Abstract
The best known outcome of horizontal gene transfer (HGT) is the introduction of novel genes, but other outcomes have been described. When a transferred gene has a homolog in the recipient genome, the native gene may be functionally replaced (and subsequently lost) or partially overwritten by gene conversion with transiently present foreign DNA. Here we report the discovery, in two lineages of plant mitochondrial genes, of novel gene combinations that arose by conversion between coresident native and foreign homologs. These lineages have undergone intricate conversion between native and foreign copies, with conversion occurring repeatedly and differentially over the course of speciation, leading to radiations of mosaic genes involved in respiration and intron splicing. Based on these findings, we develop a model—the duplicative HGT and differential gene conversion model—that integrates HGT and ongoing gene conversion in the context of speciation. Finally, we show that one of these HGT-driven gene-conversional radiations followed two additional types of conversional chimerism, namely, intramitochondrial retroprocessing and interorganellar gene conversion across the 2 billion year divide between mitochondria and chloroplasts. These findings expand our appreciation of HGT and gene conversion as creative evolutionary forces, establish plant mitochondria as a premiere system for studying the evolutionary dynamics of HGT and its genetic reverberations, and recommend careful examination of bacterial and other genomes for similar, likely overlooked phenomena.
Keywords: gene duplication, recombination
Horizontal gene transfer (HGT) is well established as a major force in bacterial evolution (1, 2) and is increasingly recognized as relatively common in certain eukaryotic lineages and genomes (3, 4). Among organellar genomes, plant mitochondrial genes stand out for their relatively high frequency of HGT (4, 5), especially in parasitic plants and their hosts and most notably, in the tropical shrub Amborella (6). Apart from the special case of a highly invasive group I intron—whose frequent self-mediated horizontal acquisition is accompanied by coconversion of flanking exonic sequences (7)—only 2 of 42 cases of plant mitochondrial HGT reported to date have evidence for recombination between native and foreign homologs. The mitochondrial rps11 gene of the eudicot Sanguinaria is a 50:50 chimera: its 5′ half is of native, eudicot origin, whereas its 3′ half is of foreign, monocot origin (8). In the other chimeric case, a central region encompassing more than one-half the endogenous atp1 gene (which is itself the product of an earlier HGT event) from one population of the parasite Pilostyles thurberi has been replaced by a sequence from the parasite's host plant (9). In both cases, only one copy of the relevant gene seems to be present in the mitochondrial genome, and therefore these could reflect immediate recombination between (transiently present) foreign DNA and native homologs, which is commonly seen in bacterial genomes (10, 11).
These two chimeric genes both involve the joining of large blocks of native and foreign DNA such that the chimericism was readily apparent, even by whole-gene phylogenetic analysis (8). However, gene conversion often involves short tracts of DNA and can, therefore, be difficult to detect (12, 13). Indeed, using a recombination search algorithm developed specifically for plant mitochondrial genomes (14), nine putative cases of short-patch gene conversion of native, functional plant mitochondrial atp1 genes by homologous atpA genes of chloroplast origin were described just last year (15), despite most of the relevant gene sequences having been published years ago. In this study, we used the same search algorithm to reexamine 16 published cases of plant mitochondrial HGT for evidence of gene conversion between native mitochondrial genes and those introduced through HGT [these include all known cases of mitochondrial HGT in plants except for those in the remarkably HGT-rich genome of Amborella (6), which are subject to ongoing analysis]. This search revealed two cases of overlooked chimerism, and chimericism was also detected for a newly described HGT event. Two of these three newly discovered cases of chimeric plant mitochondrial genes are the subject of this paper, because they reveal a pathway for the evolutionary diversification of genes that has not, to our knowledge, been described before. In both cases, repeated gene conversion between coresident native and foreign homologs, occurring differentially over the course of speciation, has created highly diverse lineages of evolutionarily mosaic genes. We generalize these findings by developing a model for HGT-driven gene diversification, termed the duplicative HGT and differential gene conversion (DH-DC) model.
Results
Two Lineages of Differentially Mosaic Mitochondrial Genes.
One lineage of mosaic genes was identified through reexamination of the 16 published, non-Amborella cases of plant mitochondrial HGT (SI Appendix, Table S1). This revealed two cases of previously overlooked chimerism. One of these, like the two published cases of HGT-driven chimeric mitochondrial genes in plants (8, 9), involves simple chimerics, whereby only a single region of a gene was converted (differentially) in two descendant lineages. These two conversion events involve transfer of the atp1 gene in the genus Plantago (16) and are reported in a separate study on the horizontal acquisition of multiple foreign genes in Plantago mitochondrial genomes (17). The other overlooked case also involves the mitochondrial atp1 gene (which encodes the α-subunit of the mitochondrial ATP synthase), of Ternstroemia stahlii, a small tree from the shores of Puerto Rico. This gene sequence was published in 2005 as part of an 11-gene study (18) on Ericales phylogeny, which concluded that HGT might be responsible for the anomalous placement (within the Ericaceae) of the atp1 gene from T. stahlii, a member of the Pentaphylacaceae. A combination of algorithmic search (14) and visual inspection of currently available atp1 sequences led us to conclude that, although most of the T. stahlii gene is indeed of foreign origin, it is actually highly chimeric, with five short regions of native sequence interspersed with five mostly longer regions of foreign origin.
