Abstract
UNC-45/CRO1/She4p (UCS) proteins have variously been proposed to affect the folding, stability, and ATPase activity of myosins. They are the only proteins known to interact directly with the motor domain. To gain more insight into UCS function, we determined the atomic structure of the yeast UCS protein, She4p, at 2.9 Å resolution. We found that 16 helical repeats are organized into an L-shaped superhelix with an amphipathic N-terminal helix dangling off the short arm of the L-shaped molecule. In the crystal, She4p forms a 193-Å-long, zigzag-shaped dimer through three distinct and evolutionary conserved interfaces. We have identified She4p’s C-terminal region as a ligand for a 27-residue-long epitope on the myosin motor domain. Remarkably, this region consists of two adjacent, but distinct, binding epitopes localized at the nucleotide-responsive cleft between the nucleotide- and actin-filament-binding sites. One epitope is situated inside the cleft, the other outside the cleft. After ATP hydrolysis and Pi ejection, the cleft narrows at its base from 20 to 12 Å thereby occluding the inside the cleft epitope, while leaving the adjacent, outside the cleft binding epitope accessible to UCS binding. Hence, one cycle of higher and lower binding affinity would accompany one ATP hydrolysis cycle and a single step in the walk on an actin filament rope. We propose that a UCS dimer links two myosins at their motor domains and thereby functions as one of the determinants for step size of myosin on actin filaments.
Keywords: myosin stability, processivity, chaperone
All eukaryotic cells contain a variety of myosins. Although there are significant differences between the various classes of myosins, they all share a highly homologous N-terminal globular region that functions as the motor domain. This region contains a conical flexible cleft. A nucleotide-binding site is located at the top of the cleft and binding sites for actin filaments surround the open bottom of the cleft (1). Following the motor domain is a flexible lever arm region (also referred to as “neck”) of variable length (2), which also contains binding sites for myosin light chains with regulatory functions (3). For those classes of myosin that dimerize, a helical dimerization module follows the lever arm region. Finally, the C terminus end contains a distinct tail region for binding either to distinct cargo molecules (4, 5) or, in the case of muscle myosins, to associate into myosin filaments.
In Saccharomyces cerevisiae there are five distinct myosins that belong to three myosin subclasses (6). Each myosin functions in distinct cellular events, such as endocytosis (7), cytokinesis (7), organelle transport (7), and transport of a subclass of mRNAs from the mother to the daughter cell (8, 9).
A single protein is known to bind to the motor domain of myosin. In budding yeast there is only one representative (also a founding member) of this family of proteins, known as UCS (UNC-45/CRO1/She4p) protein. The yeast protein, She4p, is known to be capable of interacting with all five yeast myosins, which belong to three myosin classes (10, 11). The binding site of She4p has been mapped to a 100-residue-long segment in the motor domain of yeast Myo4p, which is located near the actin-filament binding region (11).
Disparate functions have been proposed for UCS. The protein was first described as a chaperone for myosin folding (12–19). Subsequent reports invoked UCS in protecting myosin from degradation (20, 21) and in enhancing the binding of the motor domain of myosin to actin filaments (21, 22).
The atomic structure of a UCS protein has so far not been reported. To better understand the function of UCS and its mode of interaction with the motor domain of myosins, we determined the crystal structure of She4p at 2.9 Å resolutions. In the crystal, She4p forms a 193-Å-long dimer. Monomer/dimer equilibrium was also detected in solution. Furthermore, we identified a 27-residue-long region of yeast Myo4p, which interacts with the C-terminal region of She4p. We suggest that the ends of the UCS dimer, like a flexible string, tether two myosin motor domains together and therefore affect processivity and step size of myosin heads along actin filaments.
Results
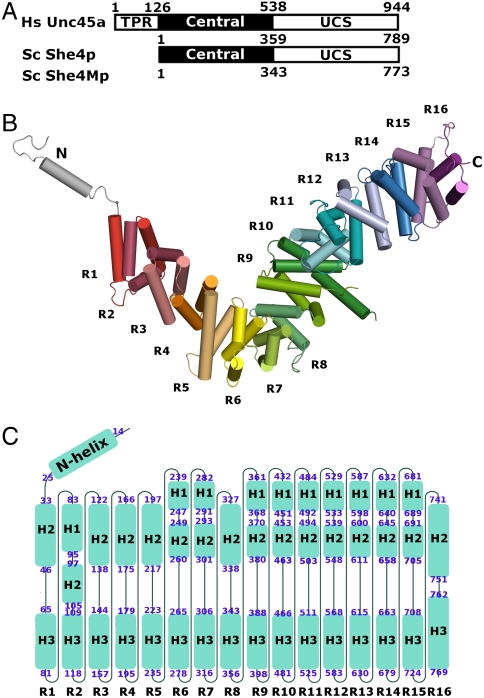
Unlike the UCS proteins of vertebrates, the fungal homologues lack an N-terminal tetratricopeptide repeat (23) (Fig. 1A) that in vertebrates has been reported to interact with hsp90 (12). The “central domain” is well conserved within some fungal species, but shows little homology between fungal and animal species (approximately 16.5% homology between She4p and humans). The UCS domain is well conserved across all species (approximately 52.7% homology between She4p and humans) (Fig. 1A and Fig. S1).
