Abstract
Gaucher disease (GD), the most common lysosomal storage disorder of humans, is caused by mutations in the gene coding for the enzyme glucocerebrosidase (GCase). Clinical manifestations vary among patients with the three types of GD, and phenotypic heterogeneity occurs even among patients with identical mutations. To gain insight into why phenotypic heterogeneity occurs in GD, we investigated mechanisms underlying the net loss of GCase catalytic activity in cultured skin fibroblasts derived from patients with the three types of GD. The findings indicate that the loss of catalytic activity of GCase correlates with its quantitative reduction, rather than a decrease in functional capacity of mutant enzyme. Use of a proteasome inhibitor, lactacystin, resulted in increased expression of GCase, suggesting a mechanism of protein degradation in GD. Furthermore, reduced binding of GCase to TCP1 ring complex (TRiC), a regulator of correct protein folding, may result in defective maturation of nascent GCase in GD cells. Additionally, increased interaction between GCase and c-Cbl, an E3 ubiquitin ligase, may be involved in the degradation and loss of GCase in GD. The findings suggest that specific molecular mediators involved in GCase maturation and degradation could be responsible for phenotypic variation among patients with the same genotypes and that these mediators could be therapeutically targeted to increase GCase activity in patients with GD.
Gaucher disease (GD), the most prevalent hereditary metabolic storage disorder of humans, is caused by a deficiency of the enzyme glucocerebrosidase (GCase) (1, 2). Typical manifestations of GD include hepatosplenomegaly, cytopenias, bone disease, and, in some patients, CNS involvement (3). Patients without CNS signs are classified as type 1 GD [Online Mendelian Inheritance in Man (OMIM) 230800], whereas those with CNS signs are categorized as either type 2 acute infantile GD (OMIM 230900) or type 3 GD with juvenile or adult onset of neurological involvement (OMIM 231000). A confounding issue is the phenotypic heterogeneity associated with identical GD genotypes. The phenotype of GD patients with identical genotypes varies significantly, sometimes spanning all three phenotypes (4). Sidransky et al. reported that the five most common GCase mutations occurred in both type 1 and type 3 patients with GD (5). Similarly, in an analysis of more than 300 mutant alleles associated with either type 1 or type 3 GD, Koprivica et al. found an inconsistent genotype–phenotype correlation with multiple alleles traditionally associated with distinct clinical presentations (6). In a systematic evaluation of the in vitro kinetics of 52 GCase mutants responsible for different subtypes of GD, Liou et al. discovered that several GCase mutants found in type 2 GD had increased enzymatic activity compared with those of type 3 GD (7). Further evidence indicating inconsistent correlation between phenotype and genotype in GD is apparent from studies demonstrating variability in the clinical manifestations between siblings (8–12).
To understand genotype–phenotype variability in GD, mechanisms potentially involved in the reduction of GCase activity were investigated in cultured skin fibroblasts derived from patients with GD. Specifically, we investigated whether variable reduction of GCase catalytic activity is caused by alterations of the amino acid sequence of the enzyme or whether GCase activity was variably reduced because of alterations in the quantity of the enzyme within cells. To resolve this uncertainty, both enzymatic activity and expression of GCase protein were evaluated in fibroblasts derived from patients with types 1, 2, and 3 GD. Based on the results from these studies, diminished cellular GCase activity was found to correlate closely with decreases in the quantity of GCase protein and GD phenotype.
Based on these findings, the effect of inhibiting protein degradation with the proteasome inhibitor lactacystin was investigated. Inclusion of this agent in the incubation medium caused an increase in the quantity of GCase protein in cells derived from patients with GD along with increased enzymatic activity. To determine whether proteomic differences occurred in GD, 2D-PAGE analysis of proteins was carried out after immunoprecipitation of GCase. This survey provided a number of candidate molecules that might be involved in cellular processing of mutant GCase, among which were TCP1, a subunit of the TCP1 ring complex (TRiC) chaperonin complex, and c-Cbl, an E3 ubiquitin ligase. The relationship of these mediators of intracellular proteasome quality control to the quantity of cellular GCase was then explored. These studies revealed that the intracellular proteasome quality-control system is a key process by which GCase degradation occurs and indicated that it is involved in the pathogenesis of GD. Manipulation of this pathway led to increased expression of mutated, yet catalytically competent, GCase.
Results
Cellular Levels of GCase Vary in GD Subtypes and Correlate with the Deficit of Enzymatic Activity.
