Abstract
Fanconi anemia (FA) is a rare genetic disease characterized by congenital defects, bone marrow failure, chromosomal instability, and cancer susceptibility. One hallmark of cells from FA patients is hypersensitivity to interstrand cross-linking agents, such as the chemotherapeutics cisplatin and mitomycin C (MMC). We have recently characterized a FANCD2/FANCI-associated nuclease, KIAA1018/FAN1, the depletion of which sensitizes human cells to these agents. However, as the down-regulation of FAN1 in human cells was mediated by siRNA and thus only transient, we were unable to study the long-term effects of FAN1 loss on chromosomal stability. We now describe the generation of chicken DT40 B cells, in which the FAN1 locus was disrupted by gene targeting. FAN1-null cells are highly sensitive to cisplatin and MMC, but not to ionizing or UV radiation, methyl methanesulfonate, or camptothecin. The cells do not display elevated sister chromatid exchange frequencies, either sporadic or MMC-induced. Interestingly, MMC treatment causes chromosomal instability that is quantitatively, but not qualitatively, comparable to that seen in FA cells. This finding, coupled with evidence showing that DT40 cells deficient in both FAN1 and FANCC, or FAN1 and FANCJ, exhibited increased sensitivity to cisplatin compared with cells lacking only FAN1, suggests that, despite its association with FANCD2/FANCI, FAN1 in DT40 cells participates in the processing of damage induced by interstrand cross-linking-generating agents also independently of the classical FA pathway.
The mismatch repair (MMR) system has evolved to control the fidelity of DNA replication and recombination. Correspondingly, MMR malfunction in all organisms is associated with an up to 1,000-fold increase in mutation frequency and a hyperrecombination phenotype. Unexpectedly, studies involving primarily knockout mice also implicated MMR genes in other processes of DNA metabolism, ranging from DNA damage signaling to hypermutation of Ig genes, where the molecular roles of MMR proteins are unclear (1). We argued that characterization of the MMR interactome might provide us with new insights into these phenomena. Among the interactors of MLH1 and PMS2 (2), we found an evolutionarily highly conserved hypothetical protein KIAA1018 (Fig. 1) predicted to contain a ubiquitin-binding zinc-finger (UBZ) domain (3), as well as a PD-(D/E)XK motif found in a superfamily of restriction and repair nucleases (4, 5). This polypeptide attracted our interest, because eukaryotic MMR has to date been shown to involve only a single exonuclease, EXO1 (6), whereas Escherichia coli deploys several such enzymatic activities. We therefore set out to characterize KIAA1018 biochemically and to study its biological role. Surprisingly, siRNA-mediated knockdown of KIAA1018 in human cells did not give rise to any phenotypic trait associated with MMR deficiency, but rather led to a hypersensitivity specific to cisplatin and mitomycin C (MMC) (7–10). These agents modify predominantly guanine residues in DNA and form, in addition to monoadducts and intrastrand cross-links, interstrand cross-links (ICLs) that block the progression of replication and transcription. The repair of ICLs at replication forks is complex, inasmuch as it involves proteins from different pathways of DNA metabolism. In higher eukaryotes, this process is coordinated by the Fanconi anemia (FA) pathway, where the ATR kinase activates the FA core complex (consisting of FANCA, B, C, E, F, G, L, and M) to monoubiquitylate the FANCD2/FANCI heterodimer and thus license it to recruit other repair factors to the damage (11). As KIAA1018 (henceforth referred to as FANCD2/FANCI-associated nuclease; FAN1) recruitment to DNA damage-induced foci was dependent on monoubiquitylated FANCD2/FANCI, FAN1 was proposed to be linked to the FA pathway of DNA damage processing (7–10). These studies also suggested that FAN1 might be involved in homologous recombination and in the maintenance of chromosomal stability. However, given the transient nature of siRNA knockdowns, we decided to disrupt the KIAA1018/FAN1 locus in chicken DT40 cells and to study the phenotype of the FAN1-deficient cells in this stable system. We also set out to establish whether the FAN1-deficient phenotype is epistatic with that of FA.