Fig. 1.
Yeast UCS (She4p) folds into helical repeats. (A) Domain organization of UCS proteins: Human Unc45a, yeast She4p, and yeast She4Mp; residue numbers at domain boundaries are indicated. All UCS proteins share central and UCS domains. Metazoan UCS proteins, exemplified here by human Unc45a, contain an additional tetratricopeptide repeat (TPR). (B) She4p assembles into helical repeats, which are shown in different colors, and are numbered from R1–R16. The repeats form an L-shaped superhelix. Extending from the short arm of the L-shaped superhelix is an N-terminal loop-helix-loop motif (silver). (C) The topology of She4p. Sixteen helical turns are arranged from left to right and labeled as R1–R16; the helices of each turn are shown as blue boxes and named according to their position in the conventional three-helical repeats as H1–H3 and in the two-helical repeat as H2–H3. The beginning and the ending residue of each helix are labeled at the ends of each box. H3 helices from 16 repeats align to form a continuous inner surface. Helix H1 of R2 and the H2 helices of the other repeats align to form a continuous outer surface in the 3D structure.
She4p Forms an L-Shaped Superhelix.
Recombinant WT She4p yielded crystals with low resolution. To improve the crystal quality we consecutively introduced several mutations. With each step we observed crystals with improved resolution (for a description of the rationale for these mutations see the legend for Fig. S1) (24). The final version resulting from this series of mutations was named She4Mp and was used in both the native and the Se-Met substituted crystals for structure determination. Importantly, She4Mp can fully replace WT She4p, indicating that the introduced mutations did not alter the function of the protein (Fig. S2). In the following text, therefore, we refer to She4Mp as She4p.
She4p crystallized in space groups P212121. The crystal structure was determined using multiwavelength anomalous dispersion (MAD) from Se-Met substituted protein crystals. The final model was refined to 2.9 Å with Rfree = 29.6% and Rwork = 23.1% (Table 1).
Table 1.
Data collection, phasing, and refinement statistics
| Native |
Crystal 1 name |
|||
| Data collection | ||||
| Space group | P212121 | P212121 | ||
| Cell dimensions | ||||
| a, b, c (Å) | 84.0, 149.9, 158.5 | 85.4, 149.7, 159.7 | ||
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 | ||
| Peak | Inflection | Remote | ||
| Wavelength | 0.9795 | 0.9790 | 0.9794 | 0.9567 |
| Resolution (Å) | 2.90 | 3.05 | 3.05 | 3.15 |
| Rsym or Rmerge | 6.3 (78.7) | 9.6 (56.5) | 7.6 (66.4) | 8.5 (66.2) |
| I/σI | 28.5 (1.4) | 26.7 (3.1) | 20.0 (1.3) | 27.4 (2.5) |
| Completeness (%) | 95.6 (83.0) | 99.9 (99.6) | 99.1 (92.8) | 99.8 (98.9) |
| Redundancy | 7.1 (6.4) | 13.1 (11.2) | 7.1 (5.5) | 11.1 (9.6) |
| Refinement | ||||
| Resolution (Å) | 50–2.90 | |||
| No. reflections | 45,409 | |||
| Rwork/Rfree | 23.1/29.6 | |||
| No. atoms | ||||
| Protein | 11874 | |||
| Ligand/ion | N/A | |||
| Water | 145 | |||
| B factors (Å2) | ||||
| Protein | 97.12 | |||
| Ligand/ion | N/A | |||
| Water | 79.02 | |||
| Rms deviations | ||||
| Bond lengths (Å) | 0.008 | |||
| Bond angles (°) | 1.38 | |||
| Ramachandran | ||||
| Most favored regions (%) | 80.8 | |||
| Additional allowed regions (%) | 15.8 | |||
| Generously allowed regions (%) | 2.3 | |||
| Disallowed regions (%) | 1.2 |
Values in parentheses are for highest resolution shell.
