cytoplasmic accumulation (see Figs. 3 and 5). Notably, reduced RPAP4/GPN1 activity also resulted in growth arrest and cell death (see Fig. 2 and data not shown) likely as a consequence of a blockade in the nuclear import of POLR2A/RPB1 and POLR2B/RPB2 and the resulting deficiency in RNAPII assembly.
Our results also indicate that RPAP4/GPN1 shuttles between the nucleus and the cytoplasm as treatment with LMB, a specific inhibitor of nuclear export mediated by the XPO1/CRM1 protein (45, 46), resulted in sequestration of RPAP4/GPN1 in the nuclear compartment (see Fig. 4). Notably, our protein-protein interaction network revealed that XPO1/CRM1 associates with RPAP4/GPN1 (see Fig. 1B), supporting the notion that RPAP4/GPN1 is transported from the nucleus to the cytoplasm in an XPO1/CRM1-dependent manner under normal conditions. The observation that a blockade in RPAP4/GPN1 nuclear export resulted in the accumulation of POLR2A/RPB1 and POLR2B/RPB2 in the cytoplasm suggests that the two processes are coupled.
The nuclear import mechanisms of proteins that contain nuclear localization signals (NLSs) were shown to involve NLS recognition by cytoplasmic importins, translocation to the nucleus via the nuclear pore complex, and release through the action of the GTPase Ran and its associated factors and cofactors (37, 38). Binding of importins to NLS sequences is mediated by the α subunit of importin α/β dimers. Of note, KPNA2, an α-importin (47), is present in our high confidence interaction data set (see Fig. 1B). As examination of the POLR2A/RPB1 and POLR2B/RPB2 amino acid sequences did not reveal the presence of an NLS, it is possible that the role of RPAP4/GPN1 is to mediate the interaction of POLR2A/RPB1, POLR2B/RPB2, and import factors.
A number of proteins were shown to be imported to the nucleus through cytoskeletal transport processes (38). Transcription factors such as p53 (48, 49), Rb (50), and the parathyroid hormone-related protein (51) are imported through a mechanism involving microtubule assembly. As the CCT (28, 29) and RPAP3/R2TP/prefoldin-like complexes (30, 31) contain chaperones previously shown to specialize in microtubule formation, we suspected that POLR2A/RPB1 and POLR2B/RPB2 nuclear import also proceeds through a mechanism involving microtubule assembly. This notion gained further support from our experiment showing that (i) yeast and human cells treated with concentrations of benomyl that produce microtubule dismantling accumulated POLR2A/RPB1 in the cytoplasm and (ii) yeast strains expressing NPA3 variants with non-lethal amino acid substitutions were hypersensitive to benomyl (see Fig. 6). Of note, human cells treated with a RPAP4/GPN1-specific siRNA and yeast cells with slow growth phenotypes resulting from mutations in the NPA3 gene presented altered morphologies, supporting the notion that the organization of microtubules is altered in cells that accumulate POLR2A/RPB1 in the cytoplasm (see Fig. 5 and data not shown). Whether the CCT complex is directly involved in nuclear import of RNAPII subunits cannot be inferred from our results.
In conclusion, our work identified a number of proteins that are components of the RNAP biogenesis machinery. We show here that RPAP4/GPN1 is an RNAPII-associated protein that is essential for cell growth in eukaryotes and a member of a new family of GTPases that requires both an intact GTP-binding domain and a GPN loop element for activity. Changes in the expression level or integrity of RPAP4/GPN1 in yeast and human cells interfere with nuclear import of the RNAPII largest subunits POLR2A/RPB1 and POLR2B/RPB2, a key step in RNAP biogenesis (see Fig. 7B for a model). The functional characterization of other RPAPs and their interacting partners should help to further define other aspects of the mechanism of nuclear RNAP biogenesis in eukaryotes.
Supplementary Material
Acknowledgments
We are grateful to the members of our laboratories and Jacques Archambault for helpful discussions and comments.
Footnotes
* This work was supported by grants from the Canadian Institutes for Health Research (CIHR), Genome Canada, Génome Québec, the Natural Sciences and Engineering Research Council of Canada (NSERC), and the Canadian Foundation for Innovation.
 This article contains supplemental Figs. S1–S4 and Tables S1–S5.
