Abstract
Oligodendrocyte precursor cells (OPCs) are lineage-restricted progenitors generally limited in vivo to producing oligodendrocytes. Mechanisms controlling genesis of OPCs are of interest because of their importance in myelin development and their potential for regenerative therapies in multiple sclerosis and dysmyelinating syndromes. We show here that the SoxE transcription factors (comprising Sox8, 9, and 10) induce multipotent neural precursor cells (NPCs) from the early postnatal subventricular zone (SVZ) to become OPCs in an autonomous manner. We performed a chromatin immunoprecipitation-based bioinformatic screen and identified Suppressor of Fused (Sufu) as a direct target of repression by Sox10. In vitro, overexpression of Sufu blocked OPC production, whereas RNAi-mediated inhibition augmented OPC production. Furthermore, mice heterozygous for Sufu have increased numbers of OPCs in the telencephalon during development. We conclude that Sox10 acts to restrict the potential of NPCs toward the oligodendrocyte lineage in part by regulating the expression of Sufu.
Neural tube patterning is controlled by the interplay between homeobox genes and basic helix–loop–helix (bHLH) transcription factor families regulating cell fate, positional identity, and the secreted morphogens that regulate them (1, 2). The transcriptional regulation of oligodendrocyte development is of increasing interest because they are the primary sources of CNS myelin and a target of attack in multiple sclerosis (3). Early oligodendrocytes are produced from the ventral progenitor domain in the spinal cord that also produces cholinergic motor neurons, and from a ventral telencephalic domain that produces GABAergic interneurons and cholinergic projection neurons (3), whereas later waves of oligodendrocytes are specified from the dorsal neural tube (both in the spinal cord and cortex) (3, 4). Thus, progenitor cells from several distinct dorsal-ventral domains have the ability to adopt oligodendrocyte fate despite dramatic differences in regional specification by morphogenic signals.
Two transcription factor families: the bHLH proteins Olig1 and 2 (5–9) and the SoxE family of transcription factors are implicated in the transcriptional control of oligodendrocyte development. Sox proteins contain a high-mobility–group (HMG) domain essential for interaction with and recruitment of other transcription factors (10, 11) and have a fundamental role in oligodendrocyte genesis. Upon commitment to the oligodendrocyte lineage, expression of Sox1–3 is turned off and Sox8, 9, and 10 (classified as the SoxE group) are induced. Sox9 is important for oligodendrocyte specification, because there are reduced oligodendrocytes and increased numbers of motor neurons in the Sox9−/− ventral spinal cord (12). Sox10 is only expressed by OPCs and oligodendrocytes in the CNS (13) and essential for the generation of mature oligodendrocytes (14, 15). Overexpression of Sox10 in the chick spinal cord induces ectopic oligodendrocytes (16); furthermore, neurospheres isolated from Sox10 mutants do not myelinate axons after injection into the brains of dysmyelinating mice (15). Sox10 also directly regulates the expression of myelin basic protein cooperatively with Olig1 (17) and may regulate the expression of PDGFRα, another marker of the oligodendrocyte lineage (18). Thus, SoxE transcription factors are superb candidates for regulating oligodendrocyte fate (19) but whether they work to instructively induce oligodendrocyte fate and restrict the ability of multipotential cells to produce alternate fates remains unclear.
Here, we demonstrate that expression of SoxE transcription factors drives multipotential neural precursor cells (NPCs) to become oligodendrocyte precursor cells (OPCs) in vitro. Moreover, an important Sox10 transcriptional effect is decreased expression of Sufu, a newly identified modulator of oligodendrocyte production.
Results
Expression of Sox8, 9, and 10 Induce Oligodendrocyte Lineage Markers in Subventricular Zone (SVZ) Progenitors.
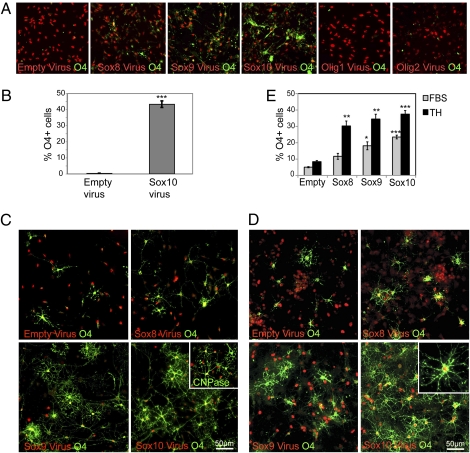
Multipotential NPCs from the postnatal cortical SVZ produce OPCs in vivo (20). These progenitors should pass through a similar OPC-like state during differentiation into oligodendrocytes in vitro, and this transition should allow for a study of factors that can influence OPC specification. Undifferentiated NPC cultures did not express the OPC marker O4 (Fig. 1A). However, within 2 d after induction to differentiate with serum, cells with all of these markers were present as a significant, but minority population (12% O4+ cells indicative of OPCs, 17% Tuj1+ cells indicative of neurons, 38% GFAP+ cells indicative of astrocytes). To determine whether forced overexpression of the SoxE or Olig transcription factors is sufficient to drive multipotential NPCs toward oligodendrocyte fate, retroviruses expressing Sox8, Sox9, Sox10, Olig1, or Olig2 (together with a monomeric red fluorescent protein fused to histone H2B to track infected cells) were used to infect dissociated neurospheres. These cells were plated onto laminin 24 h after viral infection and maintained in EGF/FGF growth conditions for an additional 48 h with no differentiation stimulus added (Fig. 1A).