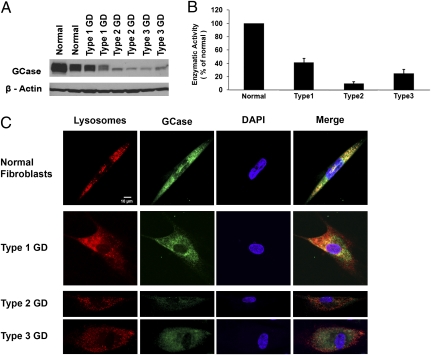
We sought to determine whether the clinical manifestations in GD were associated with reduced levels of GCase protein compared with the amount in normal individuals. Fibroblast cell lines were obtained from four type 1, three type 2, four type 3 GD patients, and four normal individuals (Table 1). Semiquantitative analysis of intracellular GCase levels by SDS/PAGE revealed marked heterogeneity of GCase levels between GD phenotypes and within genotypes (Fig. 1A). The level of GCase protein in fibroblasts derived from patients with type 1 GD was 40.5 ± 5.8% (SD) of that in normal individuals, 11.3 ± 6.9% in type 2 GD, and 21.5 ± 10.2% in type 3 GD. GCase catalytic activity correlated with decreased protein expression. Type 1 GD cells had 41.8 ± 5.5% of GCase activity of normal cells, type 2 GD cells had 9.9 ± 2.4%, and type 3 GD cells had 24.7 ± 5.8% (Fig. 1B). Immunofluorescent staining revealed decreased GCase in GD patient fibroblasts as well as reduced localization of the enzyme within lysosomes in these cells (Fig. 1C), in concordance with results from previous studies (13, 14).
Table 1.
GD cell lines
| Name | Type | Genotype | Obtained From |
| NML | Normal | WT | DMNB, NINDS, NIH |
| NML | Normal | WT | DMNB, NINDS, NIH |
| NML | Normal | WT | DMNB, NINDS, NIH |
| NML | Normal | WT | DMNB, NINDS, NIH |
| DMN 00.41 | Type 1 | N370S/N370S | DMNB, NINDS, NIH |
| DMN 87.30 | Type 1 | N370S/N370S | DMNB, NINDS, NIH |
| DMN 88.17 | Type 1 | N370S/N370S | DMNB, NINDS, NIH |
| DMN 95.46 | Type 1 | N370S/N370S | DMNB, NINDS, NIH |
| GM 00877 | Type 2 | L444P/L444P | Coriell Institute |
| GM 08760 | Type 2 | L444P/L444P | Coriell Institute |
| GM012860A | Type 2 | L444P/L444P | Coriell Institute |
| DMN 84.58 | Type 3 | L444P/L444P | DMNB, NINDS, NIH |
| DMN 86.2 | Type 3 | L444P/L444P | DMNB, NINDS, NIH |
| DMN 87.33 | Type 3 | L444P/L444P | DMNB, NINDS, NIH |
| DMN 89.62 | Type 3 | L444P/L444P | DMNB, NINDS, NIH |
DMNB, Developmental and Metabolic Neurology Branch; NIH, National Institutes of Health; NINDS, National Institute of Neurological Disorders and Stroke.
Fig. 1.
Quantification of GCase expression levels and measurement of enzyme activity in GD phenotypes. (A) Western blot of expression of GCase protein in cultured fibroblast cell lines derived from normal individuals and patients with GD. (B) Enzyme activity assay for GCase using equivalent amount of total protein from normal fibroblasts and GD phenotypes with results plotted as a ratio to the average enzymatic activity of normal fibroblasts. (C) Immunofluorescence staining for lysosomes (red) and GCase (green) in cultured fibroblasts derived from patients with GD and normal individuals. Decreased intensity of staining of GCase as well as reduced localization of GCase in lysosomes is apparent in types 2 and 3 GD fibroblasts compared with that in normal cells. The nuclei of cells are counterstained with DAPI (blue).
N370S and L444P Mutations Have Minimal Impact on the Catalytic Activity of GCase.
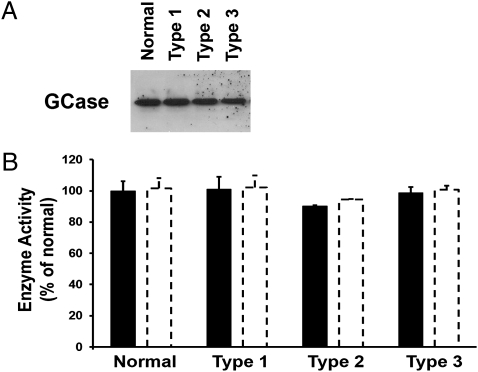
To ascertain the effects of N370S and L444P mutations on the catalytic activity of GCase without the confounding effect of the quantity of enzyme, GCase was immunoprecipitated from each cell line. GCase activity was measured after normalization to the amount of immunoprecipitated GCase protein. These experiments revealed that there was no significant difference in the catalytic activity of GCase isolated from cells derived from patients with the most prevalent mutations of GD, N370S/N370S and L444P/L444P, compared with the normal enzyme (Fig. 2).