This article contains supplemental Figs. S1–S4 and Tables S1–S5.
2 D. Forget, A.-A. Lacombe, P. Cloutier, G. Vattemi, and B. Coulombe, manuscript in preparation.
1 The abbreviations used are:
- RNAP
- RNA polymerase
- AP
- affinity purification
- CCT
- chaperonin containing TCP-1
- IR
- interaction reliability
- LMB
- leptomycin B
- NES
- nuclear export signal
- NLS
- nuclear localization signal
- RPAP
- RNAPII-associated protein
- TAP
- tandem affinity purification
- Yc
- yeast complete medium
- 5FOA
- 5-fluoroorotic acid.
REFERENCES
- 1.Ptashne M., Gann A. (1997) Transcriptional activation by recruitment. Nature 386, 569–577 [DOI] [PubMed] [Google Scholar]
- 2.Carey M. (1998) The enhanceosome and transcriptional synergy. Cell 92, 5–8 [DOI] [PubMed] [Google Scholar]
- 3.Tjian R., Maniatis T. (1994) Transcriptional activation: a complex puzzle with few easy pieces. Cell 77, 5–8 [DOI] [PubMed] [Google Scholar]
- 4.Triezenberg S. J. (1995) Structure and function of transcriptional activation domains. Curr. Opin. Genet. Dev. 5, 190–196 [DOI] [PubMed] [Google Scholar]
- 5.Orphanides G., Lagrange T., Reinberg D. (1996) The general transcription factors of RNA polymerase II. Genes Dev. 10, 2657–2683 [DOI] [PubMed] [Google Scholar]
- 6.Hampsey M. (1998) Molecular genetics of the RNA polymerase II general transcriptional machinery. Microbiol. Mol. Biol. Rev. 62, 465–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conaway R. C., Conaway J. W. (1997) General transcription factors for RNA polymerase II. Prog. Nucleic Acid Res. Mol. Biol. 56, 327–346 [DOI] [PubMed] [Google Scholar]
- 8.Coulombe B., Burton Z. F. (1999) DNA bending and wrapping around RNA polymerase: a “revolutionary” model describing transcriptional mechanisms. Microbiol. Mol. Biol. Rev. 63, 457–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roeder R. G. (2005) Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett. 579, 909–915 [DOI] [PubMed] [Google Scholar]
- 10.Marr M. T., 2nd, Isogai Y., Wright K. J., Tjian R. (2006) Coactivator cross-talk specifies transcriptional output. Genes Dev. 20, 1458–1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conaway R. C., Sato S., Tomomori-Sato C., Yao T., Conaway J. W. (2005) The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem. Sci. 30, 250–255 [DOI] [PubMed] [Google Scholar]
- 12.Kornberg R. D. (2005) Mediator and the mechanism of transcriptional activation. Trends Biochem. Sci. 30, 235–239 [DOI] [PubMed] [Google Scholar]
- 13.Kornberg R. D., Lorch Y. (1999) Chromatin-modifying and -remodeling complexes. Curr. Opin. Genet. Dev. 9, 148–151 [DOI] [PubMed] [Google Scholar]
- 14.Orphanides G., Reinberg D. (2000) RNA polymerase II elongation through chromatin. Nature 407, 471–475 [DOI] [PubMed] [Google Scholar]
- 15.Li B., Carey M., Workman J. L. (2007) The role of chromatin during transcription. Cell 128, 707–719 [DOI] [PubMed] [Google Scholar]
- 16.Hardeland U., Hurt E. (2006) Coordinated nuclear import of RNA polymerase III subunits. Traffic 7, 465–473 [DOI] [PubMed] [Google Scholar]
- 17.Zeghouf M., Li J., Butland G., Borkowska A., Canadien V., Richards D., Beattie B., Emili A., Greenblatt J. F. (2004) Sequential Peptide Affinity (SPA) system for the identification of mammalian and bacterial protein complexes. J. Proteome Res. 3, 463–468 [DOI] [PubMed] [Google Scholar]
- 18.Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. (2001) The tandem affinity purification (TAP) method: a general procedure of protein complex purification. Methods 24, 218–229 [DOI] [PubMed] [Google Scholar]
- 19.Rigaut G., Shevchenko A., Rutz B., Wilm M., Mann M., Séraphin B. (1999) A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 [DOI] [PubMed] [Google Scholar]