Fig. 1.
Overexpression of SoxE factors but not Olig genes is sufficient to drive differentiation toward the oligodendrocyte lineage. (A) Empty virus infected cells do not express O4 in EGF/FGF, whereas Sox8, Sox9, or Sox10 virus-infected cells express O4 at significant levels. In contrast, Olig1 or Olig2 expression does not lead to increased O4 staining in these culture conditions. (B) The efficiency of O4+ cell production after Sox10 expression under nondifferentiating conditions (in EGF/FGF containing medium). Results pooled from three separate experiments. Error bars are ± SEM, ***P < 0.005. (C) Cells infected with either Empty or SoxE-expressing retroviruses (red) cultured in 2% FBS for 48 h and stained with O4 antibody (green). Inset shows Sox10-infected cells also stain with CNPase, a marker of terminally differentiating oligodendrocytes. (D) Cells infected with either Empty or SoxE-expressing retroviruses (red) cultured in thyroid hormones for 48 h and stained with O4 antibody (green). Inset shows Sox10-infected cells also stain with CNPase, a marker of terminally differentiating oligodendrocytes. (E) The percentage of O4+ cells infected with Sox8, 9, or 10 cultured in serum or thyroid hormones for 48 h. Results are pooled from >3 separate experiments; error bars are ±SEM; *P < 0.05; **P < 0.02; ***P < 0.005.
Olig1 or Olig2 overexpression was insufficient to induce expression of OPC markers (Fig. 1A; Empty virus 0.3 ± 0.08% O4+ cells, Olig1 virus 0.04 oligos ± 0.03% O4+ cells, and Olig2 to 0.04 ± 0.08% O4+ cells). By contrast, expression of any of the three SoxE transcription factors induced O4 glycolipid antigen expression, despite the continued presence of EGF and FGF (Fig. 1A). Sox10 overexpression consistently generated the greatest number of O4+-infected cells (usually >40% of infected cells became O4+) (Fig. 1 A and B); however, Sox9 (≈20% O4+ cells) and Sox8 (≈15% O4+ cells) overexpression also led to a significant increase in the number of cells expressing O4 (P < 0.005). Thus, SoxE transcription factor expression is sufficient to induce rapid expression of markers of the OPC/oligodendrocyte lineage in neural progenitors without additional differentiation stimuli.
To determine whether Sox transcription factors played a role in increasing OPC specification and maturation, we also took cultures separately infected with either the SoxE factors or Empty retrovirus and induced them to differentiate by using two stimuli known to drive differentiation of neural stem cells: FBS or thyroid hormone (TH). The percentage of Sox8, 9, and 10 infected cells expressing either O4 or CNPase (a more mature OPC marker) increased dramatically by 2 d after differentiation in both FBS and TH and morphologically resembled oligodendrocytes (Fig. 1 C–E). Therefore, in the presence of a differentiation signal Sox8, 9, or 10 all effectively increase OPC production and maturation.
Sufu Is a Predicted Direct Target of Sox10 and Is Down-Regulated in Sox10-Expressing NPCs.
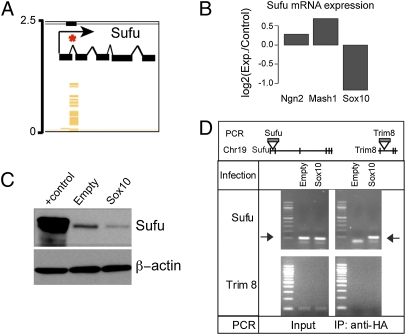
To identify novel molecular targets involved in oligodendrocyte specification we combined chromatin immunoprecipitation (ChIP) with hybridization to a promoter array after infecting SVZ progenitors with Sox10 retrovirus. We performed ChIP after infection with an equally efficient Sox10 retrovirus expressing three copies of the hemagglutinin (HA) epitope (Sox10-3×HA) and shearing of the chromatin to generate on average 1-kb fragments of genomic DNA. The immunoprecipitated DNA–protein complexes were dissociated, and the purified DNA fragments were randomly amplified by PCR and used to probe 50-bp tiled arrays made up of 5 kb of predicted upstream promoter regions for every annotated mouse gene (NimbleGen). From this analysis, we obtained a panel of genes with potential Sox10-binding sites that were statistically significant at P < 1 × 10−4 (Table S1). We focused on genes that appeared in both the ChIP-on-chip screen and an ongoing profiling study comparing the mRNA expression changes in NPCs forced to express Sox10, Mash1, or Neurogenin2. Among the genes found to have both predicted Sox10-binding sites and significant changes in mRNA levels after Sox10 expression was a well-established regulator of multiple morphogenic signaling pathways–Suppressor of Fused (Sufu) (21–25) (Fig. 2).