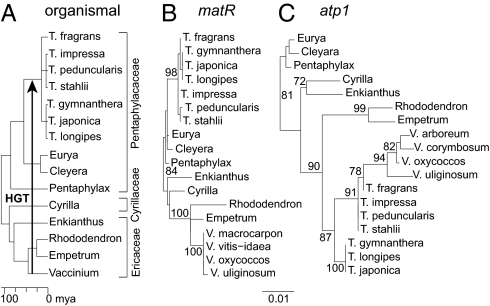
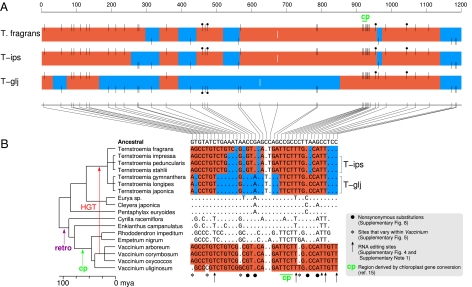
Discovery of such mosaicism prompted us to sequence atp1 from an additional 16 Ternstroemia taxa and five other members of the Ericales. To aid interpretation, we also sequenced two other mitochondrial genes (cox1 and matR) and three chloroplast genes (atpB, matK, and rbcL) from a varying subset of Ternstroemia species. With the exception of atp1, there is little molecular diversity within Ternstroemia, most of which divides the genus into a basal trichotomy (Fig. 1 A and B and SI Appendix, Figs. S1–S3). The atp1 tree (Fig. 1C) shows both unexpected levels of divergence within Ternstroemia and also an anomalous but well-supported topology, with Ternstroemia placed within the wrong family (Ericaceae) and the Ericaceous genus Vaccinium placed within Ternstroemia. The atp1 sequence alignment (SI Appendix, Fig. S4), shown boiled down to its HGT-related essentials in Fig. 2, reveals that each of the three major Ternstroemia clades possesses a differentially mosaic atp1 gene, each with multiple (four or five) foreign regions interspersed with native regions. All three types of mosaic genes evidently trace back to the same initiating HGT event, quite possibly from a donor group within the blueberry genus Vaccinium (Fig. 2B and SI Appendix, Fig. S5). This transfer is inferred to have occurred 15–50 Myr ago between donor and recipient groups that diverged about 100 Myr ago (Figs. 1A and 2B).
Fig. 1.
Aberrant phylogeny and branch lengths of mitochondrial atp1 in Ternstroemia. (A) Chronogram showing organismal relationships and divergence times of key taxa examined from the Ericales (Methods and SI Appendix, Fig. S1) (18). (B and C) Phylograms from maximum likelihood analysis of mitochondrial matR and atp1 sequences corresponding to the alignments shown in SI Appendix, Figs. S2 and S4, respectively. Bootstrap values ≥70% are shown. T, Ternstroemia; V, Vaccinium. With but a single nucleotide exception, all eight examined members from each of the two multisampled Ternstroemia subclades possess identical atp1 sequences (SI Appendix, Fig. S10); therefore, the analyses shown in this figure and elsewhere in the paper are restricted to just three members of each clade.
Fig. 2.
Differentially mosaic atp1 genes in Ternstroemia. (A) Multicolored boxes represent atp1 genes of the three subclades within Ternstroemia. Black vertical lines represent the 38 nucleotide positions inferred to have differed between donor and recipient atp1 genes at the time of atp1 transfer from Vaccinium to a Ternstroemia ancestor. Lines at the top of the boxes and red shading indicate sites and regions, respectively, of putatively foreign Vaccinium ancestry, whereas bottom lines and blue shading represent native sites and regions. White lines centered within the boxes represent the only two sites that otherwise differ within the Ternstroemia clade (SI Appendix, Text S5). (B) Nucleotide sequence of all sites marked in A for the 18 Ericales taxa shown. Dots indicate identities relative to the atp1 sequence inferred for the common ancestor of these taxa (SI Appendix, Fig. S4), whereas letters show nucleotide differences. Positions left blank represent gaps in sequence coverage. Retro indicates retroprocessing (SI Appendix, Text S1). Although the trichotomy at the base of Ternstroemia allowed us to place T. fragrans next to the T-ips subclade and thereby simplify the color-shading pattern, the best current estimate of Ternstroemia phylogeny (SI Appendix, Fig. S1) actually groups T. fragrans with T-glj to the exclusion of T-ips. If correct, this implies that the more intricately chimeric pattern shared by T. fragrans and T-ips reflects conversion events that were ancestral to the entire clade, with the T-glj clade sustaining subsequent conversions that reverted large segments of atp1 to native sequence along with a reciprocal change at position 961.