- 20.Jeronimo C., Forget D., Bouchard A., Li Q., Chua G., Poitras C., Thérien C., Bergeron D., Bourassa S., Greenblatt J., Chabot B., Poirier G. G., Hughes T. R., Blanchette M., Price D. H., Coulombe B. (2007) Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 27, 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cloutier P., Al-Khoury R., Lavallée-Adam M., Faubert D., Jiang H., Poitras C., Bouchard A., Forget D., Blanchette M., Coulombe B. (2009) High-resolution mapping of the protein interaction network for the human transcription machinery and affinity purification of RNA polymerase II-associated complexes. Methods 48, 381–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marfatia K. A., Crafton E. B., Green D. M., Corbett A. H. (2003) Domain analysis of the Saccharomyces cerevisiae heterogeneous nuclear ribonucleoprotein, Nab2p. Dissecting the requirements for Nab2p-facilitated poly(A) RNA export. J. Biol. Chem. 278, 6731–6740 [DOI] [PubMed] [Google Scholar]
- 24.Lee T. H., Lwu S., Kim J., Pelletier J. (2002) Inhibition of Wilms tumor 1 transactivation by bone marrow zinc finger 2, a novel transcriptional repressor. J. Biol. Chem. 277, 44826–44837 [DOI] [PubMed] [Google Scholar]
- 25.Gietz R. D., Woods R. A. (2002) Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 350, 87–96 [DOI] [PubMed] [Google Scholar]
- 26.Jeronimo C., Langelier M. F., Zeghouf M., Cojocaru M., Bergeron D., Baali D., Forget D., Mnaimneh S., Davierwala A. P., Pootoolal J., Chandy M., Canadien V., Beattie B. K., Richards D. P., Workman J. L., Hughes T. R., Greenblatt J., Coulombe B. (2004) RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol. Cell. Biol. 24, 7043–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krueger B. J., Jeronimo C., Roy B. B., Bouchard A., Barrandon C., Byers S. A., Searcey C. E., Cooper J. J., Bensaude O., Cohen E. A., Coulombe B., Price D. H. (2008) LARP7 is a stable component of the 7SK snRNP while P-TEFb, HEXIM1 and hnRNP A1 are reversibly associated. Nucleic Acids Res. 36, 2219–2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leroux M. R., Hartl F. U. (2000) Protein folding: versatility of the cytosolic chaperonin TRiC/CCT. Curr. Biol. 10, R260–R264 [DOI] [PubMed] [Google Scholar]
- 29.Martín-Benito J., Boskovic J., Gómez-Puertas P., Carrascosa J. L., Simons C. T., Lewis S. A., Bartolini F., Cowan N. J., Valpuesta J. M. (2002) Structure of eukaryotic prefoldin and of its complexes with unfolded actin and the cytosolic chaperonin CCT. EMBO J. 21, 6377–6386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao R., Kakihara Y., Gribun A., Huen J., Yang G., Khanna M., Costanzo M., Brost R. L., Boone C., Hughes T. R., Yip C. M., Houry W. A. (2008) Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 180, 563–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sardiu M. E., Cai Y., Jin J., Swanson S. K., Conaway R. C., Conaway J. W., Florens L., Washburn M. P. (2008) Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc. Natl. Acad. Sci. U.S.A. 105, 1454–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gstaiger M., Luke B., Hess D., Oakeley E. J., Wirbelauer C., Blondel M., Vigneron M., Peter M., Krek W. (2003) Control of nutrient-sensitive transcription programs by the unconventional prefoldin URI. Science 302, 1208–1212 [DOI] [PubMed] [Google Scholar]
