Abstract
Proteins that can be tailored to bind desired DNA sequences are key tools for molecular biology. Previous studies suggested that DNA-binding specificity of transcription activator-like effectors (TALEs) from the bacterial genus Xanthomonas is defined by repeat-variable diresidues (RVDs) of tandem-arranged 34/35-amino acid repeat units. We have studied chimeras of two TALEs differing in RVDs and non-RVDs and found that, in contrast to the critical contributions by RVDs, non-RVDs had no major effect on the DNA-binding specificity of the chimeras. This finding suggests that one needs only to modify the RVDs to generate designer TALEs (dTALEs) to activate transcription of user-defined target genes. We used the scaffold of the TALE AvrBs3 and changed its RVDs to match either the tomato Bs4, the Arabidopsis EGL3, or the Arabidopsis KNAT1 promoter. All three dTALEs transcriptionally activated the desired promoters in a sequence-specific manner as mutations within the targeted DNA sequences abolished promoter activation. This study is unique in showing that chromosomal loci can be targeted specifically by dTALEs. We also engineered two AvrBs3 derivatives with four additional repeat units activating specifically either the pepper Bs3 or UPA20 promoter. Because AvrBs3 activates both promoters, our data show that addition of repeat units facilitates TALE-specificity fine-tuning. Finally, we demonstrate that the RVD NK mediates specific interaction with G nucleotides that thus far could not be targeted specifically by any known RVD type. In summary, our data demonstrate that the TALE scaffold can be tailored to target user-defined DNA sequences in whole genomes.
Keywords: zinc-finger proteins, AvrBs4, AvrXa27
The ability to achieve site-specific manipulation of genomes has widespread implications for basic and applied research, and much effort has been devoted to identify protein folds that can be tailored to bind user-defined DNA sequences (1). Recent studies uncovered that transcription activator-like effector proteins (TALEs) from the plant pathogenic bacterial genus Xanthomonas contain a unique type of DNA-binding domain (2, 3). TALEs are modular proteins that are composed of (i) an N-terminal translocation signal, (ii) a central DNA binding domain, and (iii) a C-terminal region containing nuclear localization signals and a transcriptional activation domain (4, 5). The TALE DNA binding domain is composed of a variable number of tandemly arranged, imperfect 34/35 amino acid repeat units. Repeat unit polymorphisms are found predominantly at positions 12 and 13, referred to as the repeat-variable diresidues (RVDs), but occur also within non-RVD repeat unit positions (4, 5). Recent studies showed that repeats with different RVDs recognize different DNA base pairs and that consecutive RVDs in a TALE correspond directly to the sequence of nucleotides in the binding site, one-to-one in a linear and contiguous fashion. This TALE code explains the specificity of TALEs and enables the prediction of corresponding DNA target sequences (6–8).
Plant pathogenic xanthomonads inject TALEs into host cells via a syringe-like type III secretion system to promote bacterial growth or spread. Within the host cell, TALEs translocate to the nucleus, bind via their DNA binding domain to corresponding UPT (Up-regulated by TALE) boxes and activate downstream host genes. The primary function of TALEs is to transcriptionally activate host-susceptibility (S) genes that, by mostly unknown means, promote growth or spread of the bacterial pathogen (4, 5). However, in the course of plant-pathogen coevolution, some plants evolved disease resistance (R) genes that mediate the perception of individual TALEs. AvrBs3 from the pepper (Capsicum spec.) and tomato (Solanum lycopersicum) pathogenic strain Xanthomonas campestris pv. vesicatoria (Xcv) is one TALE for which corresponding plant S and R genes have been studied. In the context of a susceptible plant genotype, AvrBs3 has been shown to interact with a corresponding UPTAvrBs3 box present in the promoter of the pepper transcription factor UPA20 (2). Expression of the host S gene UPA20 induces hypertrophy of mesophyll cells, which is believed to promote the release of Xcv from infected leaves (2, 9). If the given pepper plant contains the R gene Bs3, AvrBs3 triggers a programmed cell death, referred to as the hypersensitive response (HR) (3, 10). Certain pepper lines have an allele of Bs3 known as Bs3-E, which confers resistance to strains carrying the AvrBs3 derivative AvrBs3Δrep16 that has a deletion of repeat units 11 to 14, compared with AvrBs3 (3, 10). AvrBs3 and AvrBs3Δrep16 interact specifically with distinct UPT boxes in the Bs3 and Bs3-E promoters, respectively, and activate transcription of the corresponding coding sequences, leading to HR (10).
The Xcv TALE protein AvrBs4 is 97% identical to AvrBs3 (11), yet it does not trigger either the pepper Bs3 or the Bs3-E gene because its distinct RVD composition does not support interaction with these promoters (3). AvrBs4 does, however, trigger an HR in the Capsicum pubescens accession PI 235047 that contains the matching R gene Bs4 (12). AvrBs4 is also recognized in tomato plants that contain the corresponding nucleotide binding site leucine-rich repeat type R protein Bs4 (13). Importantly, the tomato R gene Bs4 is distinct from all other known plant R genes that mediate recognition of TALEs because it is not transcriptionally activated by matching TALEs (14). Furthermore, the tomato Bs4 gene is currently the only TALE-specific R gene that mediates recognition of several TALEs with distinct RVD composition (14). For clarity, we refer in this article to the above-described R genes as Bs4S (S. lycopersicum) and Bs4C (C. pubescens).