Fig. 2.
Suppressor of Fused (Sufu) is a direct target of Sox10. (A) Schematic of the Sufu locus depicting the Sox10-binding site identified in the first intron of the Sufu gene (SI Methods). (B) Quantitative mRNA profiling using whole mouse transcriptome Affymetrix chips. Each value is shown as a ratio of the experimental value over Empty virus-infected controls. Shown are the relative expression of Sufu mRNA 48 h after infection with Sox10 virus or 24 h after infection with Mash1 or Ngn2 retroviruses that drive neurogenic differentiation in EGF/FGF containing medium. Note Mash1 and Ngn2 drive modest increases in Sufu expression, whereas Sox10 leads to a twofold decrease in expression of Sufu. Normalized intensity values of triplicate chip hybridization experiments are presented. Each sample was prepared from pools of mRNA derived from less than three separate experiments. (C) Sox10 expression leads to decreased Sufu protein levels in NPCs 24 h after plating. Note the slightly higher and broader band in the positive control lane was obtained from extracts of HEK-293T cells expressing both human and mouse HA-tagged Sufu. In contrast, NPCs only express endogenous untagged mouse Sufu. (D) Targeted ChIP for Sox10 bound to the Sufu gene at the first intronic site. Positions of the PCR primers are shown in relationship to the Sufu and Trim8 genes. The PCR yields the expected products for both Sufu and Trim8 after infection with either Empty or Sox10 virus (Input), but only Sox10-infected cells yielded a specific band for Sufu after ChIP (IP: anti-HA). Empty virus-infected cells showed no signal, and Sox10 showed no binding to the closely linked Trim8 gene. The arrows indicate the size of the expected Sufu product.
In addition to the reduction in Sufu mRNA after Sox10 expression (Fig. 2B), Sufu protein levels were also significantly reduced 24 h after expression of Sox10 in NPCs (Fig. 2C). Directed ChIP confirmed the identification of a Sox10-binding site in the first intron of the Sufu gene (Fig. 2 A and D). The Trim8 gene, which is adjacent to Sufu on the same chromosome, was used as a control, and we found no evidence of nonspecific Sox10 binding to this gene, consistent with Sox10 modulating the expression of this important regulatory protein directly and rapidly after expression in NPCs.
Manipulation of Sufu Expression Regulates Specification of OPCs.
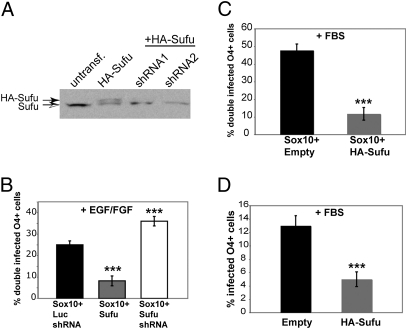
We asked whether increased or decreased expression of Sufu in NPCs alters the specification of OPCs by Sox10. Retroviruses expressing either HA-tagged mouse Sufu or shRNAs targeting Sufu (26) were used to regulate expression (Fig. 3A). To determine whether manipulation of Sufu expression affects the ability of Sox10 to drive OPC specification, we infected cells with Sox10 virus combined with one of three additional viruses: (i) a control shRNA virus (directed against luciferase) to address nonspecific off target shRNA effects; (ii) HA-Sufu virus; or (iii) Sufu shRNA1 virus. Sufu overexpression dramatically decreased the production of double-infected O4+ cells by Sox10 to ≈7.5% (Fig. 3B). Conversely, a 1.5-fold increase in the production of OPCs in EGF/FGF was observed with Sox10 and Sufu shRNA coinfection of NPCs (Fig. 3B). Infection with Sufu shRNA viruses alone in EGF/FGF was insufficient to drive OPC specification, indicating that there are other targets of Sox10 that are required for OPC specification and may include PDGFRα and MBP genes (17, 18).
Fig. 3.
