Abstract
The Clone 13 (Cl13) strain of lymphocytic choriomeningitis virus is widely studied as a model of chronic systemic viral infection. Here, we used reverse genetic techniques to identify the molecular basis of Cl13 persistence and immunosuppression, the characteristics differentiating it from the closely related Armstrong strain. We found that a single-point mutation in the Cl13 polymerase was necessary and partially sufficient for viral persistence and immunosuppression. A glycoprotein mutation known to enhance dendritic cell targeting accentuated both characteristics but when introduced alone, failed to alter the phenotype of the Armstrong strain. The decisive polymerase mutation increased intracellular viral RNA load in plasmacytoid dendritic cells, which we identified as a main initial target cell type in vivo, and increased viremia in the early phase of infection. These findings establish the enhanced replicative capacity as the primary determinant of the Cl13 phenotype. Viral persistence and immunosuppression can, thus, represent a direct consequence of excessive viral replication overwhelming the host's antiviral defense.
Keywords: persistent viral infections, viral polymerase, plasmacytoid dendritic cell, viral tropism, acquired immunodeficiency syndrome
Systemic persistent infection with lymphocytic choriomeningitis virus (LCMV) has been studied for almost a century (1), providing important concepts of virus–host interaction that subsequently have been extended to HIV and hepatitis C virus (HCV) infection in humans (2). One particularly useful feature of the LCMV model is the existence of virus strains that are genetically closely related but differ in their ability to establish persistent infection in vivo (3, 4). Thereby, self-limiting infection (successful immune response) and persistent infection (unsuccessful immune defense, which is often associated with acquired immunodeficiency) can be compared side by side to investigate underlying mechanisms.
Much progress has recently been made to understand the subversion of the antiviral immune defense in the context of persistent infection. Signaling through inhibitory receptors such as programmed death 1 (5) and lymphocyte-activation gene 3 (6) and exposure to the antiinflammatory cytokines TGF-β (7) and IL-10 (8, 9) render antiviral cytotoxic T-lymphocytes (CTLs) dysfunctional. Additionally, it has been proposed that persistent infection with continuous exposure to antigen may by itself cause T-cell exhaustion (10–12). Accordingly, investigations in LCMV and simian immunodeficiency virus infection have indicated that viral outpacing of the ensuing T-cell response is a hallmark of persistent infection (12). These two general mechanisms likely represent complementary elements of a complex series of events leading to immune paralysis and virus persistence (13, 14). However, the early events leading to this outcome and the molecular determinants endowing viruses with the capacity to subvert the host defense remain insufficiently defined.
LCMV has a bisegmented negative-strand RNA genome (Fig. 1A). The long segment (L) encodes the RNA-dependent RNA polymerase protein L and the matrix protein Z, which is responsible for particle formation. The glycoprotein (GP) is expressed from the short segment (S) and mediates receptor binding and cell fusion, whereas the nucleoprotein (NP) encapsidates the viral RNA for recognition by the viral polymerase complex (15–18).
Fig. 1.
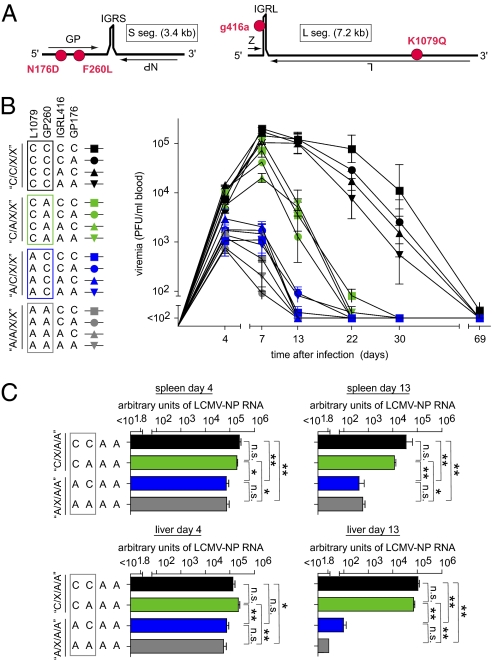
Reverse genetic mapping of LCMV Cl13 persistence. (A) Schematic of the LCMV genome consisting of the S (GP and NP genes) and L segments (Z and L genes), with intergenic regions (IGR) separating the respective two genes. Three coding mutations (N176D and F260L in GP and K1079Q in L) as well as the noncoding mutation in the IGR of the L segment (g416a) are indicated, differentiating the Cl13 and ARM strains (red). (B and C) C57BL/6 mice were infected i.v. with 2 × 106 pfu of the viruses indicated in the chart. The individual virus nomenclature (e.g., C/C/C/C) and denomination of virus groups (e.g., C/C/X/X) according to the four analyzed genome positions reflect the combination of Cl13 and ARM mutations and are described in detail in Results. At the indicated time points after infection, viral titers and viral RNA loads were determined in blood (B), spleen, and liver (C) as indicated. Each symbol and bar represents the mean ± SEM of three to five mice. In B, representative results from one of three experiments are shown.