Previously, it was shown that the TALE code could be used to predict DNA target sequences of Xanthomonas wild-type TALEs, as well as in vitro-assembled TALEs with arbitrary RVD composition (6–8). In the present study we intended to use the TALE code in a reverse manner to generate designer TALES (dTALEs) that would activate user-defined chromosomal target genes. We demonstrate that the scaffold of the TALE protein AvrBs3 can be designed with different consecutive RVDs to target desired DNA sequences. In addition, we show that variation of the repeat unit number facilitates fine-tuning of TALE target specificity. Finally, we show that the RVD NK facilitates specific targeting of G nucleotides, which thus far could not be addressed by a specific RVD.
Results
Codon-Optimized TALE Genes of AvrBs3 and AvrBs4 Specifically Trigger the Corresponding Capsicum R Genes Bs3 and Bs4C.
The molecular dissection of TALE DNA-target specificity requires TALE genes that can be modified by PCR mutagenesis in a defined manner. However, because of the high overall GC content of Xanthomonas TALE genes and their nearly identical, tandemly arranged 102-bp repeat sequences, PCR mutagenesis of Xanthomonas TALE genes is challenging. For example, the TALE gene avrBs3 has an overall GC content of 67% and is composed of 17.5 tandemly arranged 102-bp repeats that are >90% identical to each other at the nucleotide level (15). To overcome these limitations, we generated codon-optimized versions of the Xanthomonas TALE genes avrBs3 and avrBs4, herein defined as avrBs3* and avrBs4*, respectively (SI Text). Both avrBs3* and avrBs4* are codon-optimized for expression in tobacco, have a GC content of only 50%, and show on average 72% homology between the 102-bp repeats. To functionally test codon-optimized TALE genes, we made use of the Capsicum R genes Bs3 and Bs4C that mediate specific recognition of AvrBs3 and AvrBs4, respectively (3, 12). To do so, we cloned avrBs3* and avrBs4* into plant-expression vectors under transcriptional control of the constitutive cauliflower mosaic virus 35S (35S) promoter. Subsequently we used Agrobacterium tumefaciens T-DNA transfer to deliver the codon-optimized TALE genes in Capsicum plants containing the Bs3 or Bs4C genes.
As shown in Fig. S1, in planta expression of avrBs3* triggered an HR only in genotypes that contain the Bs3 gene. Reciprocally, in planta expression of avrBs4* triggered an HR only in genotypes that contain the Bs4C gene. Thus, codon-optimized avrBs3* and avrBs4* genes showed in planta the expected specificity and triggered HR only in plants containing the corresponding plant R gene.
Repeat-Polymorphisms in Non-RVD Residues Have No Major Impact on RVD Specificity.
Previous studies of Xanthomonas TALEs and matching UPT boxes suggested that RVD composition determines recognition specificity (6–8, 16). However, repeat units differ not only in RVD but also in non-RVD residues and circumstantial evidence suggests that non-RVDs that are polymorphic between repeat units might have coevolved with defined RVD motifs. For example, in the Xcv TALE AvrBs3, all seven repeat units with the RVD motif HD contain an arginine at position 24, but the three repeat units with the RVD motif NI contain an alanine at position 24 (SI Text). To clarify if distinct RVD motifs function only in the context of matching non-RVD residues, we generated chimeric proteins consisting of the Xcv TALEs AvrBs3 and AvrBs4 that both contain 17.5 repeat units (11, 15). AvrBs3 and AvrBs4 differ in 38 repeat-unit residues and 18 of those differences are located within the RVDs (SI Text). To test if only the RVDs but not other repeat unit residues define DNA binding-specificity, we replaced all RVDs in AvrBs3 by the RVDs from AvrBs4, thereby generating AvrBs3RVD4*. Reciprocally we replaced all RVDs in AvrBs4 by the RVDs from AvrBs3, thereby generating AvrBs4RVD3* (Fig. S2A). Both, avrBs3RVD4* and avrBs4RVD3* were cloned in T-DNA vectors under transcriptional control of the 35S promoter and were transiently expressed in pepper plants containing the Bs3 or the Bs4C gene. AvrBs3* and AvrBs4RVD3* triggered an HR in Bs3 but not in Bs4C genotypes (Fig. S3). Reciprocally, AvrBs4* and AvrBs3RVD4* triggered an HR in Bs4C- but not in Bs3-containing plants. In summary, these data indicate that only the RVDs of AvrBs3 and AvrBs4 dictate recognition specificity in their interaction with the pepper R genes Bs3 and Bs4C, respectively, but non-RVDs have no or only a minor impact on TALE specificity.
To compare the biological activity of wild-type and chimeric TALEs in a more quantitative fashion, we generated Bs3-promoter derivatives that contain either the UPTAvrBs3 box (Bs3PUPTAvrBs3) or the UPTAvrBs4 box (Bs3PUPTAvrBs4 ΔUPTAvrBs3), and cloned these promoters in front of an uidA reporter gene (Fig. S2B and SI Text). The UPTAvrBs4 box was deduced using the TALE code and was cloned into a Bs3 promoter in which the UPTAvrBs3 box had been deleted. The promoter:uidA reporter constructs were delivered into Nicotiana benthamiana leaves in combination with the 35S promoter-driven TALE genes avrBs3*, avrBs3RVD4*, avrBs4*, and avrBs4RVD3*, respectively. GUS (β-glucuronidase) assays showed that the UPTAvrBs3 box-containing Bs3 promoter is transcriptionally activated only by AvrBs3* and AvrBs4RVD3*, whereas the UPTAvrBs4 box containing Bs3-promoter derivative is transcriptionally activated only by AvrBs4* and AvrBs3RVD4*, respectively (Fig. S2C). The chimeras showed in both cases a specificity that was equal to the Xanthomonas wild-type proteins with identical RVD composition. This finding indicates that, as long as the general structure of the 34-aa TALE repeat is used, DNA specificity is predominantly controlled by RVD rather than non-RVD residues. Quantitative GUS assays showed that the chimeric AvrBs4RVD3* and AvrBs3RVD4* proteins produced slightly lower GUS activity than the corresponding AvrBs3* and AvrBs4* proteins, respectively (Fig. S2D). These data suggest that certain non-RVDs may have coevolved with defined RVD residues and may contribute a minor role in DNA binding. However, overall we conclude that non-RVD residues have insufficient impact on the DNA-binding specificity of TALEs to warrant factoring them into the design of de novo-engineered TALEs.