A single She4p molecule folds into 16 helical Armadillo (ARM) repeats (see the legend for Fig. S1) that are arranged in a L-shaped superhelix (Fig. 1 B and C). An amphipathic N helix extends from the short arm of the L-shaped molecule. The short arm of the L-shaped superhelix comprises repeats R1–R4, the “bend” region repeats R5–R8, and the long arm repeats R9–R16. The previously termed central domain (Fig. 1A) comprises the short arm and the bend of the L-shaped molecule, extending from repeats R1–R8 (Fig. 1B). The highly conserved UCS domain (Fig. 1A) comprises the long arm of the L-shaped superhelix and extends from repeats R8–R16 (Fig. 1 B and C). The shape of the short arm is more flattened, whereas that of the long arm is more cylindrical.
She4p Forms an 193-Å-Long Dimer.
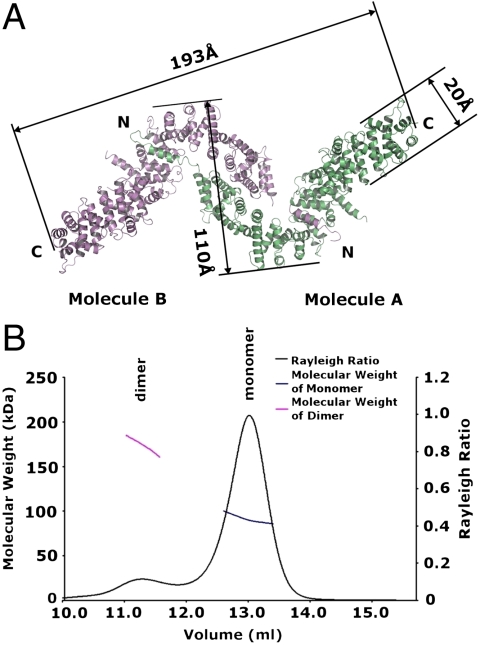
In the crystal, two L-shaped superhelices (termed A and B) pack into one asymmetric unit to form a zigzag-shaped dimer, in which the short arms of each L associate with each other in an antiparallel fashion, and in which the amphipathic N-terminal helices cross-over and connect to the long arm of the contralateral L (Fig. 2A). The overall length of the dimer is 193 Å with a bulky middle region of 110 Å and two slender ends with a width of 20 Å (Fig. 2A). Size-exclusion chromatography and multiangle light scattering showed that in solution She4p exists in monomer/dimer equilibrium (Fig. 2B). The solution and crystallographic data indicate that the She4p dimer is biologically relevant.
Fig. 2.
She4p forms an elongated dimer. (A) Two L-shaped molecules (green and purple) form a zigzag-shaped dimer; dimerization is through antiparallel association of the short arms of the Ls and the association of the two N-terminal helices with the long arms of the contra-lateral Ls. The dimer is 193-Å-long with a middle portion measuring 110 Å and two ends with a width of 20 Å. N and C indicate N and C termini, respectively. (B) Gel filtration and multiangle light scattering of She4p. The Rayleigh ratio is shown as a black curve and the molecular weight (Mr) of major and minor peaks are shown in blue and magenta, respectively. The molecular weight of the major peak was 91.09 ± 0.42 kDa (theoretical Mr = 89.9 kDa) and of the minor peak was 174.77 ± 2.99 kDa.
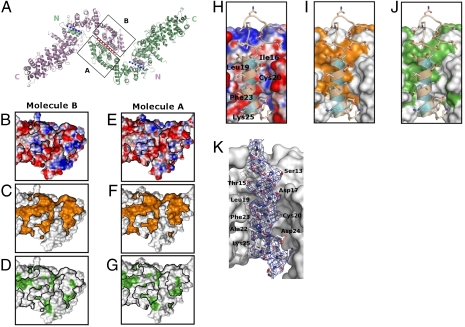
The two molecules in one asymmetric unit are related to each other by pseudo-twofold rotational symmetry (Fig. 3A) and are superimposable (rmsd ≈ 1.8 Å) except for variations at some flexible surface loops and at the far C-terminal region. The zigzag-shaped dimer forms three interfaces. A central interface is formed by antiparallel association of the two short arms of the L-shaped molecules, involving helical repeats R1–R4, and covering a total surface area of 2,271 Å2. Two lateral interfaces are formed between the amphipathic N-terminal helices and the bend of the contra-lateral L-shaped molecule involving helical repeats R7–R9 and covering a total surface area of 1,690 Å2 for each of the two interfaces. The symmetry axis is located in the central interface. Two lateral interfaces are related by a pseudo twofold rotational symmetry (Fig. 3A).