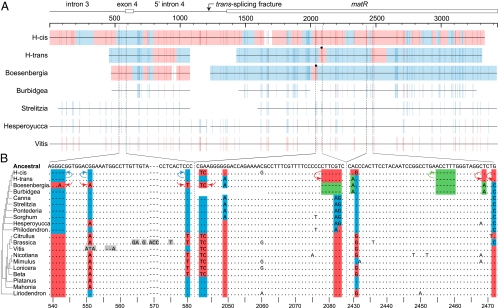
The second case of HGT-driven differential mosaicism arose from a Southern blot study (19), which suggested that Hedychium coronarium contains a second, potentially foreign (i.e., cis-spliced) copy of the fourth intron of the respiratory gene nad1 in addition to the expected form (i.e., trans-spliced) of the intron. To investigate this, we sequenced a ∼3.4-kb region encompassing most of this large intron from Hedychium, two other members of the ginger family Zingiberaceae, and eight other phylogenetically diverse angiosperms. Hedychium alone among these taxa contains two detectable copies of this region, one (H-trans) with a trans-spliced configuration of intron 4, as found in all related monocots (ref. 19 and this study), and the other (H-cis) with a cis-spliced configuration. Phylogenetic analysis (SI Appendix, Fig. S6) of matR, a splicing factor gene located within this intron, shows that the predominant phylogenetic signal in H-cis matR is foreign, derived from core eudicots, whereas H-trans matR is native. Inspection of the alignment (Fig. 3 and SI Appendix, Fig. S7), however, reveals that both Hedychium copies are, in fact, chimeras, with H-cis inferred to contain at least 13 mostly short regions of the native ginger-derived sequence interspersed with longer segments of foreign eudicot-derived DNA and H-trans at least three regions of foreign DNA. Boesenbergia, the sister to Hedychium in our sampling, also has a mosaic sequence, with its chimerism most evident upstream of matR (Fig. 3). One of its upstream foreign regions is present in identical form in H-trans, and thus the foreign elements in both taxa probably derive from the same transfer event. Burbidgea (a ginger that is sister to the Hedychium/Boesenbergia clade) and the nonginger monocots examined show little, if any, evidence of foreign sequence (Fig. 3A). The HGT event that underlies the chimerism in Hedychium and Boesenbergia probably occurred less than 26 Myr ago—the estimated divergence time (20) of the ginger subfamilies Zingiberoideae (represented in this study by Hedychium and Boesenbergia) and Alpinoideae (represented by Burbidgea)—between donor and recipient groups that diverged ∼145 Myr ago (21).
Fig. 3.
Mosaicism in nad1/matR genes of gingers. (A) Summary of HGT-relevant nucleotide positions for three mosaic ginger loci [H-cis, the ancestrally foreign (SI Appendix, Text S8), cis-spliced locus from Hedychium; H-trans, the ancestrally native, trans-spliced loci from Hedychium; and the Boesenbergia locus, which is also trans-spliced] and loci from four other angiosperms, which show little, if any, evidence of HGT. Background blue and red shading indicates, for the three mosaic loci, segments that seem to be of native and foreign origin, respectively, based on our interpretation of the presence of monocot- or ginger-specific nucleotide sites (blue) or eudicot-specific sites (red) (SI Appendix, Fig. S7). A transition between these two states was inferred when a blue site was followed by a red site or vice versa. Those sites deemed less convincing (i.e., more likely caused by point mutational homoplasy) were not used to define breakpoints between foreign and native ancestry. These regions of ambiguous origin are not shaded. Black circles indicate converted positions in H-trans and Boesenbergia that result in amino acid changes (SI Appendix, Fig. S9) (the highly chimeric H-cis sequence is not marked, because it is clearly a pseudogene). (B) Nucleotide alignments of three short portions of a taxonomically augmented version of the full alignment (SI Appendix, Fig. S7) that underlies the summary shown in A. Dots indicate identities relative to the reference ancestral angiosperm sequence, letters show nucleotide differences, and indels are marked by dashes. Green shading indicates those nucleotide character states that are specific to the last common ancestor of the sampled gingers. Blue indicates states that are also native to the gingers but are shared more widely with other monocots. Red indicates states present in a common ancestor of core eudicots (Citrullus through Beta) and often, through HGT, in certain gingers too. Gray marks two short regions of such exceptionally high divergence that they may have arisen by conversion from unidentified sources (SI Appendix, Text S2). Longer colored arrows indicate directionality of within-Hedychium conversions; shorter arrows indicate conversions inferred for the sole detected nad1/matR locus in Boesenbergia.