Sufu inhibits Sox10-driven OPC differentiation and endogenous oligodendrocyte differentiation. (A) shRNA-mediated knock down of mouse HA-Sufu protein (solid arrow, top bands) in GP2-293 cells. Note that the endogenous human Sufu protein is unaffected (open arrow, bottom bands). (B) Percentage of O4+ cells coinfected with Sox10 and Luciferase shRNA, Sufu, or Sufu siRNA. Transduced NPCs were cultured in EGF/FGF on laminin-coated plates for 72 h. Error bars are ± SEM, ***P < 0.005, n = 3. (C) Percentage of O4+ cells coinfected with Sox10 virus, and either Empty or Sufu virus then induced to differentiate by removing EGF/FGF and addition of serum. Error bars are ±SEM, **P < 0.01, n = 3. (D) Percentage of O4+ cells infected with Empty or Sufu virus and cultured in serum on laminin-coated plates for 72 h. Error bars are ± SEM, ***P < 0.01, n = 3.
Whether Sufu overexpression is also able to block the effects of Sox10 on oligodendrocyte production after exposure to serum was determined by coinfecting cells with either Sox10 + Empty virus or Sox10 + Sufu virus and switching them into serum. Sufu dramatically blocked the ability of Sox10 induced oligodendrocyte lineage (Fig. 3C) and inhibited the ability of NPCs to adopt an oligodendrocyte fate without Sox10 overexpression (Fig. 3D). Hence, Sufu is a critical negative regulator of oligodendrocyte differentiation from NPCs.
Mice with Reduced Sufu Expression Have Enhanced Developmental Production of OPCs.
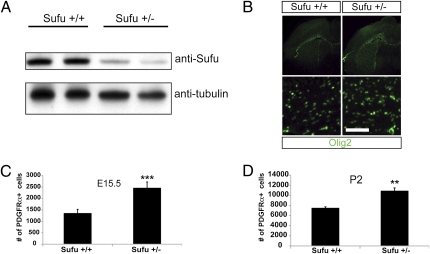
Sufu knockout mice die embryonically before OPCs are generated; however, Sufu+/− mice are viable and fertile (27) and have significantly reduced levels of Sufu protein (Fig. 4A). Coronal sections of embryonic day (E)15.5 or postnatal day (P)2 Sufu+/+ or Sufu+/− forebrains were stained with antibodies against Olig2 and PDGFRα to detect OPCs (Fig. 4B). There appeared to be a substantial increase in the numbers of OPCs (Fig. 4B), and we counted PDGFRα+ (a more stage specific marker for OPCs than Olig2) cells in brain sections at matched levels in mutant and control mice. At two developmental ages (E15.5 and P2), Sufu+/− brains had higher average numbers of PDGFRα-positive cells in the forebrain indicative of increased OPCs (E15.5: Sufu+/+, 1,359 ± 164; Sufu+/−, 2,467 ± 257; P2: Sufu+/+, 7,535 ± 202; Sufu+/−, 10,921 ± 550) (Fig. 4 C and D). These data indicate that in vivo, as in vitro, Sufu is a physiologic regulator of OPC development.
Fig. 4.
Decreased Sufu expression significantly increases the number of oligodendrocyte precursors in vivo. (A) Western blot analysis of equal amounts of protein derived from brain lysates of wild-type (Sufu+/+) mice or mice lacking one copy of Sufu (Sufu+/−). (B) Olig2 staining in brain sections of Sufu+/+ or Sufu+/− E15.5 brains show an overall increase in Olig2-positive cells in Sufu+/− brains. Higher magnifications in Lower. (Scale bar: 100 μm.) (C) The number of PDGFRα-positive cells in the forebrain is significantly increased in mice with decreased Sufu expression (Sufu+/−) at E15.5 (Error bars are ±SD, ***P < 0.005, n = 3). (D) The increased number of PDGFRα-positive cells persists in the Sufu+/− P2 forebrain. Error bars are ± SD, **P < 0.05, n = 3.
Discussion
We have shown SoxE transcription factors strongly influence multipotential neural stem cells toward an OPC fate and the oligodendrocyte lineage. Sox8 was less effective at driving cells to adopt OPC markers than either Sox9 or Sox10. These distinctions within the SoxE family are consistent with in vivo studies suggesting that Sox9 and Sox10 are redundant, whereas Sox8 is incapable of fully compensating for either Sox9 or 10 (12–16, 19, 28). We also found that Sox10 regulates the expression of Sufu, which we show is an important determinant of OPC production from forebrain precursors.
