Abstract
To cause rice blast disease, the fungus Magnaporthe oryzae breaches the tough outer cuticle of the rice leaf by using specialized infection structures called appressoria. These cells allow the fungus to invade the host plant and proliferate rapidly within leaf tissue. Here, we show that a unique NADPH-dependent genetic switch regulates plant infection in response to the changing nutritional and redox conditions encountered by the pathogen. The biosynthetic enzyme trehalose-6-phosphate synthase (Tps1) integrates control of glucose-6-phosphate metabolism and nitrogen source utilization by regulating the oxidative pentose phosphate pathway, the generation of NADPH, and the activity of nitrate reductase. We report that Tps1 directly binds to NADPH and, thereby, regulates a set of related transcriptional corepressors, comprising three proteins, Nmr1, Nmr2, and Nmr3, which can each bind NADP. Targeted deletion of any of the Nmr-encoding genes partially suppresses the nonpathogenic phenotype of a Δtps1 mutant. Tps1-dependent Nmr corepressors control the expression of a set of virulence-associated genes that are derepressed during appressorium-mediated plant infection. When considered together, these results suggest that initiation of rice blast disease by M. oryzae requires a regulatory mechanism involving an NADPH sensor protein, Tps1, a set of NADP-dependent transcriptional corepressors, and the nonconsuming interconversion of NADPH and NADP acting as signal transducer.
Keywords: fungal pathogenicity, ascomycete, cofactor
Rice blast disease represents a significant constraint on worldwide rice production, resulting in severe epidemics and overall global yield losses of 10–30% each year (1). Rice constitutes 23% of the calories consumed annually by the global human population, so understanding and controlling rice blast disease could play an important role in ensuring global food security in the future (2). To infect rice plants, the blast fungus Magnaporthe oryzae produces specialized infection cells called appressoria, which rupture the leaf cuticle and allow fungal hyphae to invade and colonize the host. The fungus is able to proliferate rapidly within rice cells, deriving nutrition from living tissue while evading or suppressing plant defenses (1). Understanding the regulatory mechanisms that allow the fungus to undergo these developmental transitions and to grow so effectively within its host may provide new means to control rice blast disease.
In this study, we set out to investigate the gene regulatory mechanisms that condition the ability of M. oryzae to respond to the nutrient status of its environment during plant infection—moving from the nutrient-free conditions of the rice leaf surface to the relatively nutrient-rich interior of the leaf. Previously, we observed a pivotal role for the biosynthetic enzyme trehalose-6-phosphate synthase (Tps1) in the regulation of carbon and nitrogen metabolism in M. oryzae (3, 4). Tps1 is required for production of the nonreducing disaccharide trehalose from glucose-6-phosphate (G6P) and uridine diphosphate (UDP)-glucose (3). Mutants lacking Tps1 are able to produce appressoria, but these cells do not function correctly and the fungus cannot colonize rice tissue (3). A Δtps1 gene deletion mutant does not produce trehalose, or the intermediate trehalose-6-phosphate (T6P), but by introducing mutations to the G6P binding pocket of Tps1, we showed that impairment of pathogenicity in M. oryzae by Δtps1 strains is not due simply to loss of trehalose biosynthesis. Introducing amino acid substitutions into Tps1, which prevent G6P binding, rendered the fungus nonpathogenic, whereas mutations that solely affected the catalytic activity of the enzyme did not affect its role in rice blast disease, suggesting that the inability of Δtps1 mutants to sense intracellular G6P may be associated with their loss of virulence (4).
Tps1 appears to integrate control of both carbon and nitrogen metabolism in M. oryzae because without the ability to sense G6P, Δtps1 mutants are unable to grow on nitrate as a sole source of nitrogen. Filamentous fungi carry out nitrate reduction to ammonium via the sequential reactions of nitrate reductase (NR) and nitrite reductase. NADPH provides the reducing power for the first step of this pathway and is a cofactor for NR. In M. oryzae, growth on nitrate normally results in increased hexokinase (Hxk1) activity and a rise in G6P production compared with growth on ammonium as nitrogen source (4). Because both G6P dehydrogenase (G6PDH) activity and NADPH levels are significantly reduced in Δtps1 mutants, we reasoned that sensing of G6P by Tps1 might lead to increased NADPH production in the oxidative pentose phosphate pathway (PPP) via activation of G6P dehydrogenase (G6PDH), thus ensuring that sufficient reducing power (in the form of NADPH) is available to NR to allow growth of the fungus on nitrate. Consistent with this idea, Δtps1 mutants are depleted for NADPH and lack the reducing power to metabolize nitrate as a nitrogen source (4). We could not, however, determine the precise mechanism that allowed Tps1 to exert such a profound effect on the establishment of rice blast disease based solely on its role as a metabolic regulator.