Fig. 3.
Evolutionary conservation of the dimerization interfaces of She4p. (A) The three dimerization interfaces are indicated with blue and red dotted lines, with molecule A in green and molecule B in purple; N and C indicate N and C termini, respectively. Boxed portions indicate apposing dimerization surfaces of the two short arms of L. (B–G) Dimerization surfaces of the two short arms of the L-shaped A and B molecules are colored according to electrostatic potential (with saturating blue and red at ± 15 kT/e). (B and E) Conservation between related fungal homologues (yellow) (C and F); or, hydrophobicity (green) (D and G); black lines encircle conserved regions. (H–J) Symmetry-related dimerization interfaces between the N-helices and the contra-lateral bend of the L-shaped A and B molecules (see Fig. 2A); the N helix of molecule A was superimposed as a ribbon with stick and ball side chains onto the surface of the bend of molecule B; conserved residues of the helix are indicated in cyan; otherwise the coloring of the electrostatic, conserved, and hydrophobic surfaces of the bend of L is as in B–G. (K) Electron density of the N helix. The composite omit map (2fo-fc) covering the N helix of molecule A is shown in blue mesh. The map was contoured at 1σ cutoff. The refined N helix is shown in stick and ball model with residue names and numbers labeled. The bend of molecule B is shown in surface representation.
The central interface involves primarily polar residues (Fig. 3 B and E) with a few scattered hydrophobic patches (Fig. 3 D and G) within a highly conserved region between fungal species (Fig. 3 C and F). This interface involves most of the previously assigned central domain (Fig. 1A). In the lateral interfaces the hydrophobic ribbon of the amphipathic N helix interacts with a largely hydrophobic surface at repeat R7–R9 of the contra-lateral L-shaped molecule (Fig. 3 H–K and Fig. S1). Both surfaces are highly conserved (Fig. 3I).
Interaction with a Conserved Myosin Motif.
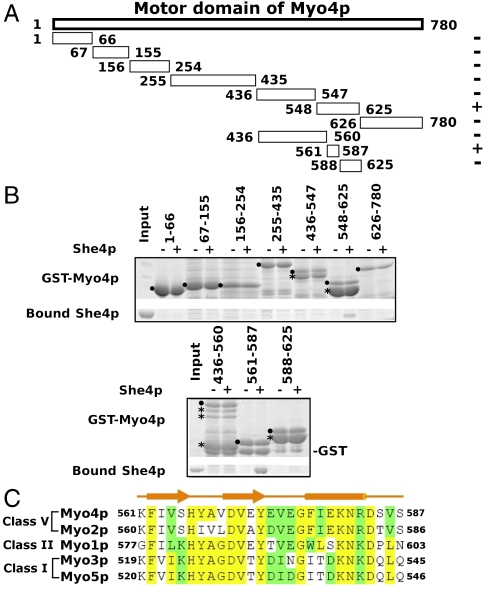
Previously She4p interactions with myosin have been mapped on all five yeast myosins. By two hybrid assays, the She4p interacting region for Myo4p has been reported to cover residues 531–631 (11). In an attempt to further fine map this interaction, we expressed various fragments of yeast Myo4p as GST fusion proteins and tested their interaction with purified She4p (Fig. 4A). We found that a 27-residue-long segment of Myo4p (residues 561–587) was able to interact with She4p ( Fig. 4B). This segment is highly conserved (Fig. 4C), not only in all five of the yeast myosins, but also in the subfamilies of myosins identified to date (25) (Fig. S3). By isothermal titration calorimetry we found that a synthetic peptide representing the 27-residue-long segment of Myo4p bound to UCS with a binding affinity of approximately 1 μM (KD) (Fig. S4).
Fig. 4.
She4p directly interacts with a highly conserved region of the myosin motor domain. (A) Fragments of yeast Myo4p were expressed as GST fusion peptides for pull-down assays. Fragments that bind to She4p are labeled with + and those that fail to bind are labeled with − at the right. (B) SDS-PAGE analysis of pull-down experiments; the fragment containing residues 561–587 of Myo4p can stably bind to purified She4p (Lower). GST fusion peptides were indicated by ● and their degradation products by *. (C) Primary structure comparison of the She4p binding sites from the five S. cerevisiae myosins. Corresponding residue numbers are indicated to the left and right. The predicted secondary structure is shown above the alignment. Identical residues are indicated in yellow, similar ones in green.