De Novo-Engineered dTALEs Specifically Activate Promoters with Matching UPT Boxes.
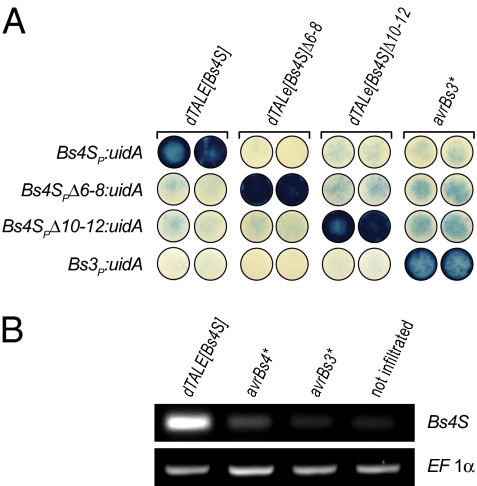
We were able to demonstrate that codon-optimized TALE genes have the expected biological function (Fig. S1) and that repeat-polymorphic non-RVDs have no significant impact on TALE DNA-binding specificity (Figs. S2 and S3). These findings suggested that dTALEs with desired specificity can be generated simply by modulation of TALE RVDs. As our first dTALE target promoter, we selected the well-studied tomato Bs4S promoter (Bs4SP), which is particularly well-suited for promoter-GUS studies because of its low basal activity (7). Thus far, no Xanthomonas TALE is known that transcriptionally activates Bs4SP. A dTALE, designated dTALE[Bs4S] (dTALE inducing the Bs4S promoter), was designed against a 19-bp sequence (TATATAACTTTGTCCAAAA; UPTdTALE[Bs4S] box) located 46-bp upstream of the ATG start codon of the tomato Bs4S coding sequence (13) (SI Text). The TALE code was used to translate the nucleotide sequence of the UPTdTALE[Bs4S] box into corresponding RVDs, and a dTALE comprised of the structural scaffold of AvrBs3 and the desired RVDs was engineered (SI Text). The function of dTALE[Bs4S] was tested by A. tumefaciens-mediated codelivery of 35S:dTALE[Bs4S] and a Bs4S promoter uidA reporter construct (Bs4SP:uidA) into N. benthamiana leaves. As hypothesized, the dTALE[Bs4S] but not the related AvrBs3* protein produced blue staining in combination with the Bs4SP:uidA, demonstrating the selective activation of the Bs4S promoter by dTALE[Bs4S] (Fig. 1A). To further test the specificity of dTALE[Bs4S] for the UPTTALE[Bs4S] box of the Bs4S promoter, we generated two promoter-deletion derivatives that lack either nucleotides 6 to 8 (Bs4SPΔ6–8) or nucleotides 10 to 12 (Bs4SPΔ10–12) of the UPTTALE[Bs4S] box and cloned these in front of an uidA reporter gene (Fig. S4). A. tumefaciens-mediated delivery of 35S:dTALE[Bs4S] with either Bs4SPΔ6–8:uidA or Bs4SPΔ10–12:uidA failed to induce the expression of the GUS reporter, indicating that dTALE[Bs4S] does indeed target the selected 19-bp motif present in the Bs4S promoter (Fig. 1A). In a reciprocal experiment, we also generated deletion derivatives of the dTALE[Bs4S] that lack repeat units 6 to 8 (dTALE[Bs4S]Δ6–8) or repeat units 10 to 12 (dTALE[Bs4S]Δ10–12). These units should be compatible with the Bs4S promoter-deletion derivatives Bs4SPΔ6–8 and Bs4SPΔ10–12, respectively (Fig. S4). Indeed, A. tumefaciens-mediated expression of dTALE[Bs4S]Δ6–8 and dTALE[Bs4S]Δ10–12 induced the GUS reporter only with the matching Bs4SPΔ6–8 and Bs4SPΔ10–12 promoter GUS constructs, respectively (Fig. 1A). In summary, these data demonstrate the high specificity of dTALE[Bs4S] and its derivatives for the selected target DNA sequence in the Bs4S promoter and the relative ease of designing highly sequence-specific transcriptional activators based on the TALE DNA-binding scaffold.
Fig. 1.