In this report, we show that the virulence-associated function of Tps1 is mediated by a family of NADP-dependent transcriptional corepressors, linking its metabolic function to a wider role in the control of gene expression. This transcriptional regulatory mechanism involves the interaction of up to three GATA-factor transcriptional activators with cognate NADP-binding corepressors and is necessary for regulation of a set of genes that are expressed during appressorium-mediated plant infection in M. oryzae. Furthermore, we demonstrate that Tps1 directly binds NADPH, consistent with the operation of a unique NADPH-dependent genetic switch that allows the rice blast fungus to regulate gene expression in response to rapidly fluctuating changes in nutrient and redox status during plant infection.
Results
Tps1 Regulates Nitrogen Source Utilization Through the Oxidative Pentose Phosphate Pathway.
We initially set out to investigate the mechanism by which M. oryzae Tps1 regulates nitrogen source utilization and to determine whether this was important for its role in rice blast disease. Tps1 is essential for M. oryzae to grow on nitrate as a sole nitrogen source and we therefore needed to establish whether nitrate metabolism was necessary for fungal virulence. Previous studies (5, 6) provided some evidence that the wide domain regulator of nitrogen source utilization was dispensable for virulence. To test this assumption in a more systematic manner, we independently generated targeted deletion mutants for the structural gene for nitrate reductase (NIA1), the pathway specific activator of NIA1 gene expression (NIR1), and the wide domain GATA factor (NUT1), as shown in Fig. S1. All of the mutants were still able to cause rice blast disease symptoms, although lesions were reduced in size compared with those caused by the isogenic wild-type strain Guy11. These results suggest that the role of Tps1 in regulating nitrate metabolism is not the primary reason for its significance in fungal pathogenicity.
Tps1 appears to exert its effect on nitrate reductase because of its function in G6P sensing by Tps1 and its modulation of NADPH production through the oxidative pentose phosphate pathway (PPP). In Δtps1 mutants, G6PDH activity is reduced during vegetative growth on nitrate compared with Guy11, which is accompanied by a significant reduction in NADPH levels (4). To investigate the relationship directly, we used quantitative real-time PCR (qPCR) and observed a 9-fold reduction in G6PDH gene expression in a Δtps1 mutant compared with the wild-type during growth on nitrate (Fig. S2A). Moreover, G6PDH was expressed 12-fold more highly in wild-type appressoria compared with conidia (Fig. S2B), suggesting that G6PDH levels might be particularly important during appressorium-mediated plant infection. To test this idea, we overexpressed the G6PDH gene in a Δtps1 mutant of M. oryzae. Using qPCR, we found that the M. oryzae β-tubulin–encoding gene was 60-fold more highly expressed than G6PDH in a Δtps1 mutant strain exposed to nitrate (Fig. S2C). We therefore fused the G6PDH gene coding sequence to the β-tubulin gene promoter and introduced this into a Δtps1 mutant. In a resulting transformant, G6PDH activity was increased 260.4 ± 5% when the fungus was switched to nitrate-containing medium.
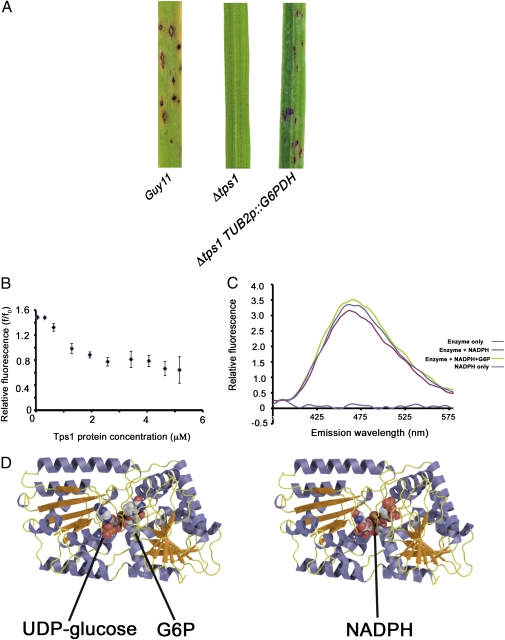
Interestingly, overexpression of G6PDH in a Δtps1 mutant background partially restored the ability to cause rice blast disease in a susceptible cultivar, CO-39, as shown in Fig. 1A. We conclude that elevation of G6PDH activity, which consumes G6P and generates NADPH, can at least partially compensate for the loss of trehalose-6-phosphate synthase in Δtps1 mutant of M. oryzae.
Fig. 1.