Provenance, Expression, and Functionality of Mosaic Genes.
We sequenced cDNAs for H-trans matR and T. gymnanthera atp1. The former is C to U edited at 10 sites, 7 of them nonsynonymous (SI Appendix, Fig. S7), whereas the latter is C to U edited at the one site of predicted editing (SI Appendix, Fig. S4). Recovery of edited transcripts is evidence of both correct expression at the RNA level and a mitochondrial location of these genes, because C to U editing is common in plant mitochondria (occurring mostly at nonsynonymous sites) but unknown for plant nuclear genes (22). Further evidence that these genes (and all chimeric genes characterized in this study) are mitochondrial is of two types. First, apart from the chimeric regions themselves, these genes show the highly conserved properties expected for genes located in the generally low mutation rate environment of the mitochondrial genome as opposed to genes transferred to the plant nuclear genome, where the synonymous substitution rate is usually 10–100 times higher (23). Second, in sharp distinction to the frequent functional intracellular transfer of many other mitochondrial genes in plants, there is no evidence that atp1 or the matR gene located in nad1 intron 4 has ever been functionally transferred to the nucleus in plants, despite survey of hundreds of diverse angiosperms (24). The only chimeric sequence discovered in this study that is probably a pseudogene is H-cis matR, because it has two frame shifts and two in-frame stop codons. All other chimeric reading frames are intact insofar as sequenced.
Additional Cases of Potential Mitochondrial Gene Conversion.
Two subsets of the many nucleotide differences (Fig. 2) introduced to Ternstroemia by HGT-associated gene conversion apparently arose themselves by prior gene conversions of two distinct types. The first was a putative retroprocessing event—conversion of the atp1 gene by a cDNA made from an edited atp1 mRNA—that changed at least four positions of C to U RNA editing from C to T in the common ancestor of Cyrilla + Ericaceae (Fig. 2 and SI Appendix, Text S1). The second is one of several cases of likely chloroplast-mediated conversion of mitochondrial genes recently reported (15, 25). This event occurred early in Ericaceae evolution and replaced four closely spaced nucleotides of mitochondrial atp1 origin with variant chloroplast-derived nucleotides (Fig. 2).
Many other potential conversion events, some compelling and others not, are evident on inspection of the mitochondrial datasets analyzed in this study. Especially clear cut are cases involving the nine Ericales cox1 sequences displayed in SI Appendix, Fig. S3. These include multiple newly discovered retroprocessing events (SI Appendix, Text S1) as well as the previously reported coconversion of a short, 3′-flanking exon region (SI Appendix, Fig. S3) that accompanied the self-insertion of a mobile homing group I intron in Symplocus, an event that has occurred through hundreds of independent HGTs during angiosperm evolution (7, 26). Fig. 3B shows (in gray) two short patches in nad1 intron 3 of such exceptional divergence that they may also have arisen by gene conversion (these two 9-nt patches contain more changes than in the whole of the remainder of the 200-nt regions in which the patches are centrally embedded). SI Appendix, Text S2 details these two cases and briefly describes several other cases of potential conversion.
Discussion
Model That Integrates HGT and Gene Conversion in the Context of Speciation.
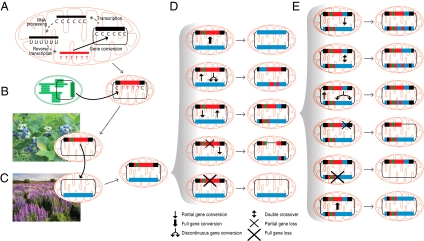
The discovery of two diverse lineages of differentially mosaic genes leads us to propose a model of gene evolution—the DH-DC model—that marries HGT, gene conversion, and speciation. The first step in this model is duplicative HGT (i.e., integration of a foreign region apart from a native homologous locus). Homologous recombination between foreign and native loci ensues, potentially occurring repeatedly and differentially (Fig. 4) over the course of cladogenesis and thereby leading to a lineage of diverse, differentially mosaic genes as found in the sampled members of Ternstroemia (Fig. 2) and Zingiberaceae (Fig. 3). The short length of these recombinations, combined with the nonreciprocal nature of all inferred events in Hedychium (the only species in this study for which both native and foreign loci were recovered), indicates that they probably occurred by gene conversion (Fig. 3) (12, 13). Gene conversion can occur either continuously or discontinuously (28, 29), the latter consistent with the highly chimeric nature of the loci described herein.
Fig. 4.