Fig. 2.
Measurement of catalytic activity of immunoprecipitated GCase from normal and GD fibroblasts. (A) Western blot of immunoprecipitated GCase demonstrating similar quantities of GCase in normal and GD and cell lines. (B) Catalytic activity of immunoprecipitated GCase from normal and GD fibroblasts measured at concentrations of 50 ng (solid bars) and 100 ng (open bars) per assay. GCase activity from each GD phenotype is plotted as a percentage of the average normal value.
Reduction of Proteasome Activity with Lactacystin Increases Cellular Levels of GCase Activity.
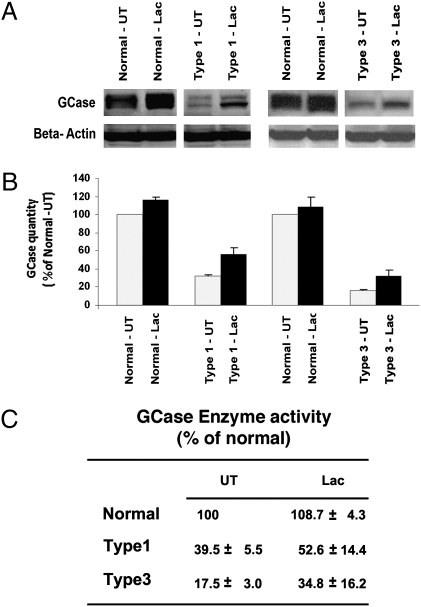
Previous studies have shown that many GD mutations lead to a delay in the exit of nascent GCase peptides from the endoplasmic reticulum (ER), subsequently inducing their degradation by the ER-associated proteasome system (15, 16). To assess whether increased proteasomal degradation occurs in GD, types 1 and 3 GD cells and normal cells were grown in the presence of 5 μM of the proteasomal inhibitor lactacystin (17) for 24 h, and the resultant GCase protein level was compared with that of untreated cells by using Western blotting. Treatment with lactacystin was associated with an increase in GCase levels in both types 1 and 3 GD cells and a small increase of GCase levels in control cells (Fig. 3 A and B). Increases in GCase catalytic activity correlated with increases in the quantity of GCase protein. GCase activity in type 1 GD increased from 39.5% to 52.6% of that in normal cells and in type 3 GD cells from 17.5% to 34.8% normal cells (Fig. 3C).
Fig. 3.
Effect of lactacystin on the expression of GCase and net enzymatic activity in control and types 1 and 3 GD fibroblasts. (A) Western blot analysis of GCase in normal and GD fibroblasts. Lac, lactacystin-treated samples; UT, untreated samples. (B) Quantification of GCase protein by densitometric analysis of Western blots. (C) GCase catalytic in normal and types 1 and 3 GD fibroblasts with (Lac) and without (UT) lactacystin (Lac) in the culture medium.
Loss of Association of Subunit TCP1 of the Molecular Chaperonin TRiC with GCase in GD.
2D-PAGE was used to identify binding partners of immunoprecipitated GCase from GD and normal cell lines. TCP1, the α-subunit of the TRiC chaperonin, was found to have altered binding with GCase in GD fibroblasts (Fig. 4A). Analysis of immunoprecipitated GCase from GD cells demonstrated minimal to no binding with TCP1 in all GD phenotypes using equivalent amounts of immunoprecipitated GCase (Fig. 4B). Similarly, when GCase was analyzed by TCP1 immunoprecipitation from GD cell lysates, binding affinity between the two proteins was reduced in GD cells compared with that of normal fibroblasts (Fig. 4C).
Fig. 4.
2D-PAGE proteomic analysis of immunoprecipitated (IP) GCase and Western blot analysis of GCase and immunoprecipitated TCP1 from normal and GD fibroblasts. (A) Analysis of binding partners from immunoprecipitated GCase in normal and types 1, 2, and 3 GD cells (Upper, left to right). Arrows indicate TCP1. (Lower) Western blot using GCase antibody on normal and types 1, 2, and 3 GD cells with GCase-immunoprecipitated proteins. (B Upper) Western blot using TCP1 antibody on GCase- immunoprecipitated proteins from GD and normal fibroblasts. (Lower) Western blot using GCase antibody on the same proteins. (C) Western blot from TCP1-immunoprecipitated proteins. (Upper) Western blot using GCase antibody on TCP1-immunoprecipitated proteins from GD and normal fibroblasts. (Lower) Western blot using TCP1 antibody on same proteins.
Interaction of c-Cbl with Mutant GCase.