Tps1 controls G6PDH activity and binds NADPH. (A) Overexpression of the G6PDH gene under control of the β-tubulin gene promoter partially restores virulence to a Δtps1 mutant. Rice seedlings were inoculated with 1 × 104 spores·mL−1. (B) Tps1 binds NADPH. The intrinsic fluorescence of 2 μM NADPH was measured in solutions containing an increasing concentration of purified Tps1 protein. Excitation was at 340 nm; emission was measured at 465 nm. A decrease in fluorescence was observed with increasing Tps1 concentration consistent with removal of NADPH from solution by Tps1 protein binding. Assays were performed in triplicate. (C) Effect of NADPH fluorescence in the presence of TPS1 and G6P. Excitation was performed at 340 nm, and the emission spectra were recorded over wavelengths spanning 375 to 575 nm. Peak emission of NADPH fluorescence occurred at 465 nm. The reaction mixture contained 1 μM Tps1, 15 μM NADPH, and 15 μM G6P, when present. NADPH alone provided the base-line level of the free solution state fluorescence. Addition of Tps1 quenched the fluorescence of NADPH. The addition of G6P caused an increase in fluorescence, indicating that the NADPH is no longer bound and, therefore, no longer being quenched through the conformational fit of the ligand within the Tps1 binding site. (D) The homology structure of Tps1 reported in ref. 4 was used to model interactions of the native substrates, G6P and UDP-glucose (Left) and NADPH (Right) with Tps1. NADPH can be modeled into the active site of Tps1, adopting a similar position within the binding site to that seen for UDP-glucose and G6P. Calculated energies of binding for UDP-glucose (−19 kcal/mol), G6P (−19 kcal/mol) and NADPH (−27 kcal/mol) suggest that NADPH binding to Tps1 is reversible and competitive.
Tps1 Can Directly Bind NADPH.
As a result of the importance of G6PDH activity in the generation of NADPH, we wondered whether Tps1 might be directly regulated by the ratio of NADP/NADPH. We therefore purified recombinant Tps1 protein and titrated it against NADPH to establish whether Tps1 was able to bind the cofactor. A specific interaction between Tps1 and NADPH was detected, as monitored by a quenching of NADPH fluorescence at 465 nm in the presence of increasing concentrations of Tps1 (Fig. 1B). Furthermore, we found evidence that NADPH could be displaced from Tps1 by the presence of G6P because the Tps1-mediated reduction in NADPH fluorescence could be reversed by the addition of excess G6P, as shown in Fig. 1C. We investigated the specificity of NADPH binding in more detail by measuring the intrinsic tryptophan fluorescence of Tps1 in the presence or absence of NAPDH. Fig. S3A demonstrates the tryptophan quenching occurs in the active site of Tps1 in the presence of NADPH. From these data, the calculated Hill coefficient, used to determine the dissociation constant for a substrate bound to a macromolecule, gave a binding constant of 3.96 ± 0.06 μM and a Hill slope of 5.18 ± 0.35. This value is consistent with cooperative binding and demonstrates that binding of NADPH by Tps1 is a specific interaction. The high Hill coefficient for Tps1 and NADPH also implies that although only one molecule is present in the active site of each protein, NADPH binding might promote higher-order protein conformations, such as Tps1 tetramers. Using our modeled structure of Tps1, based on the solution structure of the bacterial Tps1 homolog otsA (4), we sought a structural explanation of the interaction of NADPH with Tps1. The native substrates of Tps1, UDP-glucose, and G6P, when modeled into the Tps1 active site, have calculated energies of binding of −19 and −14 kcal/mol, respectively, as shown in Fig. 1D. However, NADPH can also be modeled into the active site with a calculated energy of binding of −27 kcal/mol (Fig. 1D). The structural similarity between NADPH and the native Tps1 substrates, coupled with the similarity in predicted energies of binding, is consistent with a mechanism whereby NADPH can reversibly displace G6P and UDP-glucose from the Tps1 active site to inhibit Tps1 activity during conditions of elevated NADPH levels. Conversely, when NADPH is consumed by NADPH-requiring enzymes (such as NR), G6P and UDP-glucose can displace NADPH at the active site and, through G6P sensing, activate NADPH production by G6PDH. This process is consistent with a feedback loop model controlling Tps1-dependent G6PDH activity in response to available G6P and NADPH.
Identification and Functional Characterization of NADP-Dependent Nmr Corepressors.
