Abstract
The sarcoplasmic reticulum Ca2+-ATPase transports two Ca2+ per ATP hydrolyzed from the cytoplasm to the lumen against a large concentration gradient. During transport, the pump alters the affinity and accessibility for Ca2+ by rearrangements of transmembrane helices. In this study, all-atom molecular dynamics simulations were performed for wild-type Ca2+-ATPase in the Ca2+-bound form and the Gln mutants of Glu771 and Glu908. Both of them contribute only one carboxyl oxygen to site I Ca2+, but only Glu771Gln completely looses the Ca2+-binding ability. The simulations show that: (i) For Glu771Gln, but not Glu908Gln, coordination of Ca2+ was critically disrupted. (ii) Coordination broke at site II first, although Glu771 and Glu908 only contribute to site I. (iii) A water molecule bound to site I Ca2+ and hydrogen bonded to Glu771 in wild-type, drastically changed the coordination of Ca2+ in the mutant. (iv) Water molecules flooded the binding sites from the lumenal side. (v) The side chain conformation of Ile775, located at the head of a hydrophobic cluster near the lumenal surface, appears critical for keeping out bulk water. Thus the simulations highlight the importance of the water molecule bound to site I Ca2+ and point to a strong relationship between Ca2+-coordination and shielding of bulk water, providing insights into the mechanism of gating of ion pathways in cation pumps.
Keywords: sarco(endo)plasmic reticulum Ca2+-ATPase, ion pump, mutation
The Ca2+-ATPase of skeletal muscle sarcoplasmic reticulum (SERCA1a) is an integral membrane protein that transports two Ca2+ from the cytoplasm into the sarcoplasmic reticulum (SR) lumen per ATP hydrolyzed and thereby establishes a large concentration gradient across the membrane (1). Classical E1/E2 theory postulates that the Ca2+-ATPase achieves this by alternating the affinity and accessibility of the transmembrane Ca2+-binding sites (2, 3). That is, the binding sites have high affinity and face the cytoplasm in E1, and have low affinity and face the lumen of SR in E2. This idea has been largely corroborated by crystal structures of SERCA1a in different states pertinent to the active transport cycle (4–16).
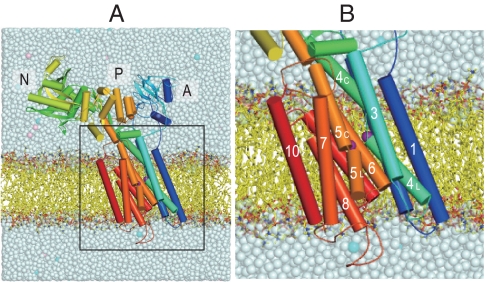
SERCA1a consists of three cytoplasmic domains designated as A (actuator), N (nucleotide-binding), P (phosphorylation), and 10 transmembrane helices (M1–M10) (Fig. 1) (4). Four transmembrane helices (M4–M6, and M8) constitute the two Ca2+-binding sites (I and II) of different characteristics (4, 17–19), although Ca2+ is coordinated in both sites by 7 oxygen atoms. In site I, all oxygen atoms come from the side chains of residues on 3 helices (M5, M6, and M8) and two water molecules. Asn768 and Glu771 (M5), Thr799 and Asp800 (M6), and Glu908 (M8), all contribute only one oxygen atom to site I Ca2+. In contrast, site II is formed almost on the M4 helix, with contributions by 3 main chain carbonyls (Val304, Ala305, and Ile307 on M4), as well as 4 side chain oxygen atoms: two from Glu309 (M4), one each from Asp800 and Asn796 (M6). The binding geometry in site II is reminiscent of the EF-hand. Asp800 on M6 is the only residue that coordinates both Ca2+. The two sites are located side-by-side but the binding process is single file, with site I Ca2+ being the first Ca2+ to bind (20). It has been demonstrated that the removal of only one coordinating oxygen atom from site I completely abolishes Ca2+-binding, yet removal of even 3 oxygen atoms from site II leaves 50% binding (21). It is important to note that mere binding of Ca2+ does not close the cytoplasmic gate and that site II Ca2+ is still exchangeable with Ca2+ in the cytoplasm (22). Closing and locking of the gate require phosphorylation of the ATPase (6, 9).