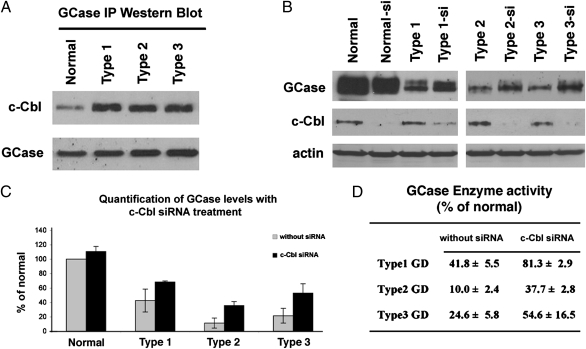
c-Cbl is a member of the highly diverse class of E3 ubiquitin ligases. Immunoprecipitation analysis revealed increased binding of c-Cbl to GCase in all GD phenotypes compared with that in normal fibroblasts (Fig. 5A).
Fig. 5.
siRNA knockdown of c-Cbl and quantification of GCase expression levels. (A) Western blot from GCase-immunoprecipitated (IP) proteins. (Upper) Western blot using c-Cbl antibody on GCase-immunoprecipitated proteins from normal and GD fibroblasts. (Lower) Western blot using GCase antibody on the same proteins. (B Top) Western blot using GCase antibody on normal fibroblasts (control) and from patients with GD before and after siRNA knockdown of c-Cbl. (Middle) Western blot using c-Cbl antibody on control and GD fibroblasts before and after siRNA knockdown of c-Cbl. (Bottom) Western blot using actin antibody on GD and normal fibroblasts. (C) Densitometric measurement of GCase expression levels before (gray bars) and after (black bars) siRNA knockdown of c-Cbl. (D) GCase activity in normal and GD fibroblasts before and after siRNA knockdown of c-Cbl shown as a percentage of GCase activity in normal fibroblasts.
Effect of siRNA Knockdown of c-Cbl.
To investigate the specificity of c-Cbl as a principal mediator of the biodegradation of GCase in GD cells, the expression of c-Cbl was suppressed with c-Cbl siRNA in normal fibroblasts and GD cells. After reduction of c-Cbl expression, GCase expression was determined by Western blot analysis. Reduction of c-Cbl caused an increase in the expression of GCase in GD cells (Fig. 5 B and C). GCase activity in GD cells was increased significantly in cells grown in the presence of c-Cbl siRNA (Fig. 5D). Treatment with negative control siRNA caused no change in GCase expression in normal and GD cells.
Discussion
The reduction of GCase activity that occurs in patients with GD could potentially be caused by a decrease in the functional capacity of GCase, or a decrease in expression of the enzyme. To assess the contribution of each of these mechanisms, the quantity of GCase protein and GCase catalytic activity were determined in extracts of cultured skin fibroblasts derived from normal controls and from patients with the three clinical phenotypes of GD. Type 1 GD patients had the N370S/N370S genotype; patients with types 2 and 3 had the L444P/L444P genotype. The investigation revealed that the decrease in catalytic activity of GCase in cultured skin fibroblasts derived from patients with the above genotypes closely paralleled reductions in the quantity of GCase protein. These mutations had little impact on catalytic activity but, rather, triggered a pathway by which the expression of GCase is reduced. Previous reports have suggested that type 1 GD patients express mutant GCase with reduced specific activity, whereas GCase isolated from type 2 and 3 GD patients correlates well with enzyme quantity (18). This discrepancy with our findings in type 1 GD patients may be the result of differing conditions used to assay GCase enzyme activity. For example, a study examining intrinsic enzyme function in GD suggests that the functional capacity of the N370S variant of GCase may be affected by variation of experimental conditions within the assay and, under certain conditions, maintains catalytic competence (14). Moreover, a study examining the x-ray crystallographic structure of GCase variants provides evidence that the N370S mutation in GCase is located in the seventh membrane helix, distant from the catalytic active site of the enzyme (19). These observations are consistent with the idea that the N370S mutation, common to type 1 GD, minimally affects the intrinsic functional capacity of GCase.
The finding that reduction of enzymatic activity in patients with GD that have the most prevalent genetic mutations is caused by a decrease of GCase expression implicates a specific pathway through which inappropriately folded polypeptides are detected and targeted for proteasomal degradation. To explore the concept that increased protein degradation is the mechanism by which net enzymatic activity is decreased in GD, type 1 and type 3 GD fibroblasts were treated with lactacystin that resulted in an elevation of cellular GCase. The increase in the quantity of enzyme was associated with a parallel increase of GCase catalytic activity. This finding suggests that proteasomal degradation of GCase may contribute significantly to GD phenotypes and progression of disease. There is a strong correlation between GCase expression levels and net enzymatic activity. Immunoprecipitation of GCase revealed that intrinsic enzyme activity was not significantly affected across the GD subtypes investigated, and the addition of a proteasome inhibitor increased GCase activity in GD fibroblasts. The results indicate that net decreases in enzyme activity in patients with these genotypes of GD can be attributed to a decrease in enzyme expression rather than to a loss of catalytic function.