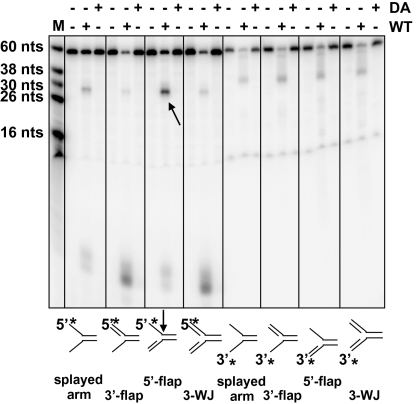
Fig. 1.
Gallus FAN1 is an endonuclease. FAN1 cleaves preferentially 5′ flaps. The wild-type (WT) enzyme or the D977A (DA) mutant was incubated with a series of oligonucleotide substrates shown below the panel. The arrow indicates the preferred cleavage product. The rapidly migrating radioactive species seen at the bottom of this 20% denaturing polyacrylamide gel correspond to DNA fragments 1–4 nucleotides in length. The position of the radiolabel is indicated by an asterisk. M, marker; 3-WJ: three-way junction.
Results
Avian FAN1 Is a Nuclease.
To test whether chicken FAN1 is indeed a nuclease as predicted by bioinformatic analysis (4, 5) and whether it shares its substrate specificity with the human polypeptide, we expressed in Sf9 cells the wild-type chicken protein as well as a mutant carrying an aspartate-to-alanine substitution (D977A) in the putative nuclease active site (Fig. S1A). Incubation of the untagged purified polypeptides (Fig. S1B) with ΦX174 DNA, single-stranded circular substrate that contains numerous secondary structures (hairpin loops and bulges) susceptible to cleavage by most structure-specific endonucleases in vitro, resulted in extensive degradation of the DNA by the wild-type enzyme but not by the D977A mutant (Fig. S1C). As the latter protein was able to bind DNA in a gel-shift assay with an affinity similar to the wild-type protein (Fig. S1D), we concluded that its inactivity was linked to the mutation of the active site, rather than to misfolding. These tests unambiguously showed that avian FAN1 is an endonuclease. By incubating the enzyme with a variety of oligonucleotide substrates, we could confirm that, like the human protein (7–10), the avian enzyme displays preference for the 5′-flap structure, even though other branched substrates were also cleaved, albeit less efficiently (Fig. 1). Interestingly, incubation of FAN1 with the 5′-labeled 3′ flap and three-way junction substrates, which were poorly addressed by the endonuclease as witnessed by the absence of an ∼30-mer product band, gave rise instead to rapidly migrating labeled species that we attributed to 1- to 4-nucleotide-long fragments. This shows that FAN1 also possesses a 5′→3′ exonuclease activity that prefers double-stranded DNA ends (Fig. 1, four left sets). We could confirm this prediction by labeling the bottom 60-mer strands at their 3′ termini. Incubation with FAN1 gave rise in all cases to degradation products, the smallest of which were ∼30 nucleotides long, which most likely resulted from a 5′→3′ degradation of the double-stranded regions of the substrates up to the junction where the strands separate (Fig. 1, four right sets).
Disruption of the DT40 FAN1 Locus Does Not Affect Cell Growth.
The chicken gene encodes a polypeptide of 1,034 amino acids (Fig. S1A) that displays 52% similarity and 65% identity at the amino acid level to its human 1,017-residue ortholog. Targeting constructs were generated which lacked parts of exons 2 and 3 (Fig. S2A) encoding the putative UBZ domain. Disruption of the FAN1 locus was confirmed by Southern blot (Fig. S2B) and RT-PCR (Fig. S2C) analyses. The FAN1−/− and wild-type DT40 cells displayed similar doubling times (Fig. S2D) and very similar cell-cycle profiles (Fig. S2E). Thus, FAN1 disruption does not affect the basal growth properties of DT40 cells.
Disruption of the FAN1 Locus Sensitizes DT40 Cells to ICL-Generating Agents.