The Strategic Binding Site of UCS on Myosin.
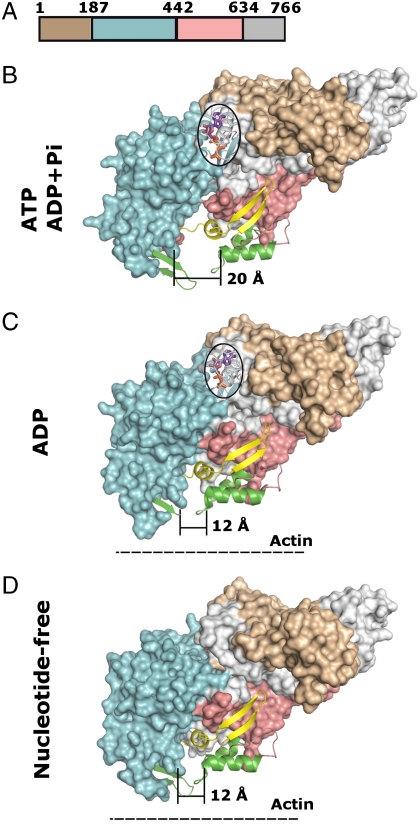
Several myosin motor domain structures are available (26). For chicken myosin V, atomic structures of nucleotide-free and -bound forms are known (27). Similar to other myosin motor domains, the chicken myosin V motor domain can be divided into four domains as indicated in Fig. 5A. The middle two domains, conventionally termed “upper” and “lower” 50 kDa subdomain, indicated in cyan and pink, respectively, surround a conical cleft (Fig. 5 B–D). The nucleotide-binding site is located on top of this cleft (bordered by an oval in Fig. 5 B and C), whereas the actin-filament binding site surrounds the bottom of the cone (colored in green in Fig. 5 B–D). When ATP or ADP/Pi occupies the nucleotide-binding site, the base of the cone measures 20 Å in diameter (Fig. 5B). The release of Pi, which causes the power stroke, narrows the conical cleft to 12 Å, and leads to a stable interaction with actin filaments (Fig. 5C). This state persists after release of ADP in the nucleotide-free form (Fig. 5D). Renewed binding of ATP completes the cycle and releases the myosin motor domain from the actin filament and, again, widens the conical cleft to 20 Å (Fig. 5B). Strikingly, the UCS binding site on myosin (indicated in yellow) is located near the conical cleft. Interestingly, two binding epitopes can be distinguished with regard to location on the myosin molecule and the hydrolysis cycle of ATP. One, consisting of two antiparallel beta strands, is located on the surface and remains accessible during the entire ATP hydrolysis cycle, whereas the other, comprised of a helix-loop motif, is located inside the cone and becomes inaccessible when the cleft narrows to 12 Å (Fig. 5 B–D; for the location of these two epitopes in the 27-residue-long peptide, see Fig. 4C).
Fig. 5.
Projection of She4p binding sites onto the myosin surface during the ATPase cycle. (A) Location of modules in the primary structure of chicken myosin V motor domain; residue numbers are indicated and domain boundaries are colored: brown, N-terminal region; cyan, Upper subdomain; pink, Lower subdomain; and gray, C-terminal region. (B–D) Surface representation of the motor domain of chicken myosin V in the ATP or ADP + Pi form [Protein Data Bank (PDB) ID code: 1w7j(B)], in the ADP form [PDB ID code: 1w7i(c)], or in the nucleotide-free form [PDB ID code: 1w8j(D)], respectively, (27); color code as in (A). Note the dramatic reduction from 20 to 12 Å in width at the base of the cleft during the ATPase cycle, concomitant with strong binding to actin filaments (dashed line). Projected on the surface of the myosin forms are nucleotides (magenta, stick and ball representation), strong actin binding sites (green, ribbon representation), and the UCS binding sites (yellow, ribbon representation). Note that the UCS binding site contains two distinct binding epitopes; One, consisting of two antiparallel beta strands, remains always exposed on the myosin surface during the ATPase cycle, while the other one, located inside the cleft (helix loop) is occluded during the reduction of the width of the cleft from 20 to 12 Å during the ATPase cycle.
Myosin Binding Site is Located at the C-Terminal Region of She4p.