To investigate how a Tps1/NADPH-dependent regulatory process might regulate fungal virulence, we next investigated how Tps1 regulates gene expression. Δtps1 mutants do not express the NR-encoding gene NIA1 when exposed to nitrate and instead adopt an ammonium-responsive status (including the expression of genes encoding ammonium transporters), irrespective of the nitrogen source to which the fungus is exposed (Fig. S4A). In filamentous fungi, such as Aspergillus nidulans and Neurospora crassa, the presence of ammonium represses the expression of genes encoding enzymes to metabolize alternative nitrogen sources (7). This is due in part to a repressor protein NmrA/Nmr1, which prevents the activity of a GATA-factor transcription factor, AreA/Nit2, the central regulator of gene expression when ammonium is depleted (4). We therefore reasoned that in M. oryzae, Tps1-dependent transcriptional regulation of NIA1 is likely to be mediated via control of the equivalent areA/Nit2-like transcription factor (TF), Nut1, by the repressor Nmr1 (ref. 4 and Fig. S4A). In contrast to previously characterized free-living fungal species, however, the genome of M. oryzae has two additional homologs of NMR1 (NMR2 and NMR3) (8), as shown in Fig. S4B. Low stringency yeast two-hybrid analysis, using β-galactosidase activity as the detection assay, provided evidence of a physical interaction between Nmr1 and Nmr3 with Nut1, suggesting that two of the M. oryzae Nmr proteins are functional homologs of NMRA (Fig. 2). To test the role of these genes in Tps1 regulation, we deleted NMR1, NMR2, and NMR3 in a Δtps1 mutant background. Strikingly, targeted deletion of NMR1, NMR2, or NMR3 all partially restored the ability of the Δtps1 mutant to cause rice blast disease (Fig. 2). No significant differences in lesion number were observed between leaves inoculated with the wild-type and Δtps1 Δnmr suppressor strains (Fig. 2B), but relative lesion sizes were reduced in Δtps1 Δnmr1 (mean relative lesion size = 49 ± 22 pixel2, n = 12 leaves), Δtps1 Δnmr2 (30 ± 19 pixel2, n = 13), and Δtps1 Δnmr3 (19 ± 7 pixel2, n = 25) compared with Guy11 (109 ± 33 pixel2, n = 12). By contrast, deletion of the NMR genes singly, or together, in a wild-type strain did not significantly affect fungal pathogenicity (Fig. S4C). Deletion of the Nmr genes also resulted in partial derepression of nitrate reductase activity in wild-type and Δtps1 mutant (see Fig. S4D and Fig. S4E) and partial remediation of sporulation of Δtps1 strains on nitrate media (Fig. S4F). However, although partially remediated for virulence, Δtps1 Δnmr3 suppressor strains were not restored for sporulation on nitrate media (Fig. S4F). This observation indicates that the regulation of virulence-associated gene expression by Nmr proteins is both complex and independent of nitrate utilization.
Fig. 2.
Fungal virulence is controlled by Tps1 via the Nmr corepessor proteins. (A) Partial restoration of virulence to Δtps1Δnmr1, Δtps1Δnmr2, and Δtps1Δnmr3 double mutants of M. oryzae. Rice seedlings were inoculated with 1 × 104 spores·mL−1. (B) Bar charts of lesion densities of Guy11, Δtps1, Δtps1Δnmr1, Δtps1Δnmr2, and Δtps1Δnmr3 mutants. Results are the average of three independent replicates, and error bars are the SD. Bars with the same letter are not significantly different (Student's t test, P ≤ 0.05). (C) Low-stringency yeast two-hybrid analysis of physical interactions among Nut1, Nmr1, Nmr2, Nmr3, and Tps1 compared with the empty vector controls. –BD are strains carrying an empty GAL4 DNA binding domain vector. –AD are strains carrying an empty GAL4 activation domain vector. X-Gal is supplemented in the media, and low stringency protein–protein interactions result in β-galactosidase gene expression and formation of blue colonies. (D) Rice infections were carried out with Δtps1, Δtps1Δnmr1, Δtps1Δnmr1:NMR1, Δtps1Δnmr1:NMR1T13T, and Δtps1Δnmr1:NMR1T13V. Expression of the NMR1T13V allele does not complement the Δtps1Δnmr1 mutant phenotype. Seedlings were inoculated with 1 × 104 spores·mL−1.
We can therefore conclude that the three Nmr proteins act as corepressors to control virulence-associated gene expression downstream of Tps1. Low-stringency yeast two-hybrid studies did not detect a physical interaction between the Nmr proteins and Tps1 (Fig. 2C), suggesting that direct modulation of Nmr activity by Tps1 is unlikely. However, each Nmr protein possesses a putative NADP-binding Rossman fold, which has been shown in A. nidulans NMRA to bind NADP (9) (Fig. S4B). We therefore investigated the role of NADP binding in Nmr function and Tps1 regulation in M. oryzae. Focusing on Nmr1, we found that introducing a native copy of NMR1 into a Δtps1 Δnmr1 double mutant complemented the Δnmr1 gene deletion, as expected, and resulted in strains that, like Δtps1 single mutants, were nonpathogenic (Fig. 2). However, we also constructed a mutant allele of NMR1 encoding a T13V substitution. This change has been shown in A. nidulans NmrA and Dictyostelium discoideum PadA to impair NADP binding (10, 11). Expression of the NMR1T13V allele in a Δtps1 Δnmr1 double mutant did not restore the Δtps1 nonpathogenic phenotype (Fig. 2D). We conclude that the nonpathogenic mutant phenotype of Δtps1 mutants is associated with the role of Nmr1, Nmr2, and Nmr3 in repressing virulence-associated gene expression. We can also infer that the function of Nmr1 to carry out this role depends on its ability to bind NADP.