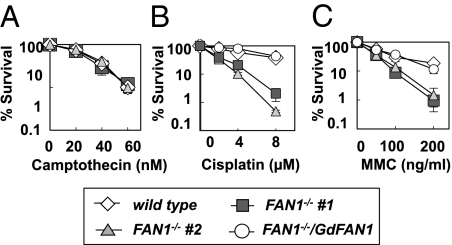
We then measured the sensitivity of the FAN1−/− DT40 cells to a range of DNA-damaging agents. DT40 wild-type and two independent clones of FAN1−/− cells showed similar sensitivities to γ-rays, UV, methyl methanesulfonate (MMS) (Fig. S2 F–H), and camptothecin (Fig. 2A). In contrast, FAN1−/− cells were more sensitive to cisplatin or MMC than wild-type DT40 cells (Fig. 2 B and C). The hypersensitivity to the latter reagents could be rescued by a stable transfection of FAN1−/− clone 1 with a vector carrying chicken wild-type FAN1 cDNA (GdFAN1) (Fig. 2 B and C). These results also agree with those obtained by siRNA-mediated knockdown of FAN1 in human cells (7–10). In contrast, the knockout cells could not be rescued by the expression FAN1 cDNA carrying a D977A substitution in the nuclease domain (Fig. S2I). Taken together, these data show that the observed hypersensitivity to ICL-inducing agents was caused solely by disruption of the FAN1 gene and that the nuclease domain is essential for the function of this protein.
Fig. 2.
FAN1-deficient DT40 cells are hypersensitive to DNA cross-linking agents. DT40 cells of the indicated genotypes were exposed to γ-rays, UV, MMS (Fig. S2 F–H), and camptothecin (A), cisplatin (B), and mitomycin C (C). The doses of the agents are shown on the x axes on a linear scale, whereas the fractions of surviving colonies are displayed on the y axis on a logarithmic scale. Each data point represents an average of at least three independent experiments ±SD. #1 and #2, two independent FAN1 knockout clones. FAN1−/−/GdFAN1 denotes a FAN1 knockout clone stably transfected with an expression vector encoding a FAN1 ORF. As shown, expression of this protein rescued the hypersensitive phenotype of the FAN1 knockout cells.
FAN1 Deficiency Leads to Chromosomal Instability.
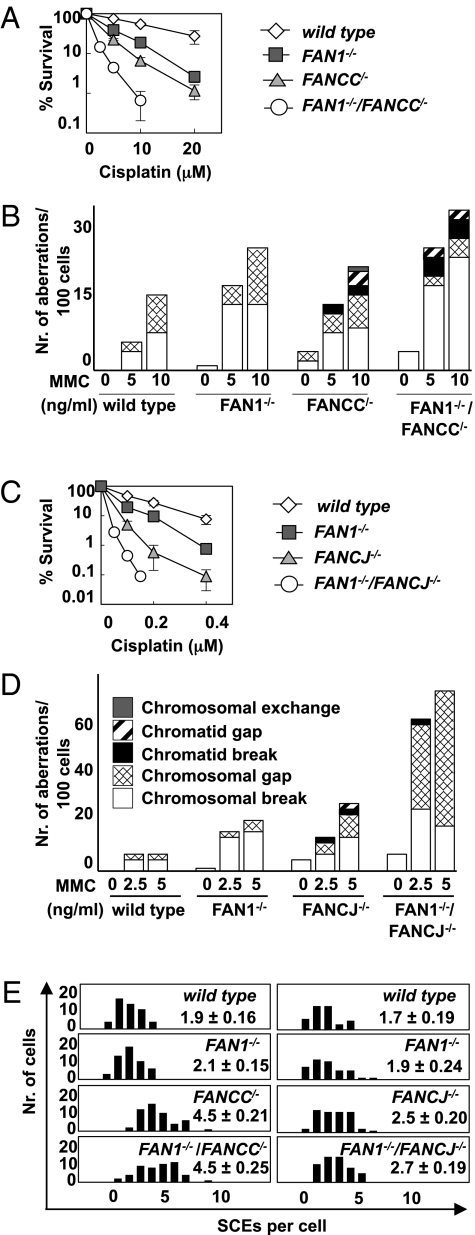
Hypersensitivity to ICL-inducing agents is a common phenotypic trait of FA cells, as is chromosomal instability, both spontaneous and damage-induced (12). We therefore studied sister chromatid exchange (SCE) frequencies in FAN1−/− DT40 cells. As shown in Fig. 3A, only very small increases in spontaneous and MMC-induced SCEs were seen in the FAN1−/− cells compared with the wild-type DT40 control. However, a very different picture emerged when we compared chromosomal aberrations in these two cell lines after MMC treatment. The number of chromosomal changes increased notably (Fig. 3B), and whereas disruption of the FAN1 locus had little effect on spontaneous aberrations, the number of induced chromosomal changes was approximately threefold higher than in wild-type cells upon treatment with 20 ng/mL MMC. Under identical conditions, cells disrupted in the FANCC and FANCJ loci displayed higher numbers of spontaneous aberrations than FAN1−/− cells, as well as a greater number and variety of aberrations after MMC treatment (Fig. 3B). Thus, whereas the changes observed in the wild-type and FAN1-deficient cells were exclusively chromosomal breaks and gaps, FANCC/− and FANCJ−/− cells displayed in addition chromatid aberrations, as could be expected from their increased SCE frequencies (Fig. 3B) (13–15).