Multiple pathways of mitochondrial gene conversion and multiple fates of native and foreign genes brought together by HGT. (A) Mitochondrial retroprocessing (cDNA-mediated gene conversion). (B) Chloroplast to mitochondrial gene conversion. (C) Mitochondrial HGT with a foreign gene (in three colors) integrating apart from a native homolog (in blue). (D) Five potential recombinational outcomes from this state of gene duplication. (E) Six potential recombinational outcomes from the state of dual mosaic genes depicted as the middle outcome in D. Not shown are single cross-over events; these are common in plant mitochondrial genomes (27) and lead to inversion for inversely oriented duplications and resolution of the master chromosome into two subgenomic circles for directly oriented duplications. Also not shown is the possibility that the two forms of the gene are present on different forms of the mitochondrial genome, one present at high copy and the other as a low-copy sublimon (27).
For only one examined species (Hedychium) (Fig. 3) is the DH-DC model supported by finding dual mosaic genes in the mitochondrion (SI Appendix, Text S3) of a given plant. Nonetheless, in both lineages, this model is far more likely than the alternative, which would require multiple independent transfers of the same locus from the same donor group to the same recipient group. This alternative is especially improbable for the atp1 transfer, given both the precision with which the donor (blueberry) group has been identified and the inference of multiple foreign/native boundaries shared between two or all three main subclades of the recipient Ternstroemia group (Fig. 2 and SI Appendix, Fig. S4). If the DH-DC model is indeed correct, then why, despite all efforts (SI Appendix, Text S3), did we fail to find a second distinct form of the transferred locus in most species examined? Biological explanations (Fig. 4) include (i) loss or truncation/fragmentation of one copy or the other, rendering it no longer detectable by PCR, (ii) partition of the two copies into the master and subliminal forms of variant genomes known to occur in plant mitochondria (27), rendering the subliminal copy difficult to detect, and (iii) endgame homogenization/concerted evolution of the two copies through pervasive gene conversion, leading them to become identical and therefore indistinguishable. The homogenizing face of gene conversion might create a ratchet—a slippery slope—leading more or less divergent homologs brought together by HGT to become more similar with time; increasing similarity promotes higher rates of gene conversion (30), ultimately leading to complete homogenization in some fraction of outcomes.
Evolution under the DH-DC model creates novel molecular variation, some fraction of it potentially adaptive. The sequence changes introduced by gene conversion, as with recombination in general, differ from those that arise by de novo mutational mechanisms in two key ways: the mutational spectrum has already been filtered by a history of selection, and gene conversion allows for simultaneous introduction of multiple, possibly coadapted, changes. As expected for functional protein-coding genes (Results), most of the fixed nucleotide changes in atp1 (Fig. 2) and matR (Fig. 3) introduced by gene conversion are probably neutral or nearly so, falling at synonymous and/or RNA editing sites. The limited extent of conversion in the functional matR gene of Hedychium (H-trans) compared with its nonfunctional locus (H-cis) (Fig. 3) may also reflect purifying selection. Nonetheless, conversion has generated a modicum of protein diversity, causing two to four amino acid changes among the Ternstroemia atp1 genes (Fig. 2 and SI Appendix, Fig. S8) and a single amino acid change each for the functional matR genes of Hedychium and Boesenbergia (Fig. 3 and SI Appendix, Fig. S9). Furthermore, certain intron-specific changes introduced by DH-DC may also have functional consequences (SI Appendix, Text S4).
Gene Conversion in Plant Mitochondrial Genomes.
The mitochondrial datasets explored in this study illustrate the diversity of sources and pathways by which gene conversion sculpts the sequence landscape of plant mitochondrial genes. Most notably, the mosaic genes in Ternstroemia seem to have arisen by a complicated series of gene conversions (Fig. 4) involving nucleic acids of different chemical type (DNA and RNA), deeply divergent evolutionary origin (organelles of ca. 2 billion year common ancestry), or different organismal residency (blueberry donor and Ternstroemia recipient). These findings reinforce other recent studies (15, 31) in suggesting that gene conversion, through converting sequences of endogenous (retroprocessed) or foreign origin (both horizontal and intracellular), might play a much bigger role in plant mitochondrial genome evolution than generally realized.
Obtaining accurate estimates of the frequency and extent of gene conversion in plant mitochondrial genomes—overall and with respect to different types of converting sequences—will require far more data than are currently available as well as more effective tools to detect conversion (Methods). With respect specifically to HGT, clear evidence for gene conversion between native and foreign homologs has now been obtained for 5 of 17 cases of non-Amborella HGT listed in SI Appendix, Table S1. Importantly, conversion may have either gone undetected in some of the other 12 cases or been effectively impossible in the first place. The latter is represented by the first three cases of HGT listed in SI Appendix, Table S1, in which genes previously lost from the mitochondrial genome (owing to functional transfer to the nucleus) were reacquired from an unrelated mitochondrial donor (8). In several other cases (SI Appendix, Table S1), PCR failed to recover a native-like gene, raising the possibility that opportunities for ongoing gene conversion were limited by short coresidency times of foreign and native homologs. At the extreme, coresidency times will be zero and chimerics will be nearly impossible to detect when HGT occurs by direct conversion of transiently present foreign DNA so as to replace either most of the native locus or conversely, only a small portion of it. The chimeric genes in Sanguinaria (8) and Pilostyles (9) thus represent the “Goldilocks” or “just right” zone.