To identify mediators involved in proteasomal degradation of GCase, the immunoprecipitated proteins from GD and normal cells were subjected to 2D-PAGE and Western blot analyses. Potential regulators of this process included TCP1 and c-Cbl, which are thought to play important roles in protein folding and maturation (20, 21). TCP1, a subunit of the TRiC chaperonin, is known to function in protein folding and quality control (22). The observation that there is a reduction of the interaction of the TRiC chaperonin with mutated GCase provides a potential mechanism underlying the reduction of these proteins. The TRiC chaperonin is distinguished by its ring-shaped architecture, which includes a central cavity thought to serve as a “folding chamber” for nascent polypeptides. The structure is similar to that of the GroES and GroEL chaperonins in prokaryotes (20, 23–25). c-Cbl, a member of the highly diverse family of E3 ubiquitin ligases, has been shown to play an important role in protein degradation in cells (21, 26). However, little is known about the role of TCP1 and c-Cbl in the pathogenesis of GD. Therefore, potential roles of TCP1 and c-Cbl in nascent protein folding, maturation, and quality control in GD were investigated. Decreased interaction between GCase and TCP1 is consistent with a model in which inappropriately folded GCase is the result of reduced interaction between GCase protein and a specific chaperonin. Expression of TCP1 was found to be normal in the phenotypes of GD we examined. This finding is consistent with the deduction that reduced interaction between GCase and TCP1 is caused by decreased binding, rather than decreased expression, of TCP1 (Fig. 4C).
The findings indicate that proper enzyme maturation is impaired when there is a loss of interaction between GCase and its specific chaperonin. This deduction supports the concept that TCP1/TRiC-mediated folding of proteins may represent a late stage in quality-control pathways governing the maturation of nascent polypeptides, including GCase. Before a nascent protein can enter the ER for modification, it likely interacts with folding mediators to maintain a single-length polypeptide conformation (27). Evidence is presented that interaction with the chaperonin mediator is reduced, which may represent a quality-control checkpoint at which misfolded proteins cannot enter the ER and are instead shuttled to the proteasome degradation pathway.
To analyze the mechanism of degradation of GCase in GD cells caused by decreased interaction with TCP1, a potential role of c-Cbl, a member of the family of E3 ubiquitin ligases, was examined. Increased binding of c-Cbl to mutated GCase was observed while total expression levels of GCase remained at a normal level. These results support a model in which mutated GCase is selectively targeted for the proteasomal degradation pathway and implicates c-Cbl as a mediator of this process. To substantiate the hypothesis that increased c-Cbl binding to GCase in GD cells leads to GCase degradation, siRNA knockdown of c-Cbl was performed. The results showed that GCase activity was significantly increased in GD and normal fibroblasts after siRNA knockdown of c-Cbl. The findings strongly suggest that c-Cbl is one of the mediators of the proteasomal degradative fate of GCase in GD cells and demonstrate an increase in GCase levels with its inhibition. Such enzyme–E3 ligase interaction may be restricted to specific mutations in GD as a similar up-regulation of mutated α-galactosidase A (GLA) was not observed with siRNA knockdown of c-Cbl in fibroblasts derived from patients with Fabry disease (Fig. S1). It has also been suggested that another E3 ligase, parkin, may play a role in the degradation of mutant GCase and thus may explain the uniquely high concurrence of GD and Parkinson disease (28). Although c-Cbl appears to play an important role in the degradation of variant GCase, evidence suggests that there is more than one such mediator of protein quality control (29).
The therapeutic implications of these findings are significant. For example, the use of a small-molecule inhibitor of c-Cbl could increase enzyme function in GD analogous to our knockdown model. Further experiments are needed to address the implications of these findings in an in vivo model. Nevertheless, the observations suggest an important mechanism of enzyme regulation that might be manipulated to treat patients with certain inborn errors of metabolism.
In summary, cellular levels of GCase vary significantly between phenotypes of GD and within the same GD genotype, attributable in part to the sensitivity of mutant GCase to protein quality control and ER-associated proteasome systems. The differences in quantitative levels of GCase appear to underlie decreases in enzymatic activity, whereas intrinsic enzyme function is not significantly affected in the two most prevalent genotypes of patients with GD.
Materials and Methods
Cell Culture and Treatment.
Cell lines were obtained from the Developmental and Metabolic Neurology Branch of the National Institute of Neurological Disorders and Stroke (NINDS) and the Coriell Institute (Table 1). All Type 1 GD cell lines were homozygous for the N370S/N370S mutation. Type 2 and type 3 GD cell lines were homozygous for the L444P/L444p mutation. Two cell lines derived from patients with Fabry disease were obtained from the Developmental and Metabolic Neurology Branch with genotypes A143P and N215S. Cells were cultured in Eagle's minimum essential medium (Invitrogen) supplemented with 10% FBS, penicillin, and streptomycin.