De novo-engineered dTALEs specifically activate promoters with matching UPT boxes. (A) In planta functional analysis of different TALE-promoter combinations. The uidA reporter constructs under transcriptional control of the promoters indicated at left were codelivered via A. tumefaciens into N. benthamiana leaves with the 35S promoter-driven TALE genes indicated above leaf discs. GUS assays were carried out 40 hpi. Leaf discs were stained with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, cyclohexylammonium salt (X-Gluc) to visualize activity of the GUS reporter. Asterisks (*) indicate TALE genes with optimized codon usage and corresponding gene products. (B) The dTALE[Bs4S] transcriptionally activates the endogenous Bs4S gene in tomato. Semiquantitative RT-PCR was carried out on RNA from tomato leaf tissue 36 h after A. tumefaciens-mediated delivery of the depicted 35S promoter-driven TALE genes. Isolated RNA was used for cDNA synthesis and semiquantitative RT-PCR was performed with primers specific for the tomato Bs4S gene. The constitutively expressed gene elongation factor 1α (EF1α) served as a normalization control.
To demonstrate that the dTALE[Bs4S] is also capable of transcriptionally activating the Bs4S endogene in tomato, 35S promoter-driven TALE genes dTALE[Bs4S], avrBs4*, and avrBs3* were delivered into tomato leaves via A. tumefaciens-mediated T-DNA transfer. Subsequent semiquantitative RT-PCR analysis demonstrated the transcriptional activation of the Bs4S endogene by the dTALE[Bs4S] in tomato leaves (Fig. 1B). In contrast, AvrBs4* and AvrBs3* did not transcriptionally activate the Bs4S endogene, consistent with previous studies done with AvrBs3 and AvrBs4 (14). These data are unique in demonstrating that a dTALE can be designed and delivered to effectively target a user-defined chromosomal gene.
Designer TALEs Specifically Activate Promoters of Targeted Arabidopsis thaliana Endogenes.
To further support our hypothesis that the TALE code can be used to create dTALEs with desired target specificity, we designed two additional dTALEs targeted to the A. thaliana genes EGL3 (17) and KNAT1 (18). We choose these as targets because previous studies had shown that constitutive overexpression of EGL3 or KNAT1 results in an increase of trichomes (19) or dramatically altered leaf morphology (18), respectively. Designer TALE[EGL3] and dTALE[KNAT1] were raised against distinct 19-bp sequences present in each of the EGL3 and KNAT1 promoters, respectively (SI Text). The nucleotide sequences of the UPTdTALE[EGL3] (TATATACAGTGTACACACA) and UPTdTALE[KNAT1] (TATATACCTAGTTCGTTTT) box were translated into corresponding RVDs and incorporated into the structural scaffold of AvrBs3 (SI Text).
To test functionality of these two dTALEs, 35S promoter-driven T-DNA constructs with C-terminal GFP tags were generated and tested via transient promoter-GUS reporter assays in N. benthamiana leaves. To avoid problems in the GUS assay because of potentially leaky A. thaliana EGL3 and KNAT1 promoters, we introduced the two UPT boxes into the Bs4S promoter yielding the Bs4S-promoter derivatives UPTdTALE[EGL3]inBs4SP:uidA and UPTdTALE[KNAT1]inBs4SP:uidA, respectively (Fig. S5). These Bs4S promoter derivatives were then codelivered with the 35S promoter-driven dTALE[EGL3] gene by A. tumefaciens into N. benthamiana leaves. GUS staining was apparent with the matching UPTdTALE[EGL3]inBs4SP but not with the UPTdTALE[KNAT1]inBs4SP promoter construct (Fig. S5C). Reciprocally, dTALE[KNAT1] produced GUS activity only in combination with the promoter containing the matching UPTdTALE[KNAT1] box and not with the promoter that contains the UPTdTALE[EGL3] box. These data show that each of the two dTALEs specifically activated the promoter containing the corresponding UPT box.
Next, dTALE[EGL3] and dTALE[KNAT1] were tested to see if they can function as transcriptional activators of A. thaliana endogenes. We generated stable transgenic lines of the A. thaliana ecotype Col-0 by introducing the 35S promoter driven dTALE[EGL3] and dTALE[KNAT1] genes by floral dipping (20). Putative transformants were selected on kanamycin-supplemented medium and microscopically screened for expression of GFP. Analysis of leaf tissue of the dTALE-GFP–expressing plants by semiquantitative RT-PCR showed that transgenic lines expressing dTALE[EGL3] or dTALE[KNAT1] have in comparison with a nontransgenic A. thaliana plant increased transcript levels of EGL3 and KNAT1, respectively (Fig. 2). However, the expected morphological changes were not detected, which might be because of counterselection against the potentially detrimental phenotypes in the recovered transgenic lines or a posttranscriptional regulatory process.
Fig. 2.
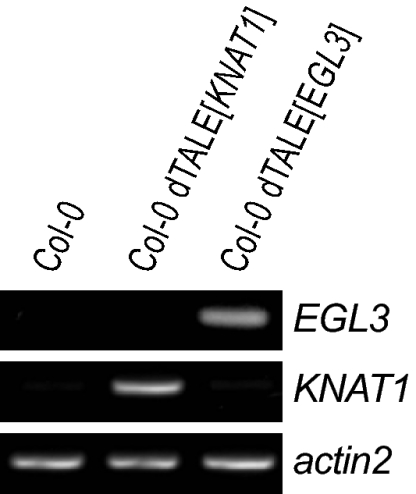
The dTALE[EGL3] and dTALE[KNAT1] transcriptionally activate the EGL3 and KNAT1 endogenes in A. thaliana, respectively. Semiquantitative RT-PCR was carried out on RNA from the Arabidopsis ecotype Columbia (Col-0) and corresponding lines containing 35S promoter-driven dTALE[EGL3] or dTALE[KNAT1] transgenes. RNA was isolated from leaves and used for cDNA synthesis. Semiquantitative RT-PCR was performed with EGL3 and KNAT1 specific primers. The constitutively expressed gene actin2 served as an internal normalization control.