Fig. 1.
A snapshot of the Ca2+-ATPase embedded in the DOPC membrane after 1-ns equilibration. (A) The full model in the simulation. (B) The transmembrane region. Purple spheres represent Ca2+; cyan and pink spheres in salt solution show K+ and Cl-, respectively. Three cytoplasmic domains (A, N, and P), and 10 transmembrane helices (M1–M10) are marked. Several phospholipids and solvent molecules are removed for clarity.
Here we focus on two glutamates in site I: Glu771 and Glu908. Both provide only one carboxyl oxygen for coordination. The Glu908Gln mutation somewhat decreases the affinity for Ca2+, but Glu771Gln is much more deleterious (21). Why this difference arises is the subject of this study. Simulations demonstrated that Glu908 is likely protonated even when it coordinates site I Ca2+ (23), and therefore the small effect of the Glu908Gln mutation is understandable. To explore why Glu771Gln is so deleterious, we apply all-atom molecular dynamics (MD) simulations to the E1·2Ca2+ state. In this state, Ca2+-ATPase could be regarded as a high-affinity Ca2+-binding protein. However, the ATPase is special in that it needs entry and exit pathways for both Ca2+ and water (4), and, being a pump, the access needs to be opened and closed; i.e. a control gate needs to be in place. It might appear that the regulation of affinity and that of water accessibility are two different matters. The simulations described here, however, demonstrate that substitution of one oxygen atom with nitrogen in the Glu771 carboxyl critically disrupts both.
Results
Simulations for Wild-Type, Glu771Gln, and Glu908Gln Mutants of Ca2+-ATPase in the Ca2+-Bound Form.
In this study, we performed three 10-ns MD simulations for each of the wild-type (WT1, WT2, and WT3), Glu771Gln (E771Q1, E771Q2, and E771Q3), and Glu908Gln mutants (E908Q1, E908Q2, and E908Q3). The root mean square deviations (RMSD) of Cα atoms relative to the crystal structure exceeded 4 Å in all the simulations (Fig. S1). This was due to large rigid body movements of the N and A domains, as the individual domains did not change much (RMSD < 2.0 Å). The transmembrane domain was particularly stable (RMSD of ∼1.0 Å for wild-type and < 1.5 Å for the mutants), suggesting that the bound Ca2+ ions tie the transmembrane helices together.
Ca2+ Coordination and Hydrophobic Shielding on the Lumenal Side of Wild-Type.
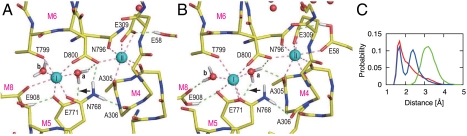
In three simulations for wild-type, the configuration of the Ca2+-binding sites remained stable with fluctuations between two major forms (Fig. 2 and Fig. S2). In one of the major forms (Fig. 2A), Ca2+ coordination was almost identical to that in the crystal structure (4), where one of the carboxyl oxygen in Glu771 coordinated site I Ca2+ and the other formed a hydrogen bond with a water (water a; Fig. 2). In the other stable structure (Fig. 2B), the Glu771 carboxyl made a bidentate coordination, replacing the Glu908 carboxyl, which instead formed a hydrogen bond with the other water molecule coordinating site I Ca2+ (water b). The seven coordination of Ca2+ was, thus, maintained in both forms. The coordinating distance of the Glu908 carboxyl (2.8 Å) was, on average, longer than other coordinations (e.g., 2.3 Å for Glu771), suggesting weaker Ca2+-binding ability of this residue (Fig. S3A).
Fig. 2.
Snapshots of the Ca2+-binding sites in a MD simulation for wild-type (WT1). (A) Atomic model at 5.4 ns simulation; (B) at 8.0 ns. Viewed from the cytoplasm. Spheres represent Ca2+ (cyan) and water (red). Dashed lines indicate Ca2+-coordinations (pink) and hydrogen bonds (light green). The lines representing Ca2+ coordination are drawn for oxygen atoms within 3.0 Å from Ca2+, those for hydrogen bonds are drawn when the donor-acceptor and hydrogen-acceptor distances are within 3.25 and 2.50 Å, respectively. Hydrogen atoms potentially participating in hydrogen bonds are shown explicitly. (C) Distance distribution between the Glu771 carboxyl oxygen (amide nitrogen in Gln771) and the hydrogen atom in water a involved initially in the hydrogen bond between them (arrows in A and B). Three simulation trajectories were averaged for each of the wild-type (red), Glu771Gln (green), and Glu908Gln (blue) mutants.