Treatment with Proteasome Inhibitor.
A total of 2.5 × 104 cells were seeded in 24-well plates, allowed to attach for at least 24 h, and then treated with lactacystin (L6785; Sigma-Aldrich) at a concentration of 5 μM for 24 h (17).
Immunofluorescence Analysis.
Cultured cells were plated onto four-well chamber slides and allowed to attach overnight. After attachment, cells were incubated with primary antibodies in a solution of PBS with 1% BSA and 0.1% Triton X-100 at 4 °C overnight. Anti-GCase antibody ab55080 was obtained from Abcam. To stain lysosomes, cells were grown in the presence of 50 nM LysoTracker DND-99 (L-7528; Invitrogen). Immunoreactive GCase was detected with Alexa Fluor 488–tagged secondary antibody (A-11001; Invitrogen). Nuclei of cells were counterstained with DAPI (Sigma-Aldrich). Staining was viewed with a Zeiss LSM 510 confocal microscope.
Western Blotting and Quantification of GCase.
Cell pellets were lysed in T-PER Tissue Protein Extraction Reagent solution (Thermo Fischer Scientific), sonicated, and centrifuged. Protein was determined in the supernatant solution by using the Bio-Rad Protein Assay Kit. Proteins were separated by SDS/PAGE on 4–15% acrylamide gels (Invitrogen) and transferred to nitrocellulose membranes (Invitrogen). Blocking buffer solution (Thermo Fischer Scientific) was used before immunoblotting with primary antibody. Expression of GCase was determined by Western blotting using monoclonal antibody at a dilution of 1:1,000. c-Cbl rabbit antibody, TCP1 rabbit antibody, and GLA rabbit antibody (Cell Signaling) were used at dilutions of 1:1,000. As a loading control, blots were probed with a goat antibody directed against actin (sc-1616; Santa Cruz Biotechnology) at a dilution of 1:5,000. Detection of antibodies was performed with a horseradish peroxidase–conjugated species-specific secondary antibody and an enhanced chemiluminescence system. Densitometric analysis using image software (NIH ImageJ software) was used to quantify the expression of GCase and GLA.
GCase Enzyme Activity Assay.
Fluorometric enzymatic activity assays were performed as described (30). Samples were loaded into a 96-well microtiter plate, and fluorescence was measured with a Victor 3 multilabel counter at an excitation/emission setting of 355 nm/460 nm. One unit of GCase activity was designated as 1 nmol of 4-methylumbelliferone released per hour.
siRNA Knockdown of c-Cbl Expression.
GD and Fabry disease cell lines and normal human fibroblast cell lines were plated in six-well plates (50–60% confluency) with Opti-MEM medium (Invitrogen). siRNA transfection was performed as described (31). The target sequence specific for c-Cbl was (RNA) AGA GCU CAA AUG UGG AGU CCA UGG C (Invitrogen). siRNA for negative control was also used (Invitrogen). The cells were collected 48 h after transfection and used for Western blotting and analysis of GCase activity.
Immunoprecipitation.
Whole-cell lysates were prepared from GD cell lines. Supernatant solutions (300 μg of protein) were incubated with monoclonal antibodies against GCase, TCP1, and c-Cbl according to an indirect immunoprecipitation kit protocol (Millipore). The preimmune complex was incubated and shaken at room temperature for 2 h. Immunoprecipitation products were used for Western blotting and 2D-gel analysis.
2D-PAGE.
Total protein extracts and immunoprecipitated proteins from normal and types 1, 2, and 3 GD fibroblast lysates were analyzed. Isometric focusing for the 1D electrophoresis was done with a Multiphore II Electrophoresis System (GE Healthcare). The strips were subjected to voltages ranging from 300 to 3,500 V. Immobilized pH gradient (IPG) strips were equilibrated with buffer solution 1 containing 6 mol/L urea, 2% SDS, 375 mmol/L Tris·HCl (pH 8.8), 20% glycerol, and 2% (wt/vol) DTT followed by buffer solution 2 containing 6 mol/L urea, 2% SDS, 375 mmol/L Tris·HCl (pH 8.8), 20% glycerol, and 2.5% (wt/vol) iodoacetamide (Bio-Rad). Precast ExcelGel SDS gels (12–14% gradient gel, pH 4–7, 245 × 180 × 0.5 mm (GE Healthcare) were used for the 2D protein separation by Multiphor II Flated System at 700 V (32). Silver staining was used to detect proteins according to the manufacturer's instructions (GE Healthcare).
Mass Spectrometry.