Tps1-Dependent Gene Expression Is Modulated by the Presence of Nmr Corepressors.
To understand how Tps1 might control virulence-associated gene expression during plant infection, we carried out comparative gene expression analysis of the wild-type, Δtps1 mutant and the Δtps1Δnmr1, Δtps1Δnmr2, and Δtps1Δnmr3 suppressor strains. During vegetative growth on nitrate compared with ammonium, we found that two known virulence genes MPG1 and ALB1 (12, 13), six genes encoding putative NADPH-dependent enzymes (NIA1, ALD1, KEF1, SDY1, OXD1, and OYE1), and a putative metalloprotease (MPT1) were expressed in a Tps1-dependent manner in response to nitrate (Fig. 3 A and B, Table S1, and Fig. S3A). Expression of these genes was at least partially restored in the Δtps1Δnmr1, Δtps1Δnmr2, or Δtps1Δnmr3 suppressor strains compared with the Δtps1 mutant, suggesting they are controlled via NADP-dependent Nmr inhibition of transcription (Fig. 3C). In addition, we found that G6PDH gene expression was also elevated in the Δtps1Δnmr1, Δtps1Δnmr2, and Δtps1Δnmr3 suppressor strains, compared with the Δtps1 mutant, during growth on nitrate (Fig. 3D). G6PDH and a subset of the genes analyzed in Fig. 3C were elevated in expression during appressorium development, consistent with the operation of this regulatory mechanism during plant infection (Fig. S2B and Fig. 3E). Moreover, some genes, like ALB1 and G6PDH, were derepressed in the Δtps1Δnmr1, Δtps1Δnmr2, and Δtps1Δnmr3 suppressor strains compared with Δtps1, whereas other genes are derepressed in some suppressor strains but not others. The expression of ALD1, for instance, was only affected in the Δtps1Δnmr2 mutant. This result suggests that Nmr corepressors may work either individually or cooperatively to regulate gene expression, which may result from their regulation of additional GATA factors. Consistent with this idea, a subset of the genes was expressed in a NUT1-dependent manner (Fig. 3A), whereas others were expressed independently of NUT1 (Fig. 3B), indicating that the Nmr proteins may regulate other GATA family transcription factors. To test this idea, we carried out high stringency yeast two-hybrid experiments to determine whether Nmr proteins can interact with other GATA family transcription factors including Asd4, a putative homolog of a Neurospora crassa morphogenetic regulator (14); Pas1, a GATA factor containing a pas-fold domain involved in redox (15) and/or light sensing, which is a putative homolog of white collar-2 from N. crassa (8), and Sre1, a GATA factor required for siderophore metabolism (16). We found evidence that the Nmr1, Nmr2, and Nmr3 can physically interact with Asd4, whereas Nmr2 interacts with Pas1, as shown in Fig. 4A. No interaction was observed with Sre1. Based on their potential interaction with Nmr corepressors, we decided to investigate the function of ASD4 and PAS1. We carried out targeted gene replacements and found that Δasd4 mutants displayed reduced hyphal growth (Fig. S5A) and sporulation. By contrast, Δpas1 mutants showed enhanced conidiation (Fig. S5B). Significantly, Δasd4 mutants were unable to cause rice blast disease (Fig. 4B), which was associated with their inability to form appressoria (Fig. S5C).
Fig. 3.