Fig. 3.
FAN1-deficient cells display an increased rate of chromosomal aberrations. (A) Sister chromatid exchange frequencies in wild-type and FAN1−/− DT40 cells, either untreated or treated with 20 ng/mL MMC. The graph shows the number of metaphases (y axis) containing a given number of SCEs (x axis). The mean number of SCEs is indicated. At least 50 metaphase spreads were counted for each experiment and the graph shows the mean number of SCEs. (B) Occurrence of chromosomal aberrations in wild-type, FAN1−/−, FANCC/−, and FANCJ−/− DT40 cells, either untreated or treated with the indicated doses of MMC.
In contrast to FANCC/− cells, which displayed a lower frequency of gene targeting than wild-type DT40 cells (Fig. S3A) (13), gene-targeting frequencies measured at two different loci in FAN1−/− cells were similar to those observed in the wild-type controls (Fig. S3A). In addition, no change in the efficiency of homologous recombination for FAN1-deficient cells was detected in a reporter assay which scores for neomycin resistance following the recombination between two disrupted neomycin ORFs integrated in DT40 genomic DNA that is triggered by the induction of a sequence-specific double-strand break in one of the neomycin inserts (Fig. S3B) (16). These results imply that FAN1 is not involved in the control of homologous recombination in DT40 cells.
FA and FAN1 Deficiencies Are Additive Rather than Epistatic.
Although lack of FAN1 or FA proteins leads in both cases to hypersensitivity to cisplatin and MMC, the differences in the subtypes of chromosomal aberrations described above suggested that the enzymes might not process identical lesions or intermediates generated by these agents. To test this prediction, we generated double mutant DT40 cells. The FAN1−/−/FANCC/− double knockout cells grew more slowly than wild-type DT40 cells, but at the same rate as FANCC/− cells (Fig. S4A). Importantly, they displayed higher sensitivity to cisplatin than either single mutant (Fig. 4A), and the number of MMC-induced chromosomal aberrations was also higher than in the single mutants (Fig. 4B). However, the number of spontaneous SCEs in the double knockout cells was not greater than in the FANC/− line (Fig. 4E Left). Similarly to the FANCC/− cells, the FANCJ−/− knockout cell line also proliferated with slower growth kinetics than wild-type DT40 or FAN1−/− cells, and FAN1 inactivation reduced their growth rate even further (Fig. S4B). Expression of wild-type FAN1 in these cells reverted the growth defect back to the level seen in FANCJ−/− cells, which showed that the difference was indeed due to the disruption of the FAN1 locus. Like the FAN1−/−/FANCC/− clones, the FAN1−/−/FANCJ−/− cells were also more sensitive to cisplatin than either of the single knockouts (Fig. 4C) and the number of MMC-induced chromosomal aberrations was likewise increased (Fig. 4D). As in the case of FANCC, SCE frequency in FAN1−/−/FANCJ−/− cells was similar to that seen in FANCJ−/− cells (Fig. 4E Right). This further confirmed our hypothesis that although FAN1 is clearly involved in the cellular response to ICL-inducing agents, it does not appear to be involved in the processing of spontaneous DNA damage together with FA proteins.