The persisting LCMV variant Clone 13 (Cl13) infects CD11c+ dendritic cells (DCs) more efficiently than the nonpersistent strain Armstrong (ARM) owing to a point mutation in the GP (F260L) that alters receptor tropism (19, 20). Whether this DC targeting concerns myeloid DCs (mDCs) and/or plasmacytoid DCs (pDCs) remains to be investigated. Moreover, LCMV infection modulates the DC phenotype and antigen presentation capacity (21). Based on these observations, the differential ability of ARM and Cl13 to persist has been accredited to the superior ability of Cl13 to target DCs through GP-mediated receptor tropism (19, 21). It is, therefore, commonly assumed that viral targeting to DCs causes altered antigen presentation and costimulation and thereby, results in defective T-cell responses and ultimately, viral persistence. However, it has remained unclear whether, apart from DC targeting, the establishment of persistence may depend on the differential replicative capacity of specific viral isolates.
HIV as well as HCV can replicate in DCs (22–26). Moreover, the parameters governing persistence or clearance of hepatitis C and B virus remain incompletely defined (27–29). Hence, a better general understanding of these mechanisms in murine LCMV infection may have general implications for the development of refined strategies for the prevention of chronicity of viral infections.
Results
Reverse Genetic Mapping of LCMV Cl13 Persistence.
Using reverse genetic tools (30), we set out to map the genetic basis of Cl13 persistence. We revisited the ARM and Cl13 genome sequences (GenBank numbers AY847350, AY847351, DQ361065, and DQ361066), which revealed coding and noncoding mutations as outlined in Tables 1 and 2 and Fig. 1A. Three coding differences were identified (denominated as ARM → Cl13). F → L at amino acid 260 of the viral GP (GP260) is known to account for increased receptor binding affinity (19). Furthermore, we found a previously unknown N → D mutation at amino acid 176 of GP (GP176) and K → Q at amino acid 1,079 of the viral polymerase gene L (L1079). This latter change is commonly found in persistence-prone LCMV variants (19, 32). Noncoding mutations were also identified, but only one nucleotide transition in the intergenic region (IGR; a → g at nt 416) of the L segment was considered for further analysis owing to the known structural role of the IGR in LCMV transcription termination and particle formation (33).
Table 1.
Comparison of the LCMV-ARM with LCMV-Cl13 genomes: S segment
Table 2.
Comparison of the LCMV-ARM with LCMV-Cl13 genomes: L segment
We generated a panel of 16 recombinant LCMVs representing all possible combinations of the Cl13 or ARM sequences at the above four positions. Each recombinant virus will, thus, be denominated according to the origin of its genomic sequence at positions L1079/GP260/IGRL416/GP176 in this order: A indicating ARM-derived and C indicating Cl13-derived sequences (Fig. 1B). Thus, the WT Cl13 genotype is referred to as C/C/C/C, whereas A/A/A/A stands for a Cl13 genome wherein all four studied positions are mutated to the sequence found in ARM. Furthermore, we will use X to summarize groups of viruses with either Cl13 or ARM sequence at a given position (e.g., the four viruses sharing the Cl13-derived amino acid residues at L1079 and GP260 will be summarized as C/C/X/X) (Figs. 1B, 2, and 3A). All 16 recombinant viruses displayed indistinguishable growth in BHK-21 cells used to amplify viral stocks. This was not unexpected because of the known identical growth kinetics of ARM and Cl13 in standard immortalized cell lines (34).
Fig. 2.