Using the Patmatch algorithm (http://is.gd/g31n9), we scanned the Arabidopsis genome for sequences with homology to the boxes in the EGL3 and KNAT1 promoters that were targeted by our dTALEs. We identified no perfect match in the Arabidopsis genome but three sequences, AT4G27740, AT3G15640, and AT3G59220, and one sequence, AT1G77980, containing two mismatches from the given EGL3 and KNAT1 target sequences, respectively (Fig. S5D). RT-PCR analysis demonstrated that AT3G59220 was the only off-target site with increased transcript levels in the transgenic lines (Fig. S5D). These data imply that some mismatches strongly impair DNA recognition, whereas others seem to have no obvious effect and are in accordance with previous findings (10).
In summary, these data demonstrate that custom dTALEs reliably activate user-defined target genes. However, these data also demonstrate that potential off-sites need to be routinely analyzed, as with other DNA binding scaffolds.
De Novo-Engineered AvrBs3 Derivatives with Four Additional Repeat Units Specifically Activate Either the Bs3 or the UPA20 Promoter.
A central question in the design of dTALEs is the correlation between repeat number and DNA-binding specificity. We have used the 17.5 repeat-unit TALE AvrBs3 as a structural scaffold for our dTALEs. This repeat-unit number is most predominant among naturally occurring Xanthomonas strains, although TALE proteins do exist with a range of repeat-unit numbers, including some with more than 30 repeat units (4). We wished to examine how the number of repeat units in a given TALE correlates with its degree of target specificity.
To address this question, we made use of the pepper Bs3 and UPA20 promoters, two well-studied AvrBs3 target promoters (2, 3, 10, 21) that are almost identical within their UPTAvrBs3 boxes but that differ significantly up- and downstream of this sequence motif (Fig. S6). Based on the assumption that the addition of repeat units might alter the DNA-binding specificity of AvrBs3*, we extended AvrBs3* by four C-terminal repeat units with RVD motifs matching to either the Bs3 promoter (target box: UPTAvrBs3Bs3P) or the UPA20 promoter (target box: UPTAvrBs3UPA20P) resulting in the AvrBs3* derivatives AvrBs3+4RBs3* and AvrBs3+4RUPA20*, respectively (Fig. S6). To study targeting of the UPTAvrBs3Bs3P and the UPTAvrBs3UPA20P boxes in the same promoter context, we introduced both UPT boxes into the Bs4S promoter and fused these promoter constructs in front of the uidA reporter gene. A tumefaciens-mediated delivery of the 35S promoter-driven avrBs3+4RBs3* TALE gene in N. benthamiana leaves produced GUS activity only in combination with the promoter containing the matching UPTAvrBs3Bs3P box but not with the promoter containing the UPTAvrBs3UPA20P box (Fig. S6D). Reciprocally, the avrBs3+4RUPA20* TALE gene induced GUS expression only in combination with the promoter containing the matching UPTAvrBs3UPA20P box and not with the promoter containing the UPTAvrBs3Bs3P box (Fig. S6D). AvrBs3* induced both promoters containing either the UPTAvrBs3Bs3P or the UPTAvrBs3UPA20P box. To further test specificity of AvrBs3* and its derivatives, we also analyzed the rice Xa27 promoter, which was previously shown to be transcriptionally activated by the matching Xanthomonas oryzea pv. oryzae TALE AvrXa27 (22) but not by the Xcv TALE AvrBs3 (23). A. tumefaciens-mediated delivery of the 35S promoter-driven dTALE genes avrBs3+4RBs3* or avrBs3+4RUPA20* showed that neither of those produced GUS activity in combination with the rice Xa27 promoter uidA reporter construct (Fig. S6D).
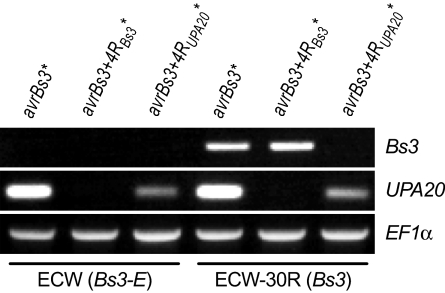
To analyze the specificity of the elongated TALEs on the UPA20 and Bs3 endogenes, we expressed the corresponding TALE genes via A. tumefaciens-mediated delivery in pepper. Semiquantitative RT-PCR demonstrated that expression of AvrBs3+4RBs3* resulted in increased Bs3 but not UPA20 transcript levels (Fig. 3). Reciprocally, expression of AvrBs3+4RUPA20* resulted in increased UPA20 but not Bs3 transcript levels. In contrast, expression of AvrBs3 resulted in increased levels of both Bs3 and UPA20 (Fig. 3).
Fig. 3.
AvrBs3 derivatives with four additional C-terminal repeat units discriminate between the pepper Bs3 and UPA20 promoter. The depicted 35S promoter-driven TALE genes were delivered via A. tumefaciens into the pepper genotypes ECW or ECW-30R. Semiquantitative reverse-transcription RT-PCR was carried out on RNA isolated from leaf material 36 h upon infection with primers specific for the Bs3 or UPA20 gene. The constitutively expressed gene EF1α served as an internal normalization control.
In summary, our data show that the addition of repeat units to the TALE protein AvrBs3 alters its target specificity.
RVD NK Mediates Specific Targeting of G Nucleotides.