SoxE expression in the CNS is carefully orchestrated beginning with Sox9 expression throughout the spinal cord neuroepithelium, persisting in OPCs and maintained in adult astrocytes (12), followed by Sox8 expression, which persists in maturing oligodendrocytes and astrocytes (14, 29, 30). Consistent with this finding, Sox8 knockouts have delayed oligodendrocyte terminal differentiation (14) and mice lacking Sox9 have significant reductions in both OPC and astrocyte precursors (12). Sox10 is expressed as OPCs are specified and is required for terminal differentiation of OPCs into oligodendroctyes (15). More recently, studies using combinations of SoxE knockouts demonstrate significant redundancy between the SoxE members. For example, Sox8 and 9 double knockout animals lack oligodendrocytes, entirely suggesting a redundant role for Sox8 in early specification of OPCs. Sox9 conditional knockout mice crossed with mice lacking Sox10 display increased OPC apoptosis, indicating a role for both proteins in OPC survival (28). Our in vitro experiments using cultured neurospheres infected with Sox8, 9, or 10 under differentiating conditions such as FBS or TH demonstrate that all of the SoxE factors enhance OPC specification and maturation.
Sufu acts as a powerful negative regulator of the hedgehog signaling pathway in vertebrates (23) by binding to Gli1, 2, and 3 and affecting Gli processing or degradation (21, 23, 31, 32). Recent studies have indicated Sufu has a complex relationship with hedgehog signaling and appears to be both an inhibitor of Shh signaling but also required for the proper balance of Gli activators and repressors with both cilia-dependent and -independent aspects of its functions (31, 32). Sufu also interacts with the Wnt signaling pathway (25) and may act as a general scaffold protein for other signaling molecules (27, 33). These complex relationships of Sufu with both the hedgehog, Wnt, and perhaps other pathways make it unsurprising that there have been only moderate effects, after the initial patterning stage, of Shh on OPC production seen in previous reports (5, 34). The potential role of Sufu as a regulator of Wnt signaling is particularly relevant given recent studies showing that dysregulated Wnt signaling inhibits the progression of oligodendrocyte precursors to mature myelinating oligodendrocytes (35, 36) and that Wnt signaling has a negative regulatory role in oligodendrocyte cell fate acquisition (37, 38).
We have identified Sufu as a target of Sox10 repression and identified a Sox10-binding site in the first intron of Sufu. Moreover, we have demonstrated that Sufu plays a role in vitro and in vivo in OPC specification. In the peripheral nervous system, Sox10 controls Schwann cell myelination by acting in synergy with other transcription factors such as Krox20, NFATc, and Oct6 to regulate myelin genes (39–41). For example, Sox10, Krox20, and NFATc synergistically bind to the first intron of Myelin protein zero and activate transcription (40). We propose a similar model whereby Sox10, perhaps in combination with a novel transcription factor(s), binds and represses Sufu transcription thereby enhancing OPC differentiation. Whether Sox10 repression of Sufu would result in modulation of the Shh or Wnt pathways that may alter OPC specification is unknown. Finally, it remains unknown whether Sox8 or Sox9 also modulate Sufu expression to induce early OPC specification or, in the case of Sox9, astrocyte maintenance in the adult. Future investigation of Sufu dysregulation in the SoxE knockouts would help to answer these questions.
Our observations also have important implications for tumorigenesis in the nervous system. Loss of Sufu by mutation leads to deregulated Shh signaling and Wnt signaling and is an important cause of medulloblastoma (a cerebellar granule precursor tumor) in children (42) and mice (27). SoxE transcription factors are abundantly expressed in gliomas (43, 44), and we predict that in these tumors Sufu expression would be reduced because of down-regulation by transcriptional effects of SoxE proteins. This possibility may provide a mechanism for altered Shh and Wnt signaling levels in such tumors and may be a key aspect of their degree of aggressiveness.
This work may have important implications for regenerative approaches promoting oligodendrocyte replacement in demyelinating diseases. Demyelination in either experimental animal models or patients with multiple sclerosis leads to brisk up-regulation of OPC production in the SVZ (20, 45, 46). However, the ubiquity of this response in patients with demyelination is not known and it remains unclear whether further augmentation of this response would be beneficial. Our studies indicate that developing methods to down-regulate the expression of Sufu in these patients may have therapeutic benefit.
Methods
Retroviral cloning and production, neurosphere infection, clonal analysis, data analysis, Western blotting, and immunohistochemistry are described in SI Methods and in published work (38).
Primary Cell Culture.