qPCR analysis of TPS1-dependent gene expression. Genes encoding putative NADP(H)-dependent enzymes (NIA1, ALD1, KEF1, SDY1, OXD1, and OYE1), known virulence-associated genes (MPG1 and ALB1), a putative metalloprotease (MPT1), and glucose-6-phosphate dehydrogenase (G6PDH) (for details, see Table S1) were analyzed for expression. Strains were grown in CM for 48 h followed by 16 h of growth in MM containing either 10 mM nitrate (filled bars) or 10 mM ammonium (open bars) as the sole nitrogen source. Gene expression results were normalized against expression of the β-tubulin gene (TUB2). Results are the average of at least three independent replicates, and error bars are the SD. (A) Bar charts showing gene expression in Guy11, Δtps1, and Δnut1 mutant. ALD1, KEF1, SDY1, OXD1, and ALB1 are nitrate-inducible and dependent on the presence of TPS1 and NUT1. Expression is relative to gene expression in the wild-type grown on nitrate. (B) Bar charts showing gene expression in Guy11, Δtps1, and Δnut1 mutant. MPT1, MPG1, and OYE1 appear nitrate-inducible and TPS1-dependent, but independent of NUT1. Expression is relative to gene expression in the wild type grown on nitrate. (C) Bar charts showing that TPS1-dependent gene expression is partially restored in a Δtps1 mutant by introduction of Δnmr1, Δnmr2, or Δnmr3 gene deletions. Expression is relative to gene expression in Δtps1 strains grown on nitrate. (D) Bar charts showing that G6PDH gene expression is partially restored in a Δtps1 mutant by introduction of Δnmr1, Δnmr2, or Δnmr3 gene deletions. (E) Bar charts showing gene expression of all nine genes in conidia (black bars) and appressoria (gray bars) of Guy11. Expression is relative to TUB2 gene expression.
Fig. 4.
A unique NADP(H)-dependent genetic switch is essential for fungal virulence in the rice blast fungus. (A) The Nmr repressors interact with three GATA transcription factors in M. oryzae. High stringency yeast two-hybrid analysis of physical interactions between Nut1, Asd4, Pas1, and Sre1 with Nmr1, Nmr2, and Nmr3. –BD are strains carrying an empty GAL4 DNA-binding domain vector. –AD are strains carrying an empty GAL4 activation domain vector. High stringency selection was on media lacking adenine, histidine, tryptophan, and leucine. (B) Role of the GATA-factors Pas1 and Asd4 in pathogenicity. Δpas1 strains are fully pathogenic, whereas Δasd4 strains are unable to infect host tissue. Rice seedlings were inoculated with 1 × 105 spores·mL−1. (C) A model for the action of Tps1 in virulence of M. oryzae. In response to G6P sensing, Tps1 activates G6PDH to produce NADPH from G6P and NADP in the oxidative pentose phosphate pathway. NADPH can bind to Tps1 to maintain a balance between G6P consumption and NADPH production. The conversion of NADP to NADPH inactivates the Nmr inhibitor proteins and results in the derepression of genes encoding at least two known virulence factors, at least six NADPH-dependent proteins, and G6PDH. NADPH is subsequently consumed by NADPH-dependent proteins, such as NR. Under starvation conditions, such as found during appressorium formation on the surface of the leaf, the homeostatic balance between G6P consumption and NADPH production is not maintained, NADP levels are elevated, and the Nmr inhibitor proteins become activated leading to repression of the genes required for virulence. Proteins are indicated as black circles. TF, transcription factor(s). The corresponding genes are shown in red. UDP-glc, UDP-glucose.
We conclude that Tps1 affects virulence-associated gene expression via its modulation of a set of NADP-dependent transcriptional repressors, which target GATA factors implicated in both fungal development and pathogenicity.
Discussion
In this study we set out to understand the role of trehalose-6-phosphate synthase as a key regulator in the establishment of rice blast disease (4). When considered together, our results are consistent with a role for Tps1 as part of an NADPH-dependent genetic switch in M. oryzae, which is essential for virulence and integrates the intracellular monitoring of nutritional and redox status with control of fungal gene expression. At the heart of this regulatory mechanism is the nonconsuming interconversion of NADPH and NADP, which bind to Tps1 and the Nmr transcriptional corepressor proteins, respectively, as shown in the model presented in Fig. 4C. Nmr proteins interact with at least three GATA family transcription factors to regulate expression of genes necessary for virulence, in addition to a number of genes encoding putative NADPH-dependent enzymes. The switch has two distinct modes, an “on” and a default “off” status, depending on the nutrient condition of the fungal cell. When G6P is available and the NADPH/NADP ratio is high, the switch is on and dynamically links G6P availability to gene expression with (i) NADPH depletion by enzymes such as NR being balanced by Tps1-dependent NADPH production in response to G6P sensing, (ii) NADPH production and G6P consumption maintained in equilibrium by competition for Tps1-binding, and (iii) the expression of virulence-associated genes and genes encoding NADPH-requiring enzymes, such as NR, induced only under NADPH replete conditions due to inactivation of the Nmr corepressors resulting from low levels of NADP. Trehalose would also be produced as a storage compound under these conditions. However, when G6P levels and the NADPH/NADP ratio are low, the system cannot maintain dynamic equilibrium. Tps1 becomes inactive because it cannot bind either NADPH or its native substrates. G6PDH activity and gene expression is reduced, and the default off position of the pathway is therefore NADP activation of the Nmr corepressor proteins. Further feedback to the system is provided by the likely control of G6PDH gene expression exerted by the GATA factor/Nmr transcriptional regulators. The default off position is also seen in Δtps1 mutants, because the Nmr proteins are constitutively active, repressing virulence-associated gene expression.