The findings from this and other recent plant mitochondrial studies (15, 31) are consistent with the overall gene conversion literature (12, 13) in suggesting that most gene conversions in plant mitochondrial genomes are relatively short, ranging from several to a few hundred nucleotides in length. This, together with the very low rates of apparent point mutations in most plant mitochondrial genomes (23), means that most plant-to-plant HGT-driven conversions will be difficult to detect with confidence. Those short conversion patches that carry a disproportionate number of differences relative to native sequences will be most readily detected. This is the criterion that we used at the end of Results to infer the possibility of several additional gene conversion events. As described in SI Appendix, Text S2, denser taxonomic sampling should help discriminate in such cases between conversion (and other processes that might produce multiple mutations in synchrony) (SI Appendix, Text S2) and the serial accumulation of single mutations.
General Implications.
The richness of the conversional landscape in plant mitochondrial genes has troubling implications for efforts to estimate mutation rates and exploit these genes as sources of phylogenetic information. The latter is less problematic, but only if plant phylogeneticists carefully examine mitochondrial sequence alignments, discarding all those regions of likely horizontal origin and counting each HGT event as but a single character. The implications for estimating mutation rates derive from the realization that single-nucleotide differences in a plant mitochondrial sequence alignment may not be the product of conventional point mutation (i.e., nucleotide misincorporation during DNA replication or repair). Instead, some unknowable fraction must have arisen by conversion from donor sequences either native or foreign to the plant and its mitochondrial genome. In this sense, the already unusually low mutation (i.e., synonymous substitution) rates estimated for most plant mitochondrial genomes (23) should be considered overestimates. The Ternstroemia atp1 gene illustrates all of these conundrums. Its variously foreign nature wreaks havoc on atp1 phylogenies (Fig. 1C) (18), and at least 32, if not all 34, nucleotide variants that arose during the roughly 50 Myr of Ternstroemia atp1 evolution (Fig. 1A) result not from conventional point mutation but from gene conversion here (coupled with HGT). The two potential exceptions here (SI Appendix, Text S5) illustrate the fundamental problem in distinguishing between these evolutionary forces.
Unusually low mutation rates and a propensity for HGT are just two of many features (SI Appendix, Text S6) that make angiosperm mitochondrial genomes fertile territory for detecting both HGT and gene conversion and exploring their evolutionary interplay through DH-DC. The cornerstone of the Ternstroemia atp1 case of DH-DC was a sequence published and recognized as likely foreign 5 years ago (18) but whose complex chimeric nature was overlooked until now. This is just part of a larger history of HGT and gene conversion being long overlooked in plant mitochondrial genomes (8, 15). With HGT so rampant in the bacterial world (1, 2) and multigene families and gene conversion so common (32, 33), including between presumptively transiently present foreign DNA and native genes (34, 35), it is surprising that no cases of DH-DC have yet been reported in bacteria. Surely, it must happen, and equally surely, it too must be overlooked. One contributing factor is the understandable practice of relying on formal programs and associated significance tests to recognize recombination, because these are largely insensitive (SI Appendix, Text S7) to detect the kinds of mosaicism reported in this study. Our findings thus pose two challenges to students of genome evolution. They should stimulate the development of more sensitive approaches for identifying gene conversion and impel careful search—regardless of genome and organism—for the kinds of genetic mosaicism that plant mitochondrial genomes so spectacularly display.
Methods
Plant DNA and RNA were prepared as in ref. 8 or obtained from various sources (Acknowledgments). Voucher information and GenBank numbers are listed in SI Appendix, Table S2. PCR and RT-PCR were carried as in ref. 8. Sequences were aligned using MUSCLE (36). Phylogenies were constructed using PhyML (37) with a GTR+Γ+I model. A dated phylogenetic tree (chronogram) of the Ericales was constructed using the BEAST program (38), and a Eurya reference fossil calibration of 86 Myr ago (39) was applied to a three-chloroplast gene dataset (atpB, rbcL, and matK) that included the taxa shown in Fig. 1A plus three not shown outgroups (Fouquieria, Marcgravia, and Pentamerista). Ancestral sequences were reconstructed using BASEML in the PAML package (40).