All animal procedures were performed according to protocols approved by the University of California, San Francisco Institutional Animal Care and Use Committee (IACUC). SVZ tissue from P5 CD1 mice was collected in and dissociated with 0.1% Trypsin (Worthington) at 37 °C water bath. After dissocation by trituration single cells were plated in Complete Media consisting of DMEM/F12 (50:50; Gibco) supplemented with 10× hormone mix [per 50 mL of stock added to 450 mL of medium: 40 mg of transferrin, 10 mg of insulin, 3.86 mg of putrescine, 4.0 mL of 3 mM selenium, 4.0 mL of 2 mM progesterone, 10 mL of 2 mg/mL heparin (all from Sigma)], 0.8 mL of 30% glucose, 0.6 mL of 7.5% NaHCO3, 10 mL of 30% glucose, 7.5 mL of 7.5% NaHCO3, 2.5 mL of 1 M Hepes, 5 mL of 200 mM glutamine, 5 mL of Pencillin-Streptomycin, 2 mL of Fungizone, EGF (Sigma; 10 ng/mL), FGF (Sigma; 20 ng/mL), and B27 (Invitrogen). Approximately 80,000 cells were plated in a 25-cm2 flask (Corning), grown for 3–4 d until spheres grew large enough to be passaged. Neurospheres were dissociated and infected with retroviruses as described in SI Methods.
ChIP Assays and DNA Microarrays.
Established protocols were used for the chromatin immunoprecipitation experiments (http://genomics.ucdavis.edu/farnham/protocols/chips.html), and hybridization to mouse promoter arrays was performed by NimbleGen. Neurosphere cultures infected with either a Sox10-3×HA expressing or an Empty control retrovirus were plated on laminin coated six-well plates (Falcon) for 48 h before the samples were collected. A monoclonal HA antibody [HA.11 (Covance), 1:150] was used for the immunoprecipitations. Hybridization intensities were expressed as a ratio of the signals from Sox10-3×HA- infected cells and Empty control-infected cells. For targeted ChIP, ChIP followed by targeted PCR to examine Sox10 binding to Sufu and the adjacent Trim8 gene, which served as a negative control. The following primers were used in a PCR with 37 amplification cycles (94 °C for 15 s, 60 °C for 15 s, and 72 °C for 30 s): Sufu-5′ gcaaatggtggtcctgaagt and Sufu-3′ cccatctccagaggcaataa, product = 128 bp; TRIM-5′ cccggcagaagttatgtttg and TRIM-3′ gcggagaattggaagaactg, product = 106 bp.
Immunocytochemistry.
Cells plated on chamber slides were fixed with 4% paraformaldehyde, washed three times with PBS, and blocked and permeabilized for 30 min with PBS containing 0.3% Triton X-100 and 10% lamb serum. Incubation with primary antibodies [O4 (mouse 1:30; Chemicon and Sigma), Nestin (rabbit 1:100; Chemicon), GFAP (rat 1:500; Zymed), Tuj1 (mouse or rabbit 1:500; Covance), PDGFRα (rabbit 1:200; Santa Cruz; and rat 1:500; BD Biosciences Pharmingen), Sox10 (rabbit 1:100; ABR), CNPase (mouse 1:500; Sternberger Monoclonals), and Olig2 (rabbit 1:1,000; D. Rowitch, University of California, San Francisco)] were at 4 °C overnight. Each experiment was performed multiple times and graphs represent three separate experiments for which six fields were quantified per condition. Similar protocols were used for histological analysis of brain sections and are described in SI Methods.
Supplementary Material
Acknowledgments
We thank Drs. David Rowitch and John Rubenstein for helpful discussions, Becca Wolsky and Trung Huynh for excellent technical assistance, and Mark Bünger and Ru-Fang Yeh for help with ChIP-on-chip data analysis. These studies were supported by funds from the Human Frontiers in Science Program Organization Fellowship and the Alberta Heritage Foundation Fellowship (to C.D.P.); the Stewart Trust (University of California, San Francisco Comprehensive Cancer Center); Developmental Project from the Brain Specialized Programs of Research Excellence Grants P50 CA97257, R01 NS039278 (to Z.W.), K02 MH074958, and R21 NS055783; the Sandler Foundation for Basic Research; a research grant from the National Multiple Sclerosis Society; a California Institute for Regenerative Medicine Comprehensive Research Grant; and a generous gift endowment from the family of Glenn W. Johnson, Jr. (to S.J.P.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016485107/-/DCSupplemental.