The involvement of dinucleotide cofactors in transcriptional regulation has not been widely reported, but it was recently predicted from a study in Saccharomyces cerevisiae where NAD(P) was shown to have the potential to act as a cofactor in the interaction between Gal80p and Gal4p during induction of galactose metabolism (17). In mammals, meanwhile, a family of histone deacetylases, the sirtuins, are involved in regulation of diverse cellular processes and appear to depend on NAD for their activity (reviewed in ref. 18), ensuring a tight link between sirtuin activity and the underlying metabolic status of the cell. However, the results presented here suggest that M. oryzae Tps1 acts as part of an NADP(H) homeostatic model, where the expression of genes encoding NADP(H)-requiring enzymes directly influences NADPH production and corresponding gene expression. We speculate that this adaptability to fluctuating nutritional environments may be key to the success of plant pathogens such as M. oryzae, which must quickly adapt from nutrient-free leaf surfaces to intracellular propagation within living plant tissue. Once in the plant, the fungus must, for instance, undergo rapid genetic reprogramming to elaborate specialized invasive hyphae from the thin penetration hyphae and evade or suppress the host defense response to establish disease (19). The Tps1 genetic switch described in this study may be responsible for postpenetration genetic reprogramming because Δtps1 strains form appressoria, but these cells are unable to develop into invasive hyphae to establish disease (3). Significantly, T6P synthases have been shown to have important roles in developmental biology in an increasing range of organisms, being necessary for plant embryonic development, flowering, and other morphogenetic processes (20). The genetic mechanism defined in this study may therefore be a more widespread mechanism for integrating metabolic and genetic regulation during eukaryotic development.
Materials and Methods
Fungal Strains, Growth Conditions, and DNA Analysis.
M. oryzae strains used in this study are derived from Guy11 (1, 3, 4). Standard procedures of M. oryzae growth, maintenance, appressorium formation, and transformation were performed, as described (12). Rice plant infections were performed as described (12). DNA and RNA extractions were as described in ref. 4. Gel electrophoresis, restriction enzyme digestions, DNA ligations, and PCR were performed by using standard procedures (21).
Transcript and Protein Analysis.
cDNA synthesis and qRT-PCR was performed as described in SI Materials and Methods. Yeast two-hybrid analysis was performed by using the Matchmaker GAL4 Two-Hybrid System 3 kit from Clontech. Purified Tps1 recombinant protein was obtained by using the Arcticexpress (Stratagene) gene expression system. Whole-cell protein extraction and nitrate reductase assays were performed as described (4). Activities were determined on a Pharmacia Biotech spectrophotometer in triplicate. All assay components were purchased from Sigma, except NADPH (Calbiochem). NADPH fluorescence was recorded on a 200 series microplate reader (Tecan Group). Protein modeling of M. oryzae Tps1 was performed as described in SI Materials and Methods. Statistics were performed by using Student's t test. Protein sequence alignments were performed by using ClustalW and illustrated using BoxShade.
Generation of M. oryzae Gene Deletion Mutants.
Targeted gene replacement of M. oryzae genes were performed based on the split marker strategy (22), as described in SI Materials and Methods and Fig. S4. M. oryzae gene sequence information was acquired from www.broad.mit.edu/annotation/genome/magnaporthe_grisea/MultiHome.html. For full experimental protocols, see SI Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by a grant to N.J.T. from the Biotechnology and Biological Sciences Research Council. C.F.Q. was supported by a graduate student assistantship from the Department of Plant Pathology, University of Nebraska, Lincoln, NE.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1006839107/-/DCSupplemental.