Supplementary Material
Acknowledgments
We thank B. Cruz and P. Fritsch (California Academy of Sciences, San Francisco), J. Kress (Smithsonian Institution, Washington, DC), K. Kron (Wake Forest University, Winston-Salem, NC), L. Prince (Rancho Santa Ana Botanic Garden, Claremont, CA), Y.-L. Qiu (University of Michigan, Ann Arbor, MI), the Plant DNA Bank in Korea (Korea University, Seoul, Korea), and the Kew DNA Bank (Royal Botanic Gardens, Kew, UK) for providing most of the DNAs used in this study; M. McKain and Y. Yuan for providing the Hesperoyucca and Mimulus nad1 sequences, respectively; Y.-L. Qiu for advice; A. Alverson, D. Sloan, and the two reviewers for critical reading of the manuscript; and E. Anderson for graphical assistance. W.H. was supported by a Natural Sciences and Engineering Research Council Postdoctoral Fellowship. A.O.R. was supported by a National Science Foundation Graduate Research Fellowship. Y.Z. was supported by the China Scholarship Council. J.D.P. was supported by National Institutes of Health Grant R01-GM-70612. W.H. and J.D.P. were supported by the METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. HQ437915–HQ437987).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016295107/-/DCSupplemental.
References
- 1.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Beiko RG, Harlow TJ, Ragan MA. Highways of gene sharing in prokaryotes. Proc Natl Acad Sci USA. 2005;102:14332–14337. doi: 10.1073/pnas.0504068102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Keeling PJ, Palmer JD. Horizontal gene transfer in eukaryotic evolution. Nat Rev Genet. 2008;9:605–618. doi: 10.1038/nrg2386. [DOI] [PubMed] [Google Scholar]
- 4.Bock R. The give-and-take of DNA: Horizontal gene transfer in plants. Trends Plant Sci. 2010;15:11–22. doi: 10.1016/j.tplants.2009.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Richardson AO, Palmer JD. Horizontal gene transfer in plants. J Exp Bot. 2007;58:1–9. doi: 10.1093/jxb/erl148. [DOI] [PubMed] [Google Scholar]
- 6.Bergthorsson U, Richardson AO, Young GJ, Goertzen LR, Palmer JD. Massive horizontal transfer of mitochondrial genes from diverse land plant donors to the basal angiosperm Amborella. Proc Natl Acad Sci USA. 2004;101:17747–17752. doi: 10.1073/pnas.0408336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sanchez-Puerta MV, Cho Y, Mower JP, Alverson AJ, Palmer JD. Frequent, phylogenetically local horizontal transfer of the cox1 group I Intron in flowering plant mitochondria. Mol Biol Evol. 2008;25:1762–1777. doi: 10.1093/molbev/msn129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergthorsson U, Adams KL, Thomason B, Palmer JD. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature. 2003;424:197–201. doi: 10.1038/nature01743. [DOI] [PubMed] [Google Scholar]
- 9.Barkman TJ, et al. Mitochondrial DNA suggests at least 11 origins of parasitism in angiosperms and reveals genomic chimerism in parasitic plants. BMC Evol Biol. 2007;7:248. doi: 10.1186/1471-2148-7-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klasson L, et al. The mosaic genome structure of the Wolbachia wRi strain infecting Drosophila simulans. Proc Natl Acad Sci USA. 2009;106:5725–5730. doi: 10.1073/pnas.0810753106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mau B, Glasner JD, Darling AE, Perna NT. Genome-wide detection and analysis of homologous recombination among sequenced strains of Escherichia coli. Genome Biol. 2006;7:R44. doi: 10.1186/gb-2006-7-5-r44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Palmer S, Schildkraut E, Lazarin R, Nguyen J, Nickoloff JA. Gene conversion tracts in Saccharomyces cerevisiae can be extremely short and highly directional. Nucleic Acids Res. 2003;31:1164–1173. doi: 10.1093/nar/gkg219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaeffer SW, Anderson WW. Mechanisms of genetic exchange within the chromosomal inversions of Drosophila pseudoobscura. Genetics. 2005;171:1729–1739. doi: 10.1534/genetics.105.041947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hao W. OrgConv: Detection of gene conversion using consensus sequences and its application in plant mitochondrial and chloroplast homologs. BMC Bioinformatics. 2010;11:114. doi: 10.1186/1471-2105-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hao W, Palmer JD. Fine-scale mergers of chloroplast and mitochondrial genes create functional, transcompartmentally chimeric mitochondrial genes. Proc Natl Acad Sci USA. 2009;106:16728–16733. doi: 10.1073/pnas.0908766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mower JP, Stefanović S, Young GJ, Palmer JD. Plant genetics: Gene transfer from parasitic to host plants. Nature. 2004;432:165–166. doi: 10.1038/432165b. [DOI] [PubMed] [Google Scholar]