Experimental analysis of TALEs and matching UPT boxes uncovered RVDs that target A, C, or T nucleotides, specifically (7). The RVD type NN binds to G and A nucleotides, but no RVD type was shown to target exclusively G nucleotides. Bioinformatic analysis of Xanthomonas TALEs and potential UPT boxes in the rice genome uncovered two interaction sites in which an NK type RVD aligned to a G nucleotide (6). To test if NK would bind G nucleotides in a specific manner, we made use of three tandem-arranged NI-type RVDs in the TALE AvrBs3 (repeat units 5–7) (Fig. S7 and SI Text) and the corresponding tandem-arranged A nucleotides in the UPTAvrBs3 box of the pepper Bs3 promoter. We generated Bs3-promoter derivatives in which the three A nucleotides were replaced by three C, G, or T nucleotides and fused these Bs3-promoter derivatives in front of the uidA reporter gene, generating UPTAvrBs3Bs3P:5–7C:uidA, UPTAvrBs3Bs3P:5–7G:uidA, and UPTAvrBs3Bs3P:5–7T:uidA, respectively (Fig. S7). Furthermore, we constructed an AvrBs3* derivative in which the three subsequent NI type RVDs were replaced by NK type RVDs, generating AvrBs3 5–7 NK*. Target specificity of AvrBs3 5–7 NK* was studied by A. tumefaciens-mediated delivery of the 35S promoter-driven avrBs3 5–7 NK* gene in combination with the different Bs3-promoter derivatives. We found that AvrBs3 5–7 NK* induced GUS expression only in combination with the Bs3-promoter derivative UPTAvrBs3Bs3P:5–7G:uidA that contains the three tandem-arranged G nucleotides (Fig. S8). In contrast, AvrBs3*, which contains three tandem-arranged NI-type RVDs, produced blue staining only in combination with the Bs3 wild-type promoter (Bs3P:uidA) that contains three corresponding tandem-arranged A nucleotides. In summary, our findings show that the NK-type RVD facilitates specific targeting of G nucleotides.
Discussion
RVD Composition of the Repeat Unit Array and dTALE Target Specificity.
In this article, we demonstrate that the TALE scaffold can be used for transcriptional activation of user-defined chromosomal loci. Target specificity is a central goal in the design and use of dTALEs, raising the question as to whether the AvrBs3 scaffold can facilitate targeting of single-copy sequences in complex genomes. Extensive screening has uncovered in total 18 AvrBs3-induced genes in the pepper genome (2, 3, 9, 21), which has an estimated genome size of 3 × 109 bp (24) and which is similar in size to the human genome (25). These findings may suggest that the AvrBs3 scaffold does not facilitate targeting of single-copy sequences in the human genome or genomes of comparable size. However, it needs to be noted that AvrBs3 contains three NS-type RVDs, which have been shown to target A, C, G, and T nucleotides with almost identical affinity (7). Hence, sequence-defined AvrBs3 scaffold-based dTALEs should have an ∼64-fold (43) higher target specificity than the Xanthomonas AvrBs3 protein if RVDs with ambiguous target specificity are replaced with RVDs with tight sequence specificity. Our finding, that the RVD NK specifically mediates targeting of G nucleotides (Fig. S8), is thus an important contribution for construction of dTALEs with increased sequence specificity, especially in the context of G-rich sequences.
Repeat-Unit Number and TALE Target Specificity.
In addition to RVD composition, we speculated that another important feature of dTALE target specificity is the number of repeat units in a given TALE or dTALE. This postulate is supported by our analysis of two distinct AvrBs3 derivatives with four additional repeat units that mediate specific targeting of the Bs3 and UPA20 promoters, although the wild-type AvrBs3 protein activates both promoters (Fig. 3). These data suggest that dTALEs with a high number of repeat units generally have superior target specificity, compared with dTALEs with fewer repeat units.
The modified target specificity of AvrBs3 derivatives with additional repeat units also raises the question of the molecular basis of this modified target specificity. Previous studies had shown that AvrBs3 physically interacts with and transcriptionally activates the pepper Bs3 and UPA20 promoters (2, 3). However, the two elongated TALEs that contain the complete repeat unit array of AvrBs3 specifically activate the Bs3 or the UPA20 promoters (Fig. 3), suggesting that these AvrBs3 derivatives bind specifically to either the Bs3 or the UPA20 promoter. We envision two molecular scenarios of why those AvrBs3-derivatives have a modified-promoter specificity compared with their progenitor AvrBs3. One possibility is that additional, nonmatching repeat units at the C terminus cause these terminal repeat units to “loop out,” which may hinder the C-terminal TALE activation domain to get in physical proximity and activate the given promoter. Alternatively, we envision that the TALE scaffold is rigid and, hence, the C-terminal, nonmatching RVDs would destabilize the structure and prevent the entire TALE from binding to a given target sequence. Further studies will have to clarify which model adequately explains our experimental findings.
TALE Repeat Units: A Promising Sequence-Specific Targeting Device for Nucleases and Other DNA-Modifying Enzymes.
Previous studies of Xanthomonas TALEs have shown that TALE-mediated promoter activation strictly correlates with physical interaction of the given promoter target sequences (2, 3, 8, 10, 21). Hence, dTALE repeat-unit arrays are likely to function as a sequence-specific targeting modules not only in the context of a transcription factor, but also when fused to other functional domains as, for example, nucleases, methylases, recombinases, or transcription-repressor domains. Most recently, two reports have indeed demonstrated that translational fusions of TALE proteins to the nonsequence-specific FokI nuclease result in sequence-specific TALE nucleases (TALENs) (26, 27), suggesting that TALENs might pose an alternative to the widely used zinc-finger nucleases (1).