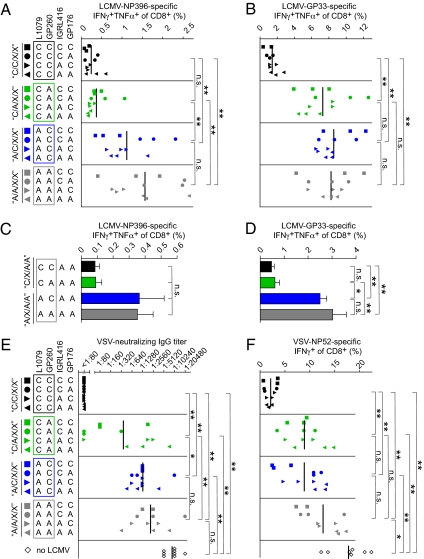
Genetic determinants of LCMV-induced CD8+ T-cell exhaustion and generalized immunosuppression. (A–D) C57BL/6 mice were infected i.v. with 2 × 106 pfu of the indicated LCMV; 22 d later, IFN-γ/TNF-α coproducing CD8+ T cells specific for NP396 (A) and GP33 (B) were enumerated by intracellular cytokine staining from blood. On day 13 after infection, analogous analyses were performed in spleen (C and D). (E and F) C57BL/6 mice were infected i.v. with 2 × 106 pfu of the indicated LCM virus and 18 d later, were given 2 × 106 pfu VSV i.p.; 8 d later (day 26 after LCMV infection), VSV-specific IgG was measured in serum (E), and VSV-NP52–specific IFN-γ–producing CD8+ T cells were enumerated in peripheral blood (F). Symbols in A, B, E, and F represent individual mice. Bars in C and D indicate the mean ± SEM of three to five mice. A, B, E, and F show representative results from two independent experiments.
Fig. 3.
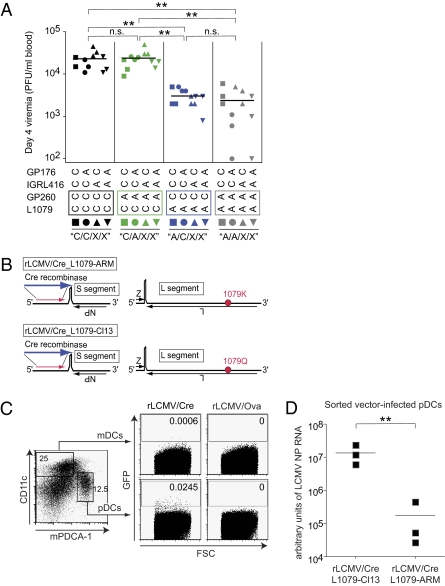
Cl13 polymerase mutation enhances replication in pDCs and increases early viremia. (A) C57BL/6 mice were infected with 2 × 106 pfu of the indicated viruses i.v., and viremia was determined 4 d later. Symbols represent individual mice. Representative results from one of two independent experiments are shown. (B) Schematic representation of the replication-deficient rLCMV/Cre_L1079-ARM (lysine at position 1079 of L) and rLCMV/Cre-L1079-Cl13 vector genomes (glutamine at position 1079 of L). Note that substitution of GP for Cre recombinase renders the resulting vector replication-deficient. (C and D) STOP-GFP reporter mice were inoculated with 107 pfu of rLCMV/Cre_L1079-Cl13 pseudotyped with GP of Cl13 from producer cells; 3 d later, pDC and mDC subsets (discriminated according to mPDCA-1 and CD11c expression) were analyzed for GFP expression. rLCMV/OVA (vector expressing ovalbumin instead of Cre recombinase) served as GFP background control. Numbers indicate the percentage of GFP-positive cells. (D) STOP-GFP reporter mice were inoculated with 107 pfu of rLCMV/Cre_L1079-Cl13 or rLCMV/Cre_L1079-ARM. Both vectors were pseudotyped with the GP of Cl13 from producer cells to assure identical cell targeting; 3 d later, GFP-positive pDCs were sorted by FACS, total cellular RNA was extracted, and LCMV S segment RNA was determined by quantitative RT-PCR. Each symbol represents a pool of sorted cells prepared from splenocytes obtained from three mice.
For in vivo mapping of the Cl13 persistence phenotype, we infected C57BL/6 mice with either of the recombinant viruses. The course of viremia (Fig. 1B) displayed a clear segregation of the 16 viruses into three main phenotypic clusters, indicative for an unbalanced contribution of the four mutations to the phenotypic difference of ARM and Cl13. Viruses with the L1079 polymerase version of Cl13 (C/X/X/X) reached considerably higher peak viremia than A/X/X/X viruses, indicating that this mutation dictates virus load differences between ARM and Cl13. Within the group of eight C/X/X/X viruses, the four C/C/X/X viruses persisted longer than the four C/A/X/X viruses, which is compatible with the known role of GP260 in facilitating DC targeting. However, infection with A/C/X/X viruses resulted in viremia of similarly short duration as A/A/X/X infections, suggesting that the GP260-mediated DC targeting effect required the L1079 mutation to become manifest in prolonged persistence. Unlike the L1079 and GP260 mutations, neither the IGRL416 nor the GP176 mutation influenced viral persistence to a measurable extent, irrespective of the Cl13 amino acid position at L1079 and GP260. When the latter two decisive mutations were further studied in the context of an ARM virus backbone, the L