- 33.Nitta M., Saijo M., Kodo N., Matsuda T., Nakatsu Y., Tamai H., Tanaka K. (2000) A novel cytoplasmic GTPase XAB1 interacts with DNA repair protein XPA. Nucleic Acids Res. 28, 4212–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lembo F., Pero R., Angrisano T., Vitiello C., Iuliano R., Bruni C. B., Chiariotti L. (2003) MBDin, a novel MBD2-interacting protein, relieves MBD2 repression potential and reactivates transcription from methylated promoters. Mol. Cell. Biol. 23, 1656–1665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gras S., Chaumont V., Fernandez B., Carpentier P., Charrier-Savournin F., Schmitt S., Pineau C., Flament D., Hecker A., Forterre P., Armengaud J., Housset D. (2007) Structural insights into a new homodimeric self-activated GTPase family. EMBO Rep. 8, 569–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dez C., Froment C., Noaillac-Depeyre J., Monsarrat B., Caizergues-Ferrer M., Henry Y. (2004) Npa1p, a component of very early pre-60S ribosomal particles, associates with a subset of small nucleolar RNPs required for peptidyl transferase center modification. Mol. Cell. Biol. 24, 6324–6337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorokin A. V., Kim E. R., Ovchinnikov L. P. (2007) Nucleocytoplasmic transport of proteins. Biochemistry 72, 1439–1457 [DOI] [PubMed] [Google Scholar]
- 38.Pouton C. W., Wagstaff K. M., Roth D. M., Moseley G. W., Jans D. A. (2007) Targeted delivery to the nucleus. Adv. Drug Deliv. Rev. 59, 698–717 [DOI] [PubMed] [Google Scholar]
- 39.Dekker C., Stirling P. C., McCormack E. A., Filmore H., Paul A., Brost R. L., Costanzo M., Boone C., Leroux M. R., Willison K. R. (2008) The interaction network of the chaperonin CCT. EMBO J. 27, 1827–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Machin N. A., Lee J. M., Barnes G. (1995) Microtubule stability in budding yeast: characterization and dosage suppression of a benomyl-dependent tubulin mutant. Mol. Biol. Cell 6, 1241–1259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Coates J. C. (2003) Armadillo repeat proteins: beyond the animal kingdom. Trends Cell Biol. 13, 463–471 [DOI] [PubMed] [Google Scholar]
- 42.Hatzfeld M. (1999) The armadillo family of structural proteins. Int. Rev. Cytol. 186, 179–224 [DOI] [PubMed] [Google Scholar]
- 43.Cliff M. J., Williams M. A., Brooke-Smith J., Barford D., Ladbury J. E. (2005) Molecular recognition via coupled folding and binding in a TPR domain. J. Mol. Biol. 346, 717–732 [DOI] [PubMed] [Google Scholar]
- 44.Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 45.Arnaoutov A., Azuma Y., Ribbeck K., Joseph J., Boyarchuk Y., Karpova T., McNally J., Dasso M. (2005) Crm1 is a mitotic effector of Ran-GTP in somatic cells. Nat. Cell Biol. 7, 626–632 [DOI] [PubMed] [Google Scholar]
- 46.Paraskeva E., Izaurralde E., Bischoff F. R., Huber J., Kutay U., Hartmann E., Lührmann R., Görlich D. (1999) CRM1-mediated recycling of snurportin 1 to the cytoplasm. J. Cell Biol. 145, 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tseng S. F., Chang C. Y., Wu K. J., Teng S. C. (2005) Importin KPNA2 is required for proper nuclear localization and multiple functions of NBS1. J. Biol. Chem. 280, 39594–39600 [DOI] [PubMed] [Google Scholar]
- 48.Giannakakou P., Sackett D. L., Ward Y., Webster K. R., Blagosklonny M. V., Fojo T. (2000) p53 is associated with cellular microtubules and is transported to the nucleus by dynein. Nat. Cell Biol. 2, 709–717 [DOI] [PubMed] [Google Scholar]
- 49.Giannakakou P., Nakano M., Nicolaou K. C., O'Brate A., Yu J., Blagosklonny M. V., Greber U. F., Fojo T. (2002) Enhanced microtubule-dependent trafficking and p53 nuclear accumulation by suppression of microtubule dynamics. Proc. Natl. Acad. Sci. U.S.A. 99, 10855–10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Roth D. M., Moseley G. W., Glover D., Pouton C. W., Jans D. A. (2007) A microtubule-facilitated nuclear import pathway for cancer regulatory proteins. Traffic 8, 673–686 [DOI] [PubMed] [Google Scholar]
- 51.Lam M. H., Thomas R. J., Loveland K. L., Schilders S., Gu M., Martin T. J., Gillespie M. T., Jans D. A. (2002) Nuclear transport of parathyroid hormone (PTH)-related protein is dependent on microtubules. Mol. Endocrinol. 16, 390–401 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.