The report by Christian et al. (26) described also custom-designed TALE repeat-unit arrays fused to FokI, targeting 12- and 13-bp sequences from the A. thaliana ADH1 or the zebrafish gridlock genes, respectively. The authors could show that homodimeric TALENs, which bind to two identical sequences from either the ADH1 or the gridlock gene, placed on opposite sides of a central spacer function in a sequence-specific manner. Although none of the two recent reports on TALENs demonstrated cleavage of chromosomal loci by TALENs, it seems possible that this goal can be achieved in the near future.
TALE DNA Binding Domain: A Promising Alternative for Zinc-Finger Arrays?
Our work on dTALEs and the most recent report on custom-designed TALENs (26) suggest that the TALE DNA-binding domain might pose an alternative to the currently used zinc-finger arrays. However, a major difference between tandem-arranged zinc fingers and TALE repeat units is that the latter modules seem to act in a context-independent manner (6). In consequence, tandem-arranged TALE repeat units have a highly predictable sequence specificity but zinc-finger arrays of desired specificity have to be identified from complex expression libraries. The laborious selection procedure for custom zinc-finger arrays and the more restrictive requirements for candidate-binding sites are limitations on the wide-spread use of this technology, which we do not expect to plague TALE scaffold proteins.
Materials and Methods
Generation of Codon-Optimized TALE Genes.
The avrBs3* gene, optimized for minimal sequence homology between the 102-bp repeats (for sequence, see SI Text), was synthesized by GenScript with a tobacco codon usage, and cloned into the MCS of pUC57 yielding pUC57 avrBs3*. To obtain the GATEWAY entry clone pENTR-D-avrBs3*, amplification was carried out with Phusion high-fidelity DNA polymerase (New England Biolabs), primers RM1 and RM2 (for primer sequences, see Table S1) and pUC57 avrBs3* as template DNA. SspI and FseI restriction sites flanking the repeat unit array were introduced into pENTR-D-avrBs3* via PCR using primers RM3 and RM4, yielding pENTR-D-avrBs3*-L. The repeat-arrays of avrBs4*, avrBs3RVD4*, and avrBs4RVD3* were synthesized in the same way as avrBs3* and cloned using PvuII and FseI into the SspI and FseI digested pENTR-D-avrBs3*-L yielding pENTR-D-avrBs4*, pENTR-D-avrBs3RVD4*, and pENTR-D-avrBs4RVD3*, respectively. To create pENTR-D-TAL[Bs4S], pENTR-D-TAL[EGL3], and pENTR-D-TAL[KNAT1], each repeat-array was synthesized partially from repeat number 6 to 17.5 flanked by XhoI and FseI restriction sites. These fragments were cloned into pENTR-D-avrBs3*-R6-XhoI, which was generated from pENTR-D-avrBs3* using primers RM4 and RM5. Repeat deletion derivatives of pENTR-D-TAL[Bs4S] were obtained using primers RM6 and RM7 or RM8 and RM9, resulting in pENTR-D-TAL[Bs4S]Δ6–8 or pENTR-D-TAL[Bs4S]Δ10–12, respectively. To generate pENTR-D-avrBs3*+4RBs3 and pENTR-D-avrBs3*+4Rupa20, an XhoI site was introduced into pENTR-D-avrBs3* using primers RM10 and RM11, yielding pENTR-D-avrBs3*-R17-XhoI. Terminal XhoI sites were introduced into synthesized four repeat-unit fragments with primers RM12 and RM13 or RM14 and RM15 and cloned via XhoI into pENTR-D-avrBs3*-R17-XhoI, yielding pENTR-D-avrBs3*+4RBs3 or pENTR-D-avrBs3*+4Rupa20, respectively. Mutations were introduced with the Phusion site directed mutagenesis kit (New England Biolabs). All entry clones were transferred by LR recombination (Invitrogen) into the expression vector pGWB5 (28) and transformed into A. tumefaciens GV3101 (29) for in planta analysis. Transgenic A. thaliana for dTALE[EGL3] and dTALE[KNAT1] were obtained as described previously (20).
Generation and Analysis of Promoter Mutants.
Sequences of all promoter constructs are provided in the SI Text. Insertion of different UPT boxes was done by Phusion site directed mutagenesis (New England Biolabs). Promoter constructs were transferred by LR recombination into the binary vector pGWB3 (28) and transformed into A. tumefaciens GV3101 (29) for in planta analysis. Analysis of promotors via semiquantitative RT-PCR and GUS measurements were carried out as described earlier (3, 10).
Supplementary Material
Acknowledgments
We thank D. Horvath, M. Bhakta, O. de Lange, and A. Strauß for helpful comments on the manuscript; T. Strauß, J. Elsaesser, B. Rosinsky, and M. Schulze for technical support; and T. Nakagawa (Shimane University, Matsue, Japan) for providing the pGWB vector series. Work in our laboratory has been supported by grants from the 2Blades Foundation, the Exzellenznetzwerk Biowissenschaften (Ministry of Culture of Saxonia-Anhalt), and the Deutsche Forschungsgemeinschaft (SPP1212, SFB 648, and LA1338/2-2).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013133107/-/DCSupplemental.