Uniquely expressed proteins from 2D-PAGE were used for in-gel digestion and were analyzed by liquid chromatography/tandem mass spectrometry (LC-MS/MS) using a ProteomeX LC/MS system (ThermoElectron) operated in the high-throughput mode. Reverse-phase HPLCy was carried out by using a BioBasic-18 column (0.18 150 mm; ThermoElectron) eluted at 1–2 L/min of solution A. Mobile phase A was H2O (0.1% formic acid), and mobile phase B was CH3CN (0.1% formic acid). The effluent from the column was analyzed on the LCQ Deca XP Plus (ThermoElectron) operating in the “Top Five” mode.
Protein Identification.
Uninterpreted MS/MS spectra were searched against a human database by using the BioWorks and SEQUEST programs (ThermoElectron). Protein identification was accepted when the MS/MS spectra of at least two peptides from the same protein exhibited the default Xcorr versus charge values set by the program (for Z = 1, 1.50; for Z = 2, 2.00; for Z = 3, 2.50) at a minimum.
Supplementary Material
Acknowledgments
We thank Dr. Paul E. Gallant of the NINDS Light Microscopy Facility for his assistance with confocal microscopy and Dr. Howard Jaffe of the National Institute of Neurological Disorders and Stroke (NINDS) Protein/Peptide Sequencing Facility for his assistance with protein identification. This research was supported by the Intramural Research Program of the NINDS and the National Cancer Institute at the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1014376107/-/DCSupplemental.
References
- 1.Brady RO, Kanfer JN, Shapiro D. Metabolism of glucocerebrosides II. Evidence of an enzymatic deficiency in Gaucher's disease. Biochem Biophys Res Commun. 1965;18:221–225. doi: 10.1016/0006-291x(65)90743-6. [DOI] [PubMed] [Google Scholar]
- 2.Brady RO, Kanfer JN, Bradley RM, Shapiro D. Demonstration of a deficiency of glucocerebroside-cleaving enzyme in Gaucher's disease. J Clin Invest. 1966;45:1112–1115. doi: 10.1172/JCI105417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutler E, Grabowski GA. In: The Metabolic & Molecular Bases of Inherited Disease. 8th Ed. Scriver CR, editor. New York: McGraw-Hill; 2001. pp. 3635–3668. [Google Scholar]
- 4.Charrow J, et al. The Gaucher registry: Demographics and disease characteristics of 1698 patients with Gaucher disease. Arch Intern Med. 2000;160:2835–2843. doi: 10.1001/archinte.160.18.2835. [DOI] [PubMed] [Google Scholar]
- 5.Sidransky E, Bottler A, Stubblefield B, Ginns EI. DNA mutational analysis of type 1 and type 3 Gaucher patients: How well do mutations predict phenotype? Hum Mutat. 1994;3:25–28. doi: 10.1002/humu.1380030105. [DOI] [PubMed] [Google Scholar]
- 6.Koprivica V, et al. Analysis and classification of 304 mutant alleles in patients with type 1 and type 3 Gaucher disease. Am J Hum Genet. 2000;66:1777–1786. doi: 10.1086/302925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liou B, et al. Analyses of variant acid β-glucosidases: Effects of Gaucher disease mutations. J Biol Chem. 2006;281:4242–4253. doi: 10.1074/jbc.M511110200. [DOI] [PubMed] [Google Scholar]
- 8.Lachmann RH, Grant IR, Halsall D, Cox TM. Twin pairs showing discordance of phenotype in adult Gaucher's disease. QJM. 2004;97:199–204. doi: 10.1093/qjmed/hch036. [DOI] [PubMed] [Google Scholar]
- 9.Beutler E. Gaucher disease as a paradigm of current issues regarding single gene mutations of humans. Proc Natl Acad Sci USA. 1993;90:5384–5390. doi: 10.1073/pnas.90.12.5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grabowski GA. Gaucher disease: Gene frequencies and genotype/phenotype correlations. Genet Test. 1997;1:5–12. doi: 10.1089/gte.1997.1.5. [DOI] [PubMed] [Google Scholar]
- 11.Amato D, Stachiw T, Clarke JT, Rivard GE. Gaucher disease: Variability in phenotype among siblings. J Inherit Metab Dis. 2004;27:659–669. doi: 10.1023/b:boli.0000042983.60840.f3. [DOI] [PubMed] [Google Scholar]
- 12.Elstein D, et al. Disease severity in sibling pairs with type 1 Gaucher disease. J Inherit Metab Dis. 2010;33:79–83. doi: 10.1007/s10545-009-9024-7. [DOI] [PubMed] [Google Scholar]
- 13.Ron I, Horowitz M. ER retention and degradation as the molecular basis underlying Gaucher disease heterogeneity. Hum Mol Genet. 2005;14:2387–2398. doi: 10.1093/hmg/ddi240. [DOI] [PubMed] [Google Scholar]