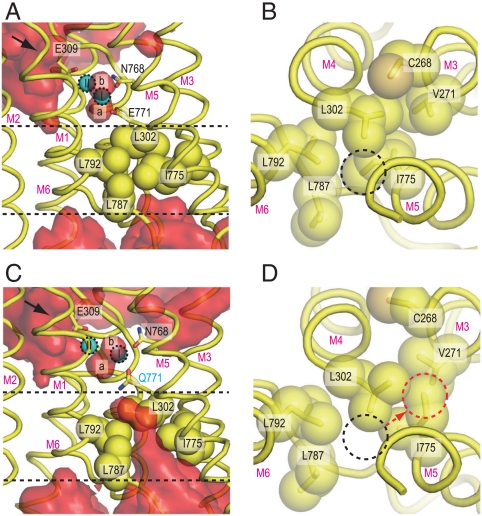
It is well established that site II Ca2+, but not site I Ca2+, can be exchanged with Ca2+ in the cytoplasm (22), and that Glu309 on the M4 helix works as the gating residue (24, 25). Simulations for wild-type were consistent with this experimental evidence (Fig. 3 and Fig. S4). In the crystal structure, a few water molecules are located outside of Glu309 (4) and, in the simulations, they exchanged rapidly with other water molecules in the cytoplasm. A clear water path was observed between the M1 and M2 helices, leading to Glu309 (the arrow in Fig. 3 and Fig. S4). On the lumenal side, a hydrophobic cluster involving Val271 (M3), Leu302 (M4), Ile775 (M5), and Leu787 and Leu792 (M6) blocked access of bulk water (Fig. 3 and Fig. S4). Thus, no water molecules from outside the membrane penetrated into the binding sites.
Fig. 3.
Hydrophobic shielding on the lumenal side. Atomic models in simulations for wild-type [WT1 at 10 ns; (A and B)] and for the Glu771Gln mutant [E771Q3 at 10 ns; (C and D)] are shown. (A and C) Views parallel to the membrane, and (B and D) those from the Ca2+-binding site toward the lumen. Hydrophobic residues, located between the two dashed lines in A and C, are shown in space fill (B and D). Red surfaces [(A) WT1 and (C) E771Q3] represent average water densities calculated with Maptools 1.0 (http://www.mpibpc.mpg.de/groups/de_groot/maptools.html) and contoured at 1.5σ using Pymol (33). The arrows in A and C indicate water paths leading to Glu309. The dashed circles in B and D specify the positions of the Ile775 Cδ in WT1 (black) and E771Q3 (red). Two bound Ca2+ (I and II) and water molecules (a and b) are marked.
Changes in Ca2+ Coordination by the Mutations.
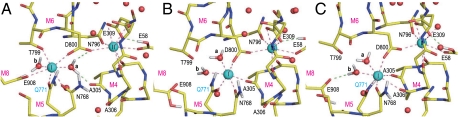
Coordination of Ca2+ in the simulations for Glu908Gln was less stable than that for wild-type (Figs. S2 E–G and S3), as expected from mutagenesis studies (21). In all three simulations, rotations of the Glu771 carboxyl frequently occurred, an event not observed in wild-type (Fig. S5A) presumably due to a hydrogen bond between the Glu771 and Glu908 carboxyls, mediated by protonation of Glu908 (23) (Fig. 2). Instead, in Glu908Gln, the substituted amide group formed a hydrogen bond with the Leu904 carbonyl, one turn below Gln908 on the M8 helix (Figs. S2 E–G). As a result, an empty space was created around the Glu771 side chain, allowing it greater freedom. The hydrogen bond between Gln908 and Leu904 pulled site I Ca2+ toward M8, creating a larger space between M4C (the cytoplasmic half of M4) and M6. As a result, in the severest case, a few water molecules penetrated from the cytoplasm to site I Ca2+ (a dashed arrow in Figs. S2 E and F) and partially destroyed Ca2+ coordination, consistent with a reduction of Ca2+ affinity (21).