Analysis of the atomic structure of She4p showed a conserved surface at the C-terminal region of the She4p molecule. Because this region was a potential candidate for myosin binding, we prepared recombinant proteins comprised of several truncated forms of the C-terminal region of UCS and tested them for binding to a fusion protein consisting of GST, a 10-residue-long linker, and the 27-residue-long UCS binding peptide of Myo4p. Only one of five tested truncated molecules, She4p 1–544, was soluble. As expected, full-length She4p bound to the Myo4p fusion protein (Fig. S5, Arrowhead Lane 8), whereas binding of the truncated form of She4p was reduced (Fig. S5, Arrowhead Lane 5). Although these data suggest that the C-terminal region of UCS does indeed contain a binding site for myosin, an atomic description of this binding site has to await analysis of a She4p/myosin peptide cocrystal.
Discussion
Our crystallographic and biochemical data here show that the UCS protein She4p of yeast is an L-shaped superhelix that dimerizes to form a 193-Å-long, zigzag-shaped structure. A C-terminal region of the She4p monomer functions as a ligand for the motor domain of myosin. Hence, a UCS dimer would act like a bivalent linker whose C-terminal regions join two myosin motor domains in a joint-like fashion.
Biochemical mapping showed that UCS binds to a highly conserved, 27-residue-long region of the myosin motor domain (Fig. 4). When projected onto the atomic structure of a chicken myosin, the 27-residue-long region maps to a nucleotide-responsive cleft that is strategically located between the nucleotide- and actin-filament-binding sites of the myosin motor domain (Fig. 5). Strikingly, the 27-residue-long binding region consists of two distinct epitopes. One is located inside the cleft surface, whereas the other one is situated on the surface surrounding the cleft. In the ATP-bound state, the myosin cleft is 20-Å wide at its base. In this open state both binding epitopes are likely accessible (Fig. 5B). However, after ATP hydrolysis and following the release of Pi, the cleft narrows to 12 Å, likely preventing access to the binding epitope located inside the cleft (Fig. 5C). Although the binding epitope outside the cleft would still be accessible, the occlusion of the binding epitope inside the cleft would reduce the overall binding affinity of UCS to myosin. Hence, each ATP hydrolysis cycle would be accompanied by an alternating cycle of higher and lower affinity binding of each UCS dimer to each of the two myosin heads as they move by one step along the actin filament (Fig. 6).
Fig. 6.
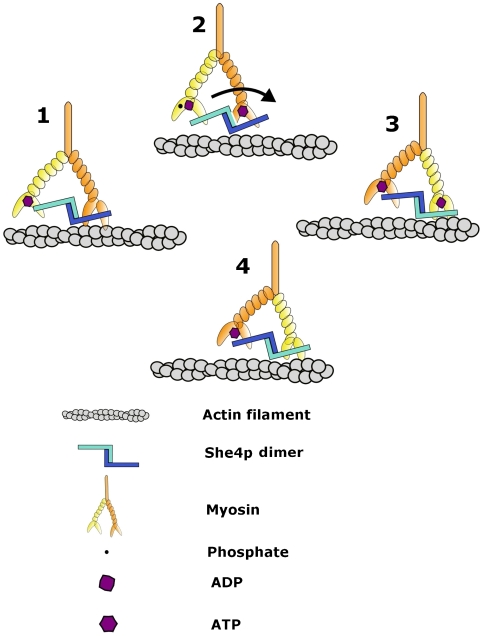
A model in which the zigzag-shaped UCS dimer (monomers are colored in light and dark blue) joins two myosin motor domains during the ATP hydrolysis cycle, during which one motor domain moves one step forward on an actin filament (indicated in gray). The prong-like structure represents the myosin motor domain (see Fig. 5); the region with bound light chains (oval) represents the lever arm (yellow and orange) and is followed by the coiled-coil dimerization module (orange). For details see text.
All myosin molecules, at least in yeast, have been shown to interact with UCS. Our data here show that UCS is a dimerization module that flexibly links two motor domains of two myosin chains. Hence, even single myosin heavy chains that do not contain a built-in dimerization module in form of a coiled coil region may be dimerized by a bivalent UCS dimer.
The stepwise movement on an actin filament of one myosin head passing the other during the power stroke is likely to cause mechanical tension on the linkage between UCS and the myosin motor domains. Alternating cycles of strong and weak binding affinities of UCS for the myosin motor domain (Fig. 5) would be one way to reduce such tension. Another way would be stretching the UCS superhelix (Figs. 3 and 4), including transient dedimerization.