References
- 1.Wilson SW, Rubenstein JL. Induction and dorsoventral patterning of the telencephalon. Neuron. 2000;28:641–651. doi: 10.1016/s0896-6273(00)00171-9. [DOI] [PubMed] [Google Scholar]
- 2.Briscoe J, Ericson J. Specification of neuronal fates in the ventral neural tube. Curr Opin Neurobiol. 2001;11:43–49. doi: 10.1016/s0959-4388(00)00172-0. [DOI] [PubMed] [Google Scholar]
- 3.Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kessaris N, et al. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tekki-Kessaris N, et al. Hedgehog-dependent oligodendrocyte lineage specification in the telencephalon. Development. 2001;128:2545–2554. doi: 10.1242/dev.128.13.2545. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Q, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109:61–73. doi: 10.1016/s0092-8674(02)00677-3. [DOI] [PubMed] [Google Scholar]
- 7.Lu QR, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109:75–86. doi: 10.1016/s0092-8674(02)00678-5. [DOI] [PubMed] [Google Scholar]
- 8.Takebayashi H, et al. The basic helix-loop-helix factor olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12:1157–1163. doi: 10.1016/s0960-9822(02)00926-0. [DOI] [PubMed] [Google Scholar]
- 9.Petryniak MA, Potter GB, Rowitch DH, Rubenstein JL. Dlx1 and Dlx2 control neuronal versus oligodendroglial cell fate acquisition in the developing forebrain. Neuron. 2007;55:417–433. doi: 10.1016/j.neuron.2007.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wissmüller S, Kosian T, Wolf M, Finzsch M, Wegner M. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res. 2006;34:1735–1744. doi: 10.1093/nar/gkl105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wegner M. From head to toes: The multiple facets of Sox proteins. Nucleic Acids Res. 1999;27:1409–1420. doi: 10.1093/nar/27.6.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stolt CC, et al. The Sox9 transcription factor determines glial fate choice in the developing spinal cord. Genes Dev. 2003;17:1677–1689. doi: 10.1101/gad.259003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, Wegner M. Sox10, a novel transcriptional modulator in glial cells. J Neurosci. 1998;18:237–250. doi: 10.1523/JNEUROSCI.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stolt CC, Lommes P, Friedrich RP, Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–2358. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- 15.Stolt CC, et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002;16:165–170. doi: 10.1101/gad.215802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z, et al. Induction of oligodendrocyte differentiation by Olig2 and Sox10: Evidence for reciprocal interactions and dosage-dependent mechanisms. Dev Biol. 2007;302:683–693. doi: 10.1016/j.ydbio.2006.10.007. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Lu Y, Smith HK, Richardson WD. Olig1 and Sox10 interact synergistically to drive myelin basic protein transcription in oligodendrocytes. J Neurosci. 2007;27:14375–14382. doi: 10.1523/JNEUROSCI.4456-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Finzsch M, Stolt CC, Lommes P, Wegner M. Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor alpha expression. Development. 2008;135:637–646. doi: 10.1242/dev.010454. [DOI] [PubMed] [Google Scholar]
- 19.Wegner M, Stolt CC. From stem cells to neurons and glia: A Soxist's view of neural development. Trends Neurosci. 2005;28:583–588. doi: 10.1016/j.tins.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Menn B, et al. Origin of oligodendrocytes in the subventricular zone of the adult brain. J Neurosci. 2006;26:7907–7918. doi: 10.1523/JNEUROSCI.1299-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barnfield PC, Zhang X, Thanabalasingham V, Yoshida M, Hui CC. Negative regulation of Gli1 and Gli2 activator function by Suppressor of fused through multiple mechanisms. Differentiation. 2005;73:397–405. doi: 10.1111/j.1432-0436.2005.00042.x. [DOI] [PubMed] [Google Scholar]
- 22.Østerlund T, Kogerman P. Hedgehog signalling: How to get from Smo to Ci and Gli. Trends Cell Biol. 2006;16:176–180. doi: 10.1016/j.tcb.2006.02.004. [DOI] [PubMed] [Google Scholar]
- 23.Ding Q, et al. Mouse suppressor of fused is a negative regulator of sonic hedgehog signaling and alters the subcellular distribution of Gli1. Curr Biol. 1999;9:1119–1122. doi: 10.1016/s0960-9822(99)80482-5. [DOI] [PubMed] [Google Scholar]
- 24.Taylor MD, et al. Failure of a medulloblastoma-derived mutant of SUFU to suppress WNT signaling. Oncogene. 2004;23:4577–4583. doi: 10.1038/sj.onc.1207605. [DOI] [PubMed] [Google Scholar]
- 25.Meng X, et al. Suppressor of fused negatively regulates beta-catenin signaling. J Biol Chem. 2001;276:40113–40119. doi: 10.1074/jbc.M105317200. [DOI] [PubMed] [Google Scholar]