Ca2+ coordination was severely altered in the simulations for Glu771Gln (Fig. 4 and Fig. S2 C and D). In all three, the Ala305 carbonyl detached first from site II Ca2+, making a hydrogen bond with water a. Then the simulations differed depending on water a, now free to move due to the lack of the hydrogen bond with the Glu771 carboxyl by the Glu to Gln substitution. In the least deleterious case (E771Q2), the Ala305-Ca2+ coordination resumed, as water a hydrogen bonded with the amide group of Gln771. Nevertheless, the system did not appear to be stable, as a water molecule was introduced to the binding site from the lumenal side (Fig. S2D).
Fig. 4.
Disruption of Ca2+-coordination in a simulation for the Glu771Gln mutant (E771Q3). (A) Atomic models at 0.8 ns, (B) at 1.0 ns, and (C) at 1.2 ns in the simulation, respectively. Presented in the same way as in Fig. 2.
In other simulations (E771Q1 and E771Q3), further changes took place, leading to more deleterious alterations in Ca2+ coordination (Fig. 4 and Figs. S2C and S3). In E771Q1, the coordination of site I Ca2+ by the Gln771 carbonyl was disrupted at 2 ns and recovered at 4 ns (Fig. S3B). Water a moved more freely than in E771Q2. During the simulation, it formed hydrogen bonds not only with the Ala305 carbonyl but also the Gln771 carbonyl, breaking Ca2+ coordination. Water molecules came from the lumenal side to site I Ca2+ (the arrow in Fig. S2C) passing the hydrophobic cluster (Fig. S4B) and formed a hydrogen bonded network that included water a. In E771Q3 (Fig. 4), water a abruptly moved to the opposite side of site I Ca2+ at 1.0 ns (Fig. 4B), pushing the Ca2+ toward the M1 and M2 helices, apparently following a water path formed between them. The Thr799 and Glu908 side chains totally detached from site I Ca2+. Instead, Ala305, a coordinating residue of site II Ca2+ in wild-type, and even Ala306 on M4 provided their carbonyls to site I Ca2+ (Fig. 4C). Glu58 now contributed a carboxyl oxygen to site II instead of Ala305. Thus, the 7 coordination of Ca2+ was always kept in both sites throughout all simulations. At the end of the simulation (10 ns), site I Ca2+ and site II Ca2+ had moved 2.7 and 3.7 Å, respectively, toward M1, as if they had been squeezed out from their normal binding positions and were on their way to the cytoplasm. The hydrogen bonds between the Asn768 amide (M5L) and the Ala306 carbonyl (M4L), and between Asn768 and the Asp800 carboxyl (M6) were disrupted in the simulation (Fig. S5), leading to detachment of the M4L, M5L and M6 helices.
Changes in Accessibility of Water to the Ca2+-Binding Sites by the Mutations.
The release of water a, a direct consequence of the Glu771Gln mutation, strongly influenced Ca2+ coordination as well as the tight packing of the transmembrane helices in the E1·2Ca2+ structure. As shown in Fig. 3C, bulk water was also allowed to approach more deeply to the Ca2+-binding sites than in wild-type. In wild-type and the Glu908Gln mutant, the hydrophobic cluster blocked penetration of water from the lumenal side (Fig. 3A and Fig. S4 A and D). In contrast, in the simulations for Glu771Gln, a cavity surrounded by M4L, M5L and M6 was created and allowed the inflow of water molecules from the SR lumen (Fig. 3C and Fig. S4 B and C). In all the simulations, water molecules penetrated the hydrophobic barrier, and in one case (E771Q1), a water molecule from the lumen reached site I and formed a hydrogen bond directly to water a (Fig. S2C). Thus, the simulations show that one of the reasons why the Glu771Gln mutation is deleterious is that it allows introduction of bulk water into the binding sites. Water molecules flooded in from the lumenal side through a cavity surrounded by M4L, M5L, M6, and M8. This pathway is different from that for Ca2+ release lined by M1, M2, M4L and M6 as observed in the  crystal structures (13, 14). This seems reasonable because opening of the release pathway critically depends on the position of the A domain (13, 14) and will not occur with the position allowed in the E1·2Ca2+ state.
crystal structures (13, 14). This seems reasonable because opening of the release pathway critically depends on the position of the A domain (13, 14) and will not occur with the position allowed in the E1·2Ca2+ state.