References
- 1.Wilson RA, Talbot NJ. Under pressure: Investigating the biology of plant infection by Magnaporthe oryzae. Nat Rev Microbiol. 2009;7:185–195. doi: 10.1038/nrmicro2032. [DOI] [PubMed] [Google Scholar]
- 2.Pennisi E. Armed and dangerous. Science. 2010;327:804–805. doi: 10.1126/science.327.5967.804. [DOI] [PubMed] [Google Scholar]
- 3.Foster AJ, Jenkinson JM, Talbot NJ. Trehalose synthesis and metabolism are required at different stages of plant infection by Magnaporthe grisea. EMBO J. 2003;22:225–235. doi: 10.1093/emboj/cdg018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson RA, et al. Tps1 regulates the pentose phosphate pathway, nitrogen metabolism and fungal virulence. EMBO J. 2007;26:3673–3685. doi: 10.1038/sj.emboj.7601795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Froeliger EH, Carpenter BE. NUT1, a major nitrogen regulatory gene in Magnaporthe grisea, is dispensable for pathogenicity. Mol Gen Genet. 1996;251:647–656. doi: 10.1007/BF02174113. [DOI] [PubMed] [Google Scholar]
- 6.Lau G, Hamer JE. Regulatory genes controlling MPG1 expression and pathogenicity in the rice blast fungus Magnaporthe grisea. Plant Cell. 1996;8:771–781. doi: 10.1105/tpc.8.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilson RA, Arst HN., Jr Mutational analysis of AREA, a transcriptional activator mediating nitrogen metabolite repression in Aspergillus nidulans and a member of the “streetwise” GATA family of transcription factors. Microbiol Mol Biol Rev. 1998;62:586–596. doi: 10.1128/mmbr.62.3.586-596.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dean RA, et al. The genome sequence of the rice blast fungus Magnaporthe grisea. Nature. 2005;434:980–986. doi: 10.1038/nature03449. [DOI] [PubMed] [Google Scholar]
- 9.Lamb HK, et al. The negative transcriptional regulator NmrA discriminates between oxidized and reduced dinucleotides. J Biol Chem. 2003;278:32107–32114. doi: 10.1074/jbc.M304104200. [DOI] [PubMed] [Google Scholar]
- 10.Lamb HK, et al. Modulation of the ligand binding properties of the transcription repressor NmrA by GATA-containing DNA and site-directed mutagenesis. Protein Sci. 2004;13:3127–3138. doi: 10.1110/ps.04958904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Núñez-Corcuera B, Serafimidis I, Arias-Palomo E, Rivera-Calzada A, Suarez T. A new protein carrying an NmrA-like domain is required for cell differentiation and development in Dictyostelium discoideum. Dev Biol. 2008;321:331–342. doi: 10.1016/j.ydbio.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 12.Talbot NJ, Ebbole DJ, Hamer JE. Identification and characterization of MPG1, a gene involved in pathogenicity from the rice blast fungus Magnaporthe grisea. Plant Cell. 1993;5:1575–1590. doi: 10.1105/tpc.5.11.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chumley FG, Valent B. Genetic analysis of melanin deficient non-pathogenic mutants of Magnaporthe grisea. Mol Plant Microbe Interact. 1990;3:135–143. [Google Scholar]
- 14.Feng B, Haas H, Marzluf GA. ASD4, a new GATA factor of Neurospora crassa, displays sequence-specific DNA binding and functions in ascus and ascospore development. Biochemistry. 2000;39:11065–11073. doi: 10.1021/bi000886j. [DOI] [PubMed] [Google Scholar]
- 15.Zhulin IB, Taylor BL, Dixon R. PAS domain S-boxes in Archaea, Bacteria and sensors for oxygen and redox. Trends Biochem Sci. 1997;22:331–333. doi: 10.1016/s0968-0004(97)01110-9. [DOI] [PubMed] [Google Scholar]
- 16.Haas H, Angermayr K, Stöffler G. Molecular analysis of a Penicillium chrysogenum GATA factor encoding gene (sreP) exhibiting significant homology to the Ustilago maydis urbs1 gene. Gene. 1997;184:33–37. doi: 10.1016/s0378-1119(96)00570-7. [DOI] [PubMed] [Google Scholar]
- 17.Kumar PR, Yu Y, Sternglanz R, Johnston SA, Joshua-Tor L. NADP regulates the yeast GAL induction system. Science. 2008;319:1090–1092. doi: 10.1126/science.1151903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finkel T, Deng CX, Mostoslavsky R. Recent progress in the biology and physiology of sirtuins. Nature. 2009;460:587–591. doi: 10.1038/nature08197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kankanala P, Czymmek K, Valent B. Roles for rice membrane dynamics and plasmodesmata during biotrophic invasion by the blast fungus. Plant Cell. 2007;19:706–724. doi: 10.1105/tpc.106.046300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eastmond PJ, et al. Trehalose-6-phosphate synthase 1, which catalyses the first step in trehalose synthesis, is essential for Arabidopsis embryo maturation. Plant J. 2002;29:225–235. doi: 10.1046/j.1365-313x.2002.01220.x. [DOI] [PubMed] [Google Scholar]
- 21.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1989. [Google Scholar]
- 22.Kershaw MJ, Talbot NJ. Genome-wide functional analysis reveals that infection-associated fungal autophagy is necessary for rice blast disease. Proc Natl Acad Sci USA. 2009;106:15967–15972. doi: 10.1073/pnas.0901477106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.