- 26.Varjosalo M, Li SP, Taipale J. Divergence of hedgehog signal transduction mechanism between Drosophila and mammals. Dev Cell. 2006;10:177–186. doi: 10.1016/j.devcel.2005.12.014. [DOI] [PubMed] [Google Scholar]
- 27.Lee Y, et al. Loss of suppressor-of-fused function promotes tumorigenesis. Oncogene. 2007;26:6442–6447. doi: 10.1038/sj.onc.1210467. [DOI] [PubMed] [Google Scholar]
- 28.Finzsch M, Stolt CC, Lommes P, Wegner M. Sox9 and Sox10 influence survival and migration of oligodendrocyte precursors in the spinal cord by regulating PDGF receptor {alpha} expression. Development. 2008;135:637–646. doi: 10.1242/dev.010454. [DOI] [PubMed] [Google Scholar]
- 29.Sock E, Schmidt K, Hermanns-Borgmeyer I, Bösl MR, Wegner M. Idiopathic weight reduction in mice deficient in the high-mobility-group transcription factor Sox8. Mol Cell Biol. 2001;21:6951–6959. doi: 10.1128/MCB.21.20.6951-6959.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Stolt CC, Schmitt S, Lommes P, Sock E, Wegner M. Impact of transcription factor Sox8 on oligodendrocyte specification in the mouse embryonic spinal cord. Dev Biol. 2005;281:309–317. doi: 10.1016/j.ydbio.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 31.Chen MH, et al. Cilium-independent regulation of Gli protein function by Sufu in Hedgehog signaling is evolutionarily conserved. Genes Dev. 2009;23:1910–1928. doi: 10.1101/gad.1794109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jia J, et al. Suppressor of Fused inhibits mammalian Hedgehog signaling in the absence of cilia. Dev Biol. 2009;330:452–460. doi: 10.1016/j.ydbio.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jenkins D. Hedgehog signalling: Emerging evidence for non-canonical pathways. Cell Signal. 2009;21:1023–1034. doi: 10.1016/j.cellsig.2009.01.033. [DOI] [PubMed] [Google Scholar]
- 34.Kessaris N, Jamen F, Rubin LL, Richardson WD. Cooperation between sonic hedgehog and fibroblast growth factor/MAPK signalling pathways in neocortical precursors. Development. 2004;131:1289–1298. doi: 10.1242/dev.01027. [DOI] [PubMed] [Google Scholar]
- 35.Fancy SP, et al. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev. 2009;23:1571–1585. doi: 10.1101/gad.1806309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Feigenson K, Reid M, See J, Crenshaw EB, 3rd, Grinspan JB. Wnt signaling is sufficient to perturb oligodendrocyte maturation. Mol Cell Neurosci. 2009;42:255–265. doi: 10.1016/j.mcn.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 37.Ye F, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Langseth AJ, et al. Wnts influence the timing and efficiency of OPC generation in the telencephalon. J Neurosci. 2010;30:13367–13372. doi: 10.1523/JNEUROSCI.1934-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ghislain J, Charnay P. Control of myelination in Schwann cells: A Krox20 cis-regulatory element integrates Oct6, Brn2 and Sox10 activities. EMBO Rep. 2006;7:52–58. doi: 10.1038/sj.embor.7400573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peirano RI, Goerich DE, Riethmacher D, Wegner M. Protein zero gene expression is regulated by the glial transcription factor Sox10. Mol Cell Biol. 2000;20:3198–3209. doi: 10.1128/mcb.20.9.3198-3209.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bondurand N, et al. Human Connexin 32, a gap junction protein altered in the X-linked form of Charcot-Marie-Tooth disease, is directly regulated by the transcription factor SOX10. Hum Mol Genet. 2001;10:2783–2795. doi: 10.1093/hmg/10.24.2783. [DOI] [PubMed] [Google Scholar]
- 42.Taylor MD, et al. Mutations in SUFU predispose to medulloblastoma. Nat Genet. 2002;31:306–310. doi: 10.1038/ng916. [DOI] [PubMed] [Google Scholar]
- 43.Bannykh SI, Stolt CC, Kim J, Perry A, Wegner M. Oligodendroglial-specific transcriptional factor SOX10 is ubiquitously expressed in human gliomas. J Neurooncol. 2006;76:115–127. doi: 10.1007/s11060-005-5533-x. [DOI] [PubMed] [Google Scholar]
- 44.Schlierf B, et al. Expression of SoxE and SoxD genes in human gliomas. Neuropathol Appl Neurobiol. 2007;33:621–630. doi: 10.1111/j.1365-2990.2007.00881.x. [DOI] [PubMed] [Google Scholar]
- 45.Nait-Oumesmar B, et al. Activation of the subventricular zone in multiple sclerosis: Evidence for early glial progenitors. Proc Natl Acad Sci USA. 2007;104:4694–4699. doi: 10.1073/pnas.0606835104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Picard-Riera N, et al. Experimental autoimmune encephalomyelitis mobilizes neural progenitors from the subventricular zone to undergo oligodendrogenesis in adult mice. Proc Natl Acad Sci USA. 2002;99:13211–13216. doi: 10.1073/pnas.192314199. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.