Discussion
Role of Glu771 and Ile775 in the Hydrophobic Shielding.
Thus, the simulations indicate a strong relationship between Ca2+-coordination and hydrophobic shielding on the lumenal side, highlighting the critical importance of Glu771 and water a in them. Compared to the E2 state, in which the transmembrane binding sites are occupied by H+ rather than Ca2+, the arrangement of transmembrane helices M1–M4 in the E1·2Ca2+ state is rather loose, in particular between M4L and M5L (Fig. 5C). The simulations show that such a loose packing is possible because a strain due to Ca2+-coordination is imposed on the helices. In this sense, the shielding mechanism is more “dynamic” in E1·2Ca2+ than in E2.
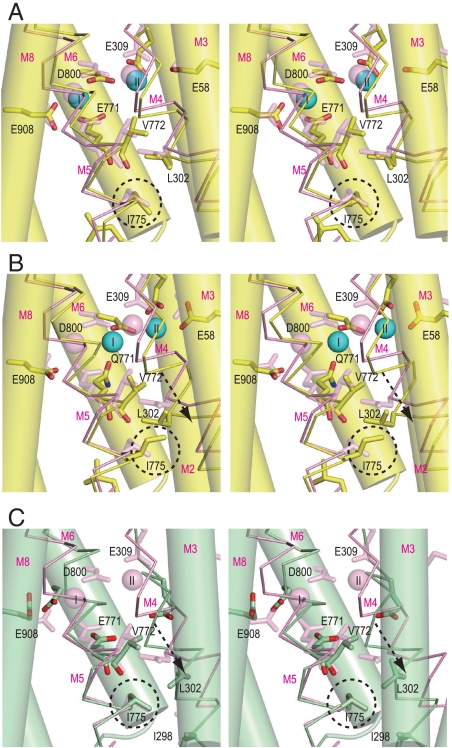
Fig. 5.
Structural changes in the Ca2+-binding sites. Viewed in stereo along the membrane plane. Superimposed on the atomic model of the crystal structure in E1·2Ca2+ (pink) are: (A) WT1 at 10 ns (atom color); (B) E771Q3 at 10 ns (atom color); and (C) crystal structure in E2 (green). The M3, M6, and M8 helices are represented with cylinders, and M4 and M5 with sticks. The arrows in B and C indicate the movements of the M4 helix relative to the rest of the transmembrane helices. Dashed circles locate the Ile775 side chain.
Of the residues that form the hydrophobic cluster, Ile775 on M5 is particularly important, as it is located at the head of the cluster (Fig. 3). Introduction of lumenal water always changed the rotamer of this residue, disrupting van der Waals contacts with the Glu771 carbonyl and the side chains of Leu302 (M4) and Leu792 (M6). The rotamers of Ile775 observed in the simulations for the mutant (Fig. S6) are prohibited for wild-type because Val772 carbonyl would come too close to the Ile775 side chain (either Cγ1 or Cγ2) (Fig. S7).
Such steric conflicts arise because the M5 helix, although a continuous helix, is bent at Gly770 toward M8 (Fig. 1B) to accommodate Ca2+ in the binding cavity, and the side of M5L that faces M4 is stretched whereas the opposite side is compressed so that the carbonyl group of Val772 points inward from the helix toward Ile775. Hence, as long as Ca2+ is securely coordinated with Glu771, Ile775 cannot change its rotamer (Fig. S7). Once Glu771 dissociates from site I Ca2+, M5L can change its orientation and the Ile775 side chain easily escapes from van der Waals contacts with the Val772 carbonyl and the Leu302 side chain to take a different conformation (Figs. S7 and S8).