- 14.Sawkar AR, et al. Chemical chaperones and permissive temperatures alter localization of Gaucher disease associated glucocerebrosidase variants. ACS Chem Biol. 2006;1:235–251. doi: 10.1021/cb600187q. [DOI] [PubMed] [Google Scholar]
- 15.Bergmann JE, Grabowski GA. Posttranslational processing of human lysosomal acid β-glucosidase: A continuum of defects in Gaucher disease type 1 and type 2 fibroblasts. Am J Hum Genet. 1989;44:741–750. [PMC free article] [PubMed] [Google Scholar]
- 16.Schmitz M, Alfalah M, Aerts JM, Naim HY, Zimmer KP. Impaired trafficking of mutants of lysosomal glucocerebrosidase in Gaucher's disease. Int J Biochem Cell Biol. 2005;37:2310–2320. doi: 10.1016/j.biocel.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 17.Fenteany G, et al. Inhibition of proteasome activities and subunit-specific amino-terminal threonine modification by lactacystin. Science. 1995;268:726–731. doi: 10.1126/science.7732382. [DOI] [PubMed] [Google Scholar]
- 18.Fuller M, Lovejoy M, Hopwood JJ, Meikle PJ. Immunoquantification of β-glucosidase: Diagnosis and prediction of severity in Gaucher disease. Clin Chem. 2005;51:2200–2202. doi: 10.1373/clinchem.2005.053538. [DOI] [PubMed] [Google Scholar]
- 19.Dvir H, et al. X-ray structure of human acid-β-glucosidase, the defective enzyme in Gaucher disease. EMBO Rep. 2003;4:704–709. doi: 10.1038/sj.embor.embor873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yam AY, et al. Defining the TRiC/CCT interactome links chaperonin function to stabilization of newly made proteins with complex topologies. Nat Struct Mol Biol. 2008;15:1255–1262. doi: 10.1038/nsmb.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmidt MH, Dikic I. The Cbl interactome and its functions. Nat Rev Mol Cell Biol. 2005;6:907–918. doi: 10.1038/nrm1762. [DOI] [PubMed] [Google Scholar]
- 22.Broadley SA, Hartl FU. The role of molecular chaperones in human misfolding diseases. FEBS Lett. 2009;583:2647–2653. doi: 10.1016/j.febslet.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 23.Llorca O, et al. Eukaryotic type II chaperonin CCT interacts with actin through specific subunits. Nature. 1999;402:693–696. doi: 10.1038/45294. [DOI] [PubMed] [Google Scholar]
- 24.Klumpp M, Baumeister W, Essen LO. Structure of the substrate binding domain of the thermosome, an archaeal group II chaperonin. Cell. 1997;91:263–270. doi: 10.1016/s0092-8674(00)80408-0. [DOI] [PubMed] [Google Scholar]
- 25.Nitsch M, et al. Group II chaperonin in an open conformation examined by electron tomography. Nat Struct Biol. 1998;5:855–857. doi: 10.1038/2296. [DOI] [PubMed] [Google Scholar]
- 26.Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Annu Rev Biochem. 2009;78:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- 27.Beckmann RP, Mizzen LE, Welch WJ. Interaction of Hsp 70 with newly synthesized proteins: Implications for protein folding and assembly. Science. 1990;248:850–854. doi: 10.1126/science.2188360. [DOI] [PubMed] [Google Scholar]
- 28.Ron I, Rapaport D, Horowitz M. Interaction between parkin and mutant glucocerebrosidase variants: A possible link between Parkinson disease and Gaucher disease. Hum Mol Genet. 2010;19:3771–3781. doi: 10.1093/hmg/ddq292. [DOI] [PubMed] [Google Scholar]
- 29.Mu T, et al. Chemical and biological approaches synergize to ameliorate protein-folding diseases. Cell. 2008;134:769–781. doi: 10.1016/j.cell.2008.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee KO, et al. Improved intracellular delivery of glucocerebrosidase mediated by the HIV-1 TAT protein transduction domain. Biochem Biophys Res Commun. 2005;337:701–707. doi: 10.1016/j.bbrc.2005.05.207. [DOI] [PubMed] [Google Scholar]
- 31.Kim HS, Das A, Gross CC, Bryceson YT, Long EO. Synergistic signals for natural cytotoxicity are required to overcome inhibition by c-Cbl ubiquitin ligase. Immunity. 2010;32:175–186. doi: 10.1016/j.immuni.2010.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okamoto H, et al. Comparative proteomic profiles of meningioma subtypes. Cancer Res. 2006;66:10199–10204. doi: 10.1158/0008-5472.CAN-06-0955. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.