Single molecule measurements carried out in vitro suggest that the myosin step size is influenced by the length of the lever arm and the swing angle (1) and therefore varies with different classes of myosin. Step size ranges between 10 and 100 nm have been reported for various myosins (1). However, these measurements were carried out in the absence of UCS. We propose that a flexible linkage by an UCS dimer of two myosin heads in a joint-like fashion is one of the determinants of step size. Given the length and the likely plasticity of the UCS dimer, we estimate a minimum step size in the range between 10 and 20 nm. Moreover, as UCS appears to interact with all myosins, such a minimum step size may pertain to all classes of myosin.
This possibility is supported by recent data of in vitro motility assays whereby myosin molecules immobilized on glass move fluorescently labeled actin filaments with higher efficiency in the presence of UCS and at low filament concentrations (21, 22). As the ligand binding site of UCS on myosin is in a strategic location inside and outside a repetitively opening and closing cleft between the binding sites to nucleotide and the actin filament, and as movement of myosin along actin filaments could cause local tensions accompanied by transient deformation of the molecule, it is conceivable that UCS may minimize such deformations and thereby physically stabilize the myosin head. By doing so, it could inhibit the pathway toward ubiquitination and subsequent protein degradation and hence contribute to the stability of myosin (21).
Materials and Methods
Primers used in yeast vector construction and integration are listed in Table S1. Mutant She4p was purified as GST fusion protein and crystallized using sitting drop vapor diffusion method. Structure of She4p was determined by three wavelength MAD method. The oligomeric state of She4p in solution was determined using size-exclusion chromatography coupled to multiangle light scattering. Various fragments of Myo4p were expressed as GST fusion peptides, immobilized on glutathione sepharose beads, and incubated with She4p. The bound protein on glutathione sepharose beads was analyzed by SDS-PAGE after washing. For additional details, please see SI Materials and Methods.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Anand Saxena and Howard Robinson for beam time at X12C and X29; Haiteng Deng and David King for mass spectrometry and Edman sequencing; Sozanne Solmaz for helping with multiangle light scattering experiments; Shaun Bevers for Isothermal titration calorimetry analysis; Yuh Min Chook for many useful suggestions and discussions; and Erik Debler for critical reading of the manuscript.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3OPB).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013038107/-/DCSupplemental.
References
- 1.Spudich JA, Sivaramakrishnan S. Myosin VI: An innovative motor that challenged the swinging lever arm hypothesis. Nat Rev Mol Cell Biol. 2010;11:128–137. doi: 10.1038/nrm2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uyeda TQ, Abramson PD, Spudich JA. The neck region of the myosin motor domain acts as a lever arm to generate movement. Proc Natl Acad Sci USA. 1996;93:4459–4464. doi: 10.1073/pnas.93.9.4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trybus KM. Role of myosin light chains. J Muscle Res Cell Motil. 1994;15:587–594. doi: 10.1007/BF00121066. [DOI] [PubMed] [Google Scholar]
- 4.Catlett NL, Duex JE, Tang F, Weisman LS. Two distinct regions in a yeast myosin-V tail domain are required for the movement of different cargoes. J Cell Biol. 2000;150:513–526. doi: 10.1083/jcb.150.3.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geething NC, Spudich JA. Identification of a minimal myosin Va binding site within an intrinsically unstructured domain of melanophilin. J Biol Chem. 2007;282:21518–21528. doi: 10.1074/jbc.M701932200. [DOI] [PubMed] [Google Scholar]
- 6.Brown SS. Myosins in yeast. Curr Opin Cell Biol. 1997;9:44–48. doi: 10.1016/s0955-0674(97)80150-0. [DOI] [PubMed] [Google Scholar]
- 7.Pruyne D, Legesse-Miller A, Gao L, Dong Y, Bretscher A. Mechanisms of polarized growth and organelle segregation in yeast. Annu Rev Cell Dev Biol. 2004;20:559–591. doi: 10.1146/annurev.cellbio.20.010403.103108. [DOI] [PubMed] [Google Scholar]
- 8.Long RM, et al. Mating type switching in yeast controlled by asymmetric localization of ASH1 mRNA. Science. 1997;277:383–387. doi: 10.1126/science.277.5324.383. [DOI] [PubMed] [Google Scholar]
- 9.Takizawa PA, Sil A, Swedlow JR, Herskowitz I, Vale RD. Actin-dependent localization of an RNA encoding a cell-fate determinant in yeast. Nature. 1997;389:90–93. doi: 10.1038/38015. [DOI] [PubMed] [Google Scholar]
- 10.Wesche S, Arnold M, Jansen RP. The UCS domain protein She4p binds to myosin motor domains and is essential for class I and class V myosin function. Curr Biol. 2003;13:715–724. doi: 10.1016/s0960-9822(03)00264-1. [DOI] [PubMed] [Google Scholar]
- 11.Toi H, et al. She4p/Dim1p interacts with the motor domain of unconventional myosins in the budding yeast, Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:2237–2249. doi: 10.1091/mbc.E02-09-0616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF. Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science. 2002;295:669–671. doi: 10.1126/science.1066648. [DOI] [PubMed] [Google Scholar]
- 13.Srikakulam R, Liu L, Winkelmann DA. Unc45b forms a cytosolic complex with Hsp90 and targets the unfolded myosin motor domain. PLoS One. 2008;3:e2137. doi: 10.1371/journal.pone.0002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amorim MJ, Mata J. Rng3, a member of the UCS family of myosin co-chaperones, associates with myosin heavy chains cotranslationally. EMBO Rep. 2008;10:186–191. doi: 10.1038/embor.2008.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wohlgemuth SL, Crawford BD, Pilgrim DB. The myosin co-chaperone UNC-45 is required for skeletal and cardiac muscle function in zebrafish. Dev Biol. 2007;303:483–492. doi: 10.1016/j.ydbio.2006.11.027. [DOI] [PubMed] [Google Scholar]
- 16.Etard C, et al. The UCS factor Steif/Unc-45b interacts with the heat shock protein Hsp90a during myofibrillogenesis. Dev Biol. 2007;308:133–143. doi: 10.1016/j.ydbio.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 17.Mishra M, D'Souza VM, Chang KC, Huang Y, Balasubramanian MK. Hsp90 protein in fission yeast Swo1p and UCS protein Rng3p facilitate myosin II assembly and function. Eukaryot Cell. 2005;4:567–576. doi: 10.1128/EC.4.3.567-576.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melkani GC, Lee CF, Cammarato A, Bernstein SI. Drosophila UNC-45 prevents heat-induced aggregation of skeletal muscle myosin and facilitates refolding of citrate synthase. Biochem Biophys Res Commun. 2010;396:317–322. doi: 10.1016/j.bbrc.2010.04.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernick E, Zhang P, Du S. Knockdown and overexpression of Unc-45b result in defective myofibril organization in skeletal muscles of zebrafish embryos. BMC Cell Biol. 2010;11:70–82. doi: 10.1186/1471-2121-11-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoppe T, et al. Regulation of the myosin-directed chaperone UNC-45 by a novel E3/E4-multiubiquitylation complex in C. elegans. Cell. 2004;118:337–349. doi: 10.1016/j.cell.2004.07.014. [DOI] [PubMed] [Google Scholar]
- 21.Lord M, Sladewski TE, Pollard TD. Yeast UCS proteins promote actomyosin interactions and limit myosin turnover in cells. Proc Natl Acad Sci USA. 2008;105:8014–8019. doi: 10.1073/pnas.0802874105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lord M, Pollard TD. UCS protein Rng3p activates actin filament gliding by fission yeast myosin-II. J Cell Biol. 2004;167:315–325. doi: 10.1083/jcb.200404045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barral JM, Bauer CC, Ortiz I, Epstein HF. Unc-45 mutations in Caenorhabditis elegans implicate a CRO1/She4p-like domain in myosin assembly. J Cell Biol. 1998;143:1215–1225. doi: 10.1083/jcb.143.5.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jin L, et al. Mutation of surface residues to promote crystallization of activated factor XI as a complex with benzamidine: An essential step for the iterative structure-based design of factor XI inhibitors. Acta Crystallogr, Sect D: Biol Crystallogr. 2005;61:1418–1425. doi: 10.1107/S0907444905024340. [DOI] [PubMed] [Google Scholar]
- 25.Hodge T, Cope MJ. A myosin family tree. J Cell Sci. 2000;113:3353–3354. doi: 10.1242/jcs.113.19.3353. [DOI] [PubMed] [Google Scholar]
- 26.Houdusse A, Sweeney HL. Myosin motors: Missing structures and hidden springs. Curr Opin Struct Biol. 2001;11:182–194. doi: 10.1016/s0959-440x(00)00188-3. [DOI] [PubMed] [Google Scholar]
- 27.Coureux PD, Sweeney HL, Houdusse A. Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J. 2004;23:4527–4537. doi: 10.1038/sj.emboj.7600458. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.