In the E2 crystal structure, Ile775 takes the same rotamer as in E1·2Ca2+ (Fig. 5). In E2, the distance between Glu771 carbonyl and Ile775 is shorter (Fig. S7D), because M5 is now bent toward M1, opposite to the situation in E1·2Ca2+. Also, due to this bending, M4L is pushed toward the lumen (Fig. 5C) and, at the same time, pressed against M5L backed by a rigid V-shaped structure formed by the M1 and M2 helices. As a result, the Ile775 side chain is interdigitated with those of Ile298 and Leu302 on M4 (Fig. S7D), making the hydrophobic barrier more sturdy. In other words, bending of M5 toward M1, achieved through the 3 cytoplasmic domains forming a compact headpiece, provides a tight seal. Tighter hydrophobic clustering is needed in E2, because the transmembrane helices are not tied together by Ca2+ and the residues participating Ca2+-coordination must become free to accept incoming Ca2+.
Comparison with Mutagenesis Results.
The simulations for wild-type and the Glu771Gln and Glu908Gln mutants qualitatively reproduced the mutagenesis results of Zhang et al.(21), in which Ca2+-binding is completely lost in Glu771Gln but only weakened in Glu908Gln.
Ile775Ala and Ile775Ser mutants have approximately the same Ca2+ affinity as wild-type (26), questioning whether hydrophobic interactions here are of overriding importance for binding of Ca2+. The shielding mechanism in E1·2Ca2+ utilizes strain caused by Ca2+-coordination and such alterations of the side chain appear to be tolerated. Conversely, both mutants may have serious consequences for lumenal sealing in E2. In this regard, it is understandable that they are classified as E2P dephosphorylation block mutants (26), as stabilization of E2 requires good shielding on the lumenal side.
We expect that such a hydrophobic cluster will be important for all P-type ATPases. In fact, Na+, K+-ATPase has reduced affinity for Na+ on the cytoplasmic side, if Phe785, the residue corresponding to Ile775 in SERCA, is substituted with Leu (27). This mutation is medically important, because it is associated with familial rapid-onset dystonia parkinsonism (28). In patients carrying the mutation, a disturbance of cellular Na+ homeostasis is considered to be the major factor contributing to the development of the disease. Although the coordinating residues are essentially the same between the two pumps, they also have critical differences. A particularly important one is the substitution of Gly770 on M5 with Pro in Na+, K+-ATPase (29). Because of this difference, the pathway or inclination of M5 cannot be identical. Furthermore, electrostatic interactions in Na+, K+-ATPase cannot be as strong as in Ca2+-ATPase due to the ion charge differences. Interactions stronger than mere van der Waals contacts must be required for stabilizing the extracellular parts of the transmembrane helices of Na+, K+-ATPase. In fact, stacking interactions between 3 Phe’s (Phe301 (M3), Phe318 (M4) and Phe785 (M5) in human α1) are likely implicated in the hydrophobic shielding in the E1·3Na+ state of Na+, K+-ATPase. Then it is understandable why the Leu substitution has such a profound impact (27).
Potential Role of the Water Molecule Attached to site I Ca2+.
Finally, we would like to address whether water a bound to site I Ca2+ has a physiological role. What is interesting is that destruction of Ca2+ coordination geometry always started by dissociation of the Ala305 carbonyl, and this directly leads to the release of water a. In the physiological case, the M4 helix is pushed toward the lumen during the E1·2Ca2+ → E2 transition. This movement will dissociate the Ala305 carbonyl from water a, cause its release, and destroy Ca2+ coordination. Because coordination and hydrophobic core packing are tightly coupled, lumenal water will flood the binding sites. Water a seems to function as a kind of mobile axe of Ca2+-coordination that accelerates disintegration of the Ca2+-binding sites and causes release of bound Ca2+.
Materials and Methods
All-atom MD simulations of SR Ca2+-ATPase with two bound Ca2+, explicit solvent, and phospholipids were carried out using MARBLE software package (30). CHARMM27 force field parameters for proteins (31), phospholipids, and ions, except for Ca2+ were used. TIP3P model (32) was used for water molecules. The starting structures for wild-type Ca2+-ATPase bound with two Ca2+ were taken from a crystal structure of the enzyme (Protein Data Bank ID: 1su4) (4). Glu58 and Glu908 were treated as protonated (23). The starting structures for Glu771Gln and Glu908Gln were made by substituting the Glu in the wild-type structure with Gln keeping the side chain conformation unchanged. Detailed procedures for setting up a full simulation system including a Ca2+-ATPase, 473 dioleoylphosphatidylcholine (DOPC) phospholipids, two bound Ca2+, 150 mM salt solution were described previously (23) and in SI Text.