References
- 1.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 2.Kay S, Hahn S, Marois E, Hause G, Bonas U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science. 2007;318:648–651. doi: 10.1126/science.1144956. [DOI] [PubMed] [Google Scholar]
- 3.Römer P, et al. Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science. 2007;318:645–648. doi: 10.1126/science.1144958. [DOI] [PubMed] [Google Scholar]
- 4.Boch J, Bonas U. Xanthomonas AvrBs3 family-type III effectors: discovery and function. Annu Rev Phytopathol. 2010;48:419–436. doi: 10.1146/annurev-phyto-080508-081936. [DOI] [PubMed] [Google Scholar]
- 5.Bogdanove AJ, Schornack S, Lahaye T. TAL effectors: Finding plant genes for disease and defense. Curr Opin Plant Biol. 2010;13:394–401. doi: 10.1016/j.pbi.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Moscou MJ, Bogdanove AJ. A simple cipher governs DNA recognition by TAL effectors. Science. 2009;326:1501. doi: 10.1126/science.1178817. [DOI] [PubMed] [Google Scholar]
- 7.Boch J, et al. Breaking the code of DNA binding specificity of TAL-type III effectors. Science. 2009;326:1509–1512. doi: 10.1126/science.1178811. [DOI] [PubMed] [Google Scholar]
- 8.Römer P, et al. Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen, Xanthomonas oryzae pv. oryzae. New Phytol. 2010;187:1048–1057. doi: 10.1111/j.1469-8137.2010.03217.x. [DOI] [PubMed] [Google Scholar]
- 9.Marois E, Van den Ackerveken G, Bonas U. The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol Plant Microbe Interact. 2002;15:637–646. doi: 10.1094/MPMI.2002.15.7.637. [DOI] [PubMed] [Google Scholar]
- 10.Römer P, et al. Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 2009;150:1697–1712. doi: 10.1104/pp.109.139931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bonas U, Conrads-Strauch J, Balbo I. Resistance in tomato to Xanthomonas campestris pv. vesicatoria is determined by alleles of the pepper-specific avirulence gene avrBs3. Mol Gen Genet. 1993;238:261–269. doi: 10.1007/BF00279555. [DOI] [PubMed] [Google Scholar]
- 12.Stall RE, Jones JB, Minsavage GV. Durability of resistance in tomato and pepper to xanthomonads causing bacterial spot. Annu Rev Phytopathol. 2009;47:265–284. doi: 10.1146/annurev-phyto-080508-081752. [DOI] [PubMed] [Google Scholar]
- 13.Schornack S, et al. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 2004;37:46–60. doi: 10.1046/j.1365-313x.2003.01937.x. [DOI] [PubMed] [Google Scholar]
- 14.Schornack S, Peter K, Bonas U, Lahaye T. Expression levels of avrBs3-like genes affect recognition specificity in tomato Bs4- but not in pepper Bs3-mediated perception. Mol Plant Microbe Interact. 2005;18:1215–1225. doi: 10.1094/MPMI-18-1215. [DOI] [PubMed] [Google Scholar]
- 15.Bonas U, Stall RE, Staskawicz B. Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol Gen Genet. 1989;218:127–136. doi: 10.1007/BF00330575. [DOI] [PubMed] [Google Scholar]
- 16.Schornack S, Minsavage GV, Stall RE, Jones JB, Lahaye T. Characterization of AvrHah1, a novel AvrBs3-like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytol. 2008;179:546–556. doi: 10.1111/j.1469-8137.2008.02487.x. [DOI] [PubMed] [Google Scholar]
- 17.Koornneef M, Dellaert LW, van der Veen JH. EMS- and radiation-induced mutation frequencies at individual loci in Arabidopsis thaliana (L.) Heynh. Mutat Res. 1982;93:109–123. doi: 10.1016/0027-5107(82)90129-4. [DOI] [PubMed] [Google Scholar]
- 18.Lincoln C, Long J, Yamaguchi J, Serikawa K, Hake S. A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell. 1994;6:1859–1876. doi: 10.1105/tpc.6.12.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang F, Gonzalez A, Zhao M, Payne CT, Lloyd A. A network of redundant bHLH proteins functions in all TTG1-dependent pathways of Arabidopsis. Development. 2003;130:4859–4869. doi: 10.1242/dev.00681. [DOI] [PubMed] [Google Scholar]
- 20.Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 21.Kay S, Hahn S, Marois E, Wieduwild R, Bonas U. Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3Δrep16. Plant J. 2009;59:859–871. doi: 10.1111/j.1365-313X.2009.03922.x. [DOI] [PubMed] [Google Scholar]
- 22.Gu K, et al. R gene expression induced by a type-III effector triggers disease resistance in rice. Nature. 2005;435:1122–1125. doi: 10.1038/nature03630. [DOI] [PubMed] [Google Scholar]
- 23.Römer P, Recht S, Lahaye T. A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc Natl Acad Sci USA. 2009;106:20526–20531. doi: 10.1073/pnas.0908812106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Mol Biol Rep. 1991;9:208–219. [Google Scholar]
- 25.Consortium IHGS. International Human Genome Sequencing Consortium Finishing the euchromatic sequence of the human genome. Nature. 2004;431:931–945. doi: 10.1038/nature03001. [DOI] [PubMed] [Google Scholar]
- 26.Christian M, et al. TAL effector nucleases create targeted DNA double-strand breaks. Genetics. 2010;186:757–761. doi: 10.1534/genetics.110.120717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T, et al. TAL nucleases (TALNs): Hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq704. 10.1093/nar/gkq704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakagawa T, et al. Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng. 2007;104:34–41. doi: 10.1263/jbb.104.34. [DOI] [PubMed] [Google Scholar]
- 29.Koncz C, Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol Gen Genet. 1986;204:383–396. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.