- 17.Mower JP, et al. Horizontal acquisition of multiple mitochondrial genes from a parasitic plant followed by gene conversion with host mitochondrial genes. BMC Biol. 2010 doi: 10.1186/1741-7007-8-150. 10.1186/1471-2148-9-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schönenberger J, Anderberg AA, Sytsma KJ. Molecular phylogenetics and patterns of floral evolution in the Ericales. Int J Plant Sci. 2005;166:265–288. [Google Scholar]
- 19.Qiu YL, Palmer JD. Many independent origins of trans splicing of a plant mitochondrial group II intron. J Mol Evol. 2004;59:80–89. doi: 10.1007/s00239-004-2606-y. [DOI] [PubMed] [Google Scholar]
- 20.Janssen T, Bremer K. The age of major monocot groups inferred from 800+ rbcL sequences. Bot J Linn Soc. 2004;146:385–398. [Google Scholar]
- 21.Bell CD, Soltis DE, Soltis PS. The age and diversification of the angiosperms re-revisited. Am J Bot. 2010;97:1296–1303. doi: 10.3732/ajb.0900346. [DOI] [PubMed] [Google Scholar]
- 22.Shikanai T. RNA editing in plant organelles: Machinery, physiological function and evolution. Cell Mol Life Sci. 2006;63:698–708. doi: 10.1007/s00018-005-5449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mower JP, Touzet P, Gummow JS, Delph LF, Palmer JD. Extensive variation in synonymous substitution rates in mitochondrial genes of seed plants. BMC Evol Biol. 2007;7:135. doi: 10.1186/1471-2148-7-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adams KL, Qiu YL, Stoutemyer M, Palmer JD. Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc Natl Acad Sci USA. 2002;99:9905–9912. doi: 10.1073/pnas.042694899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sloan DB, Alverson AJ, Storchová H, Palmer JD, Taylor DR. Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol Biol. 2010;10:274. doi: 10.1186/1471-2148-10-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cho Y, Qiu YL, Kuhlman P, Palmer JD. Explosive invasion of plant mitochondria by a group I intron. Proc Natl Acad Sci USA. 1998;95:14244–14249. doi: 10.1073/pnas.95.24.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maréchal A, Brisson N. Recombination and the maintenance of plant organelle genome stability. New Phytol. 2010;186:299–317. doi: 10.1111/j.1469-8137.2010.03195.x. [DOI] [PubMed] [Google Scholar]
- 28.Santoyo G, Martínez-Salazar JM, Rodríguez C, Romero D. Gene conversion tracts associated with crossovers in Rhizobium etli. J Bacteriol. 2005;187:4116–4126. doi: 10.1128/JB.187.12.4116-4126.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Radford SJ, Sabourin MM, McMahan S, Sekelsky J. Meiotic recombination in Drosophila Msh6 mutants yields discontinuous gene conversion tracts. Genetics. 2007;176:53–62. doi: 10.1534/genetics.107.070367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Majewski J, Cohan FM. DNA sequence similarity requirements for interspecific recombination in Bacillus. Genetics. 1999;153:1525–1533. doi: 10.1093/genetics/153.4.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR. Extensive loss of RNA editing sites in rapidly evolving Silene mitochondrial genomes: Selection vs. retroprocessing as the driving force. Genetics. 2010;185:1369–1380. doi: 10.1534/genetics.110.118000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Santoyo G, Romero D. Gene conversion and concerted evolution in bacterial genomes. FEMS Microbiol Rev. 2005;29:169–183. doi: 10.1016/j.femsre.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 33.Morris RT, Drouin G. Ectopic gene conversions in bacterial genomes. Genome. 2007;50:975–984. doi: 10.1139/g07-076. [DOI] [PubMed] [Google Scholar]
- 34.Baldo L, Bordenstein S, Wernegreen JJ, Werren JH. Widespread recombination throughout Wolbachia genomes. Mol Biol Evol. 2006;23:437–449. doi: 10.1093/molbev/msj049. [DOI] [PubMed] [Google Scholar]
- 35.Chan CX, Beiko RG, Darling AE, Ragan MA. Lateral transfer of genes and gene fragments in prokaryotes. Genome Biol Evol. 2009;1:429–438. doi: 10.1093/gbe/evp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 38.Drummond AJ, Rambaut A. BEAST: Bayesian evolutionary analysis by sampling trees. BMC Evol Biol. 2007;7:214. doi: 10.1186/1471-2148-7-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bremer K, Friis EM, Bremer B. Molecular phylogenetic dating of asterid flowering plants shows early Cretaceous diversification. Syst Biol. 2004;53:496–505. doi: 10.1080/10635150490445913. [DOI] [PubMed] [Google Scholar]
- 40.Yang Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.