Supplementary Material
Acknowledgments.
We thank David McIntosh for help in improving the manuscript and Mai Usui for help in preparing several figures in the manuscript. This research was supported in part by a Creative Science Project grant and Specially Promoted Project Grant from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (C.T.), a Grant-in-Aid for Scientific Research on Innovative Areas, “Transient Macromolecular Complexes” (Y.S.), and the Development and Use of the Next-Generation Supercomputer Project of the Ministry of Education, Culture, Sports, Science, and Technology (Y.S.), CREST and BIRD, Japan Science and Technology Agency (Y.S.). We thank the RIKEN Integrated Cluster of Clusters (RICC) for providing computational resources.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015819107/-/DCSupplemental.
References
- 1.Møller JV, Juul B, le Maire M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim Biophys Acta. 1996;1286:1–51. doi: 10.1016/0304-4157(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 2.Albers RW. Biochemical aspects of active transport. Annu Rev Biochem. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- 3.Post RL, Kume S, Hegyvary C. Activation by adenoshine-triphosphate in phosphorylation kinetics of sodium and potassium ion transport adenosine-triphosphatase. J Biol Chem. 1972;247:6530–6540. [PubMed] [Google Scholar]
- 4.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 5.Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 6.Toyoshima C, Mizutani T. Crystal structure of the calcium pump with a bound ATP analogue. Nature. 2004;430:529–535. doi: 10.1038/nature02680. [DOI] [PubMed] [Google Scholar]
- 7.Toyoshima C, Nomura H, Tsuda T. Lumenal gating mechanism revealed in calcium pump crystal structures with phosphate analogues. Nature. 2004;432:361–368. doi: 10.1038/nature02981. [DOI] [PubMed] [Google Scholar]
- 8.Olesen C, Sørensen T, Nielsen RC, Møller J, Nissen P. Dephosphorylation of the calcium pump coupled to counterion occlusion. Science. 2004;306:2251–2255. doi: 10.1126/science.1106289. [DOI] [PubMed] [Google Scholar]
- 9.Sørensen T, Møller J, Nissen P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science. 2004;304:1672–1675. doi: 10.1126/science.1099366. [DOI] [PubMed] [Google Scholar]
- 10.Obara K, et al. Structural role of countertransport revealed in Ca2+ pump crystal structure in the absence of Ca2+ Proc Natl Acad Sci USA. 2005;102:14489–14496. doi: 10.1073/pnas.0506222102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jensen A, Sørensen T, Olesen C, Møller J, Nissen P. Modulatory and catalytic modes of ATP binding by the calcium pump. EMBO J. 2006;25:2305–2314. doi: 10.1038/sj.emboj.7601135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takahashi M, Kondou Y, Toyoshima C. Interdomain communication in calcium pump as revealed in the crystal structures with transmembrane inhibitors. Proc Natl Acad Sci USA. 2007;104:5800–5805. doi: 10.1073/pnas.0700979104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toyoshima C, Norimatsu Y, Iwasawa S, Tsuda T, Ogawa H. How processing of aspartylphosphate is coupled to lumenal gating of the ion pathway in the calcium pump. Proc Natl Acad Sci USA. 2007;104:19831–19836. doi: 10.1073/pnas.0709978104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Olesen C, et al. The structural basis of calcium transport by the calcium pump. Nature. 2007;450:1036–1042. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- 15.Toyoshima C, Inesi G. Structural basis of ion pumping by Ca2+-ATPase of the sarcoplasmic reticulum. Annu Rev Biochem. 2004;73:269–292. doi: 10.1146/annurev.biochem.73.011303.073700. [DOI] [PubMed] [Google Scholar]