Fig. 4.
FAN1/FANCC and FAN1/FANCJ double knockout DT40 cells are more sensitive to cisplatin than single mutants and display more chromosomal aberrations. (A) FAN1−/−/FANCC/− cells are more sensitive to cisplatin than wild-type, FAN1−/−, or FANCC/− single knockout cells. Each data point represents an average of at least three independent experiments ±SD. (B) FAN1−/−/FANCC/− cells display more and different distribution of chromosomal aberrations compared with wild-type, FAN1−/−, or FANCC/− single knockout cells. (C) FAN1−/−/FANCJ−/− cells are more sensitive to cisplatin than wild-type, FAN1−/−, or FANCJ−/− single knockout cells. Each data point represents an average of at least three independent experiments ±SD. (D) Occurrence of chromosomal aberrations in FAN1−/−/FANCCJ−/− cells compared with wild-type, FAN1−/−, or FANCJ−/− single knockout cells. (E) Sister chromatid exchange frequencies in wild type, FAN1−/−, FANCC/−, FANCJ−/−, FAN1−/−/FANCC/−, or FAN1/FANCJ−/− DT40 cells. The y axis shows the number of metaphases containing the number of SCEs indicated on the x axis. At least 50 metaphase spreads were counted for each experiment and the graph shows the mean number of SCEs.
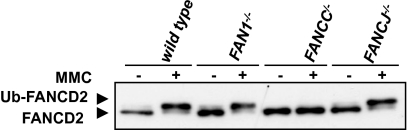
FAN1 Deficiency Does Not Affect Ubiquitylation of FANCD2.
Cells lacking constituent proteins of the FA core complex fail to monoubiquitylate the FANCD2/FANCI heterodimer upon treatment with DNA-damaging agents (11). As shown in Fig. 5, MMC-induced FANCD2 monoubiquitylation in FAN1−/− and in wild-type DT40 cells was similar, in contrast to FANCC/− cells. FAN1 is therefore not required for the activation of the ubiquitin ligase of the FA core complex. This resembles the situation in FANCJ-deficient cells (Fig. 5), and could indicate that FAN1, like FANCJ, participates in DNA damage processing downstream of the FA complex. An alternative explanation might be that FAN1 might not be directly linked to the FA pathway, as implied by the data presented above.
Fig. 5.
Damage-induced FANCD2 monoubiquitylation is unaffected by FAN1 status. In MMC-treated cells, FANCD2 is monoubiquitylated, as witnessed by its slower migration through a denaturing polyacrylamide gel. This posttranslational modification was not detected in FANCC-deficient cells, but was in cells lacking FAN1 or FANCJ.
FAN1 Targeting to DNA Damage Foci Is Dependent on the FA Complex.
Treatment of cells with DNA-damaging agents frequently leads to the accumulation of DNA damage response proteins in subnuclear foci at (or close to) the sites of damage. To find out whether FAN1 behaved similarly, we transfected DT40 cells with a vector expressing a GFP-FAN1 fusion protein. As shown in Fig. S5A, the untreated transfected cells contained a small number of nuclear foci, which might represent spontaneous DNA damage, possibly stalled replication forks. In contrast, in MMC-treated cells, the number of these foci increased substantially. Because MMC is known to induce foci containing the FA factors, we wanted to test whether the FAN1 foci colocalized with those formed by FANCD2, which, together with its heterodimeric partner FANCI, is believed to be responsible for the recruitment of repair proteins to blocked replication forks and other types of DNA damage. That FANCD2 and GFP-FAN1 almost completely colocalized (Fig. S5 A and B) was somewhat unexpected, based on the results of the experiments described above. Even more surprising was the fact that, like FANCD2, GFP-FAN1 failed to form MMC-induced foci when expressed in FANCC-deficient cells. These results indicate that FAN1 and FANCD2 are targeted to the same foci, and that this targeting depends on a functional FA complex. However, as the ubiquitylation of FANCD2 was not affected by FAN1 deficiency (Fig. 5), FAN1 appears to act downstream of FANCD2 (7–10).
Discussion
The data presented in this study confirm and extend our findings in the human system, which showed that FAN1 is a DNA repair enzyme that participates in the processing of lesions formed by ICL-generating compounds (7–10).
Hypersensitivity of FA cells to ICL-inducing agents is a clinical hallmark of Fanconi anemia. However, the slower growth and spontaneous chromosomal instability of FA cells imply that the pathway evolved to deal with endogenous damage, such as DNA adducts arising through reactions with lipid peroxidation by-products, or complex secondary structures that hinder replication, such as hairpins that might arise in regions of dyad symmetry or in triplet repeats (17, 18) and that can give rise to chromosomal instability if uncorrected. These lesions are apparently not addressed by FAN1, given that cells lacking this protein divide with similar kinetics to wild-type DT40 cells (Fig. S2D) and display normal spontaneous SCE frequency (Fig. 3A). Further evidence in support of this hypothesis came from the findings that disruption of the FAN1 locus in FANCC/− or FANCJ−/− cells gave rise to no significant increase in the frequency of SCEs (Fig. 4E). These aberrations are believed to arise through reciprocal exchange of genetic material, where Holliday junctions generated by homologous recombination at arrested or blocked replication forks are resolved to yield cross-over products. Such a situation is prevented primarily by the BLM/TopoIIIα/RMI1 complex and, consequently, BLM-deficient cells exhibit high SCE frequencies. The recruitment of the latter complex to replication forks was recently reported to be mediated (at least in part) by the FANCM protein (19), and this might explain why defects in the FA pathway give rise to SCEs. This evidence might also imply that FAN1 is not recruited or required at these spontaneous replication blocks.
On the other hand, FAN1 is recruited to foci induced by ICL-generating agents, where it most likely acts in the cleavage of cross-linked DNA molecules. It is interesting to note that the 5′-flap preference of FAN1 complements the 3′-flap selectivity of MUS81/EME1 or XPF/ERCC1 (20–22), the other nucleases implicated in ICL repair, and thus that these enzymes might act in concert to release or “unhook” the cross-linked strand to allow for bypass of the lesion.
The finding that recruitment of chicken and human FAN1 to DNA damage foci is dependent on FA complex activation implies a close association with the Fanconi anemia pathway in the processing of induced damage. Here, however, there were differences between human and chicken cells. In the former, siRNA-mediated knockdown of FANCD2 and FAN1 brought about similar hypersensitivity to ICL-generating compounds as seen in the single knockdowns (7). In contrast, the sensitivities of DT40 FAN1/FANCC and FAN1/FANCJ double mutants to cisplatin or MMC were greater than those of the single knockout cell lines (Fig. 4 A and C). Interestingly, a similar phenomenon was reported also for the FANCC/FANCJ double mutant in DT40 cells (15). This suggests that, in chicken DT40 cells, the roles of the FA pathway do not fully overlap with those of FAN1 or FANCJ. Thus, although the FA complex in human cells appears to recruit to blocked replication forks all proteins that are necessary for their rescue, the processing of damage induced by cross-linking agents in chicken cells appears to be channeled into different pathways, some of which involve the FA complex and others not. Although this scenario might appear unlikely due to the high degree of evolutionary conservation of the FA pathway in higher eukaryotes, the FAN1 nuclease motif evolved earlier, as its homologs are found already in Schizosaccharomyces pombe and Pseudomonas aeruginosa. The latter proteins contain the endonuclease domain but not the zinc-finger motif that is required for interaction with FANCD2 (8); it will therefore be interesting to learn whether these FAN1 homologs are involved in ICL processing in these monocellular organisms.
Fanconi anemia is a rare genetic disease characterized by congenital defects, bone marrow failure, chromosomal instability, and cancer susceptibility (11). Given that untreated FAN1-deficient cells do not display proliferation defects, the likelihood that individuals with inactivating mutations in FAN1 will present with an FA-like syndrome may be low. It is also possible that FAN1 is functionally at least partially redundant with other, as yet unidentified, nucleases. Functional redundancy might help explain why genes encoding XPF/ERCC1 or MUS81/EME1 (20–22) have not been found mutated in FA patients. However, it is likely that FAN1 will turn out to be clinically important, as its loss clearly sensitizes cells to ICL-generating drugs. Given that MMC and especially the platinum compounds cisplatin, oxaliplatin, and carboplatin are in increasing use in the therapy of a broad range of cancers (23, 24), elucidation of the biological role of this enzyme in the metabolism of ICLs is of substantial clinical importance. By generating multiple knockout lines, the DT40 system should allow us to dissect ICL processing further, and thus provide us with insights into this important pathway of DNA metabolism.
Methods
Accession codes, cDNA, protein purification, and standard procedures are described in SI Text.
Specific Nuclease Assay.
One nanomole of labeled substrate was incubated with 10 nmol of FAN1 wild-type or D977A mutant in 25 mM Hepes·KOH (pH 7.4), 25 mM KCl, 1 mM MgCl2 as indicated, and 0.05 mg/mL BSA for 30 min at 37 °C. The reaction was terminated with 0.1% SDS, 14 mM EDTA, and 0.1 mg/mL proteinase K and incubation at 55 °C for 15 min. Loading buffer was added and the samples were denatured and separated on a 20% denaturing polyacrylamide gel for 1 h at 40 V/cm. The gels were dried and the bands were visualized on a PhosphorImager (Typhoon 9400; GE Healthcare).
Proliferation Analysis and Sensitivity Assay.
The experimental methods for proliferation analysis and sensitivity assays were performed as described previously (16). To measure the sensitivity of DT40 double knockout FAN1/FANCC and FAN1/FANCJ cells, cells were cultured in medium containing cisplatin. The experimental methods for cell counting and cell-cycle analysis were described previously (16).
Chromosomal Aberration Analysis.
Preparation of chromosome spreads and karyotype analysis were performed as described previously (25). To measure mitomycin C-induced chromosomal aberrations, cells were incubated with 2.5, 5, 10, and 20 ng/mL mitomycin C for 24 h before harvest and fixation. Cells were incubated with colcemid (final concentration, 0.1 μg/mL) for the last 2 h to enrich for mitotic cells.
Supplementary Material
Acknowledgments
The authors thank Drs. H. Arakawa (Max Planck Institute, Martinsried, Germany) and J. M. Buerstedde (Genome Damage and Stability Centre, Brighton, UK) for generous gifts of DT40 strains and vectors. This work was supported by Swiss National Science Foundation Grant 3100A0-118158 to J.J.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011081107/-/DCSupplemental.
References
- 1.Jiricny J. The multifaceted mismatch-repair system. Nat Rev Mol Cell Biol. 2006;7:335–346. doi: 10.1038/nrm1907. [DOI] [PubMed] [Google Scholar]
- 2.Cannavo E, Gerrits B, Marra G, Schlapbach R, Jiricny J. Characterization of the interactome of the human MutL homologues MLH1, PMS1, and PMS2. J Biol Chem. 2007;282:2976–2986. doi: 10.1074/jbc.M609989200. [DOI] [PubMed] [Google Scholar]
- 3.Notenboom V, et al. Functional characterization of Rad18 domains for Rad6, ubiquitin, DNA binding and PCNA modification. Nucleic Acids Res. 2007;35:5819–5830. doi: 10.1093/nar/gkm615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kinch LN, Ginalski K, Rychlewski L, Grishin NV. Identification of novel restriction endonuclease-like fold families among hypothetical proteins. Nucleic Acids Res. 2005;33:3598–3605. doi: 10.1093/nar/gki676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kosinski J, Feder M, Bujnicki JM. The PD-(D/E)XK superfamily revisited: Identification of new members among proteins involved in DNA metabolism and functional predictions for domains of (hitherto) unknown function. BMC Bioinformatics. 2005;6:172. doi: 10.1186/1471-2105-6-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: Functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 7.Kratz K, et al. Deficiency of FANCD2-associated nuclease KIAA1018/FAN1 sensitizes cells to interstrand crosslinking agents. Cell. 2010;142:77–88. doi: 10.1016/j.cell.2010.06.022. [DOI] [PubMed] [Google Scholar]
- 8.MacKay C, et al. Identification of KIAA1018/FAN1, a DNA repair nuclease recruited to DNA damage by monoubiquitinated FANCD2. Cell. 2010;142:65–76. doi: 10.1016/j.cell.2010.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smogorzewska A, et al. A genetic screen identifies FAN1, a Fanconi anemia-associated nuclease necessary for DNA interstrand crosslink repair. Mol Cell. 2010;39:36–47. doi: 10.1016/j.molcel.2010.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu T, Ghosal G, Yuan J, Chen J, Huang J. FAN1 acts with FANCI-FANCD2 to promote DNA interstrand cross-link repair. Science. 2010;329:693–696. doi: 10.1126/science.1192656. [DOI] [PubMed] [Google Scholar]
- 11.Moldovan GL, D'Andrea AD. How the Fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Winter JP, Joenje H. The genetic and molecular basis of Fanconi anemia. Mutat Res. 2009;668:11–19. doi: 10.1016/j.mrfmmm.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 13.Hirano S, et al. Functional relationships of FANCC to homologous recombination, translesion synthesis, and BLM. EMBO J. 2005;24:418–427. doi: 10.1038/sj.emboj.7600534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niedzwiedz W, et al. The Fanconi anaemia gene FANCC promotes homologous recombination and error-prone DNA repair. Mol Cell. 2004;15:607–620. doi: 10.1016/j.molcel.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 15.Bridge WL, Vandenberg CJ, Franklin RJ, Hiom K. The BRIP1 helicase functions independently of BRCA1 in the Fanconi anemia pathway for DNA crosslink repair. Nat Genet. 2005;37:953–957. doi: 10.1038/ng1627. [DOI] [PubMed] [Google Scholar]
- 16.Yamamoto K, et al. Fanconi anemia FANCG protein in mitigating radiation- and enzyme-induced DNA double-strand breaks by homologous recombination in vertebrate cells. Mol Cell Biol. 2003;23:5421–5430. doi: 10.1128/MCB.23.15.5421-5430.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hyrien O. Mechanisms and consequences of replication fork arrest. Biochimie. 2000;82:5–17. doi: 10.1016/s0300-9084(00)00344-8. [DOI] [PubMed] [Google Scholar]
- 18.Voineagu I, Narayanan V, Lobachev KS, Mirkin SM. Replication stalling at unstable inverted repeats: Interplay between DNA hairpins and fork stabilizing proteins. Proc Natl Acad Sci USA. 2008;105:9936–9941. doi: 10.1073/pnas.0804510105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deans AJ, West SC. FANCM connects the genome instability disorders Bloom's syndrome and Fanconi anemia. Mol Cell. 2009;36:943–953. doi: 10.1016/j.molcel.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 20.Hanada K, et al. The structure-specific endonuclease Mus81-Eme1 promotes conversion of interstrand DNA crosslinks into double-strands breaks. EMBO J. 2006;25:4921–4932. doi: 10.1038/sj.emboj.7601344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Silva IU, McHugh PJ, Clingen PH, Hartley JA. Defining the roles of nucleotide excision repair and recombination in the repair of DNA interstrand cross-links in mammalian cells. Mol Cell Biol. 2000;20:7980–7990. doi: 10.1128/mcb.20.21.7980-7990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhagwat N, et al. XPF-ERCC1 participates in the Fanconi anemia pathway of cross-link repair. Mol Cell Biol. 2009;29:6427–6437. doi: 10.1128/MCB.00086-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Keys HM, et al. Cisplatin, radiation, and adjuvant hysterectomy compared with radiation and adjuvant hysterectomy for bulky stage IB cervical carcinoma. N Engl J Med. 1999;340:1154–1161. doi: 10.1056/NEJM199904153401503. [DOI] [PubMed] [Google Scholar]
- 24.Morris M, et al. Pelvic radiation with concurrent chemotherapy compared with pelvic and para-aortic radiation for high-risk cervical cancer. N Engl J Med. 1999;340:1137–1143. doi: 10.1056/NEJM199904153401501. [DOI] [PubMed] [Google Scholar]
- 25.Takata M, et al. Homologous recombination and non-homologous end-joining pathways of DNA double-strand break repair have overlapping roles in the maintenance of chromosomal integrity in vertebrate cells. EMBO J. 1998;17:5497–5508. doi: 10.1093/emboj/17.18.5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.