- 16.Toyoshima C. Structural aspects of ion pumping by Ca2+-ATPase of sarcoplasmic reticulum. Arch Biochem Biophys. 2008;476:3–11. doi: 10.1016/j.abb.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 17.Inesi G, Kurzmack M, Coan C, Lewis DE. Cooperative calcium binding and ATPase activation in sarcoplasmic reticulum vesicles. J Biol Chem. 1980;255:3025–3031. [PubMed] [Google Scholar]
- 18.Inesi G. Sequential mechanism of calcium binding and translocation in sarcoplasmic reticulum adenosine triphosphatase. J Biol Chem. 1987;262:16338–16342. [PubMed] [Google Scholar]
- 19.Clarke DM, Loo TW, Inesi G, MacLennan DH. Location of high affinity Ca2+-binding sites within the predicted transmembrane domain of the sarcoplasmic reticulum Ca2+-ATPase. Nature. 1989;339:476–478. doi: 10.1038/339476a0. [DOI] [PubMed] [Google Scholar]
- 20.Andersen JP, Vilsen B. Amino acids Asn796 and Thr799 of the Ca2+-ATPase of sarcoplasmic reticulum bind Ca2+ at different sites. J Biol Chem. 1994;269:15931–15936. [PubMed] [Google Scholar]
- 21.Zhang ZS, Lewis DE, Strock C, Inesi G. Detailed characterization of the cooperative mechanism of Ca2+ binding and catalytic activation in the Ca2+ transport (SERCA) ATPase. Biochemistry. 2000;39:8758–8767. doi: 10.1021/bi000185m. [DOI] [PubMed] [Google Scholar]
- 22.Orlowski S, Champeil P. Kinetics of calcium dissociation from its high-affinity transport sites on sarcoplasmic reticulum ATPase. Biochemistry. 1991;30:352–361. doi: 10.1021/bi00216a007. [DOI] [PubMed] [Google Scholar]
- 23.Sugita Y, Miyashita N, Ikeguchi M, Kidera A, Toyoshima C. Protonation of the acidic residues in the transmembrane cation-binding sites of the Ca2+ pump. J Am Chem Soc. 2005;127:6150–6151. doi: 10.1021/ja0427505. [DOI] [PubMed] [Google Scholar]
- 24.Vilsen B, Andersen JP. Mutation to the glutamate in the fourth membrane segment of Na+, K+-ATPase and Ca2+-ATPase affects cation binding from both sides of the membrane and destabilizes the occluded enzyme forms. Biochemistry. 1998;37:10961–10971. doi: 10.1021/bi9802925. [DOI] [PubMed] [Google Scholar]
- 25.Inesi G, Ma H, Lewis D, Xu C. Ca2+ occlusion and gating function of Glu309 in the ADP-fluoroaluminate analog of the Ca2+-ATPase phosphoenzyme intermediate. J Biol Chem. 2004;279:31629–31637. doi: 10.1074/jbc.M403211200. [DOI] [PubMed] [Google Scholar]
- 26.Rice WJ, MacLennan DH. Scanning mutagenesis reveals a similar pattern of mutation sensitivity in transmembrane sequences M4, M5, and M6, but not in M8, of the Ca2+-ATPase of sarcoplasmic reticulum (SERCA1a) J Biol Chem. 1996;271:31412–31419. doi: 10.1074/jbc.271.49.31412. [DOI] [PubMed] [Google Scholar]
- 27.Rodacker V, Toustrup-Jensen M, Vilsen B. Mutations Phe785Leu and Thr618Met in Na+, K+-ATPase, associated with familial rapid-onset dystonia parkinsonism, interfere with Na+ interaction by distinct mechanisms. J Biol Chem. 2006;281:18539–18548. doi: 10.1074/jbc.M601780200. [DOI] [PubMed] [Google Scholar]
- 28.de Carvalho Aguiar P, et al. Mutations in the Na+/K+-ATPase α3 gene ATP1A3 are associated with rapid-onset dystonia parkinsonism. Neuron. 2004;43:169–175. doi: 10.1016/j.neuron.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 29.Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 A resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 30.Ikeguchi M. Partial rigid-body dynamics in NPT, NPAT, and NPγT ensembles for proteins and membranes. J Comput Chem. 2004;25:529–541. doi: 10.1002/jcc.10402. [DOI] [PubMed] [Google Scholar]
- 31.Mackerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 32.Jorgensen WL, Chandrasekhara J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–935. [Google Scholar]
- 33.DeLano W. The PyMOL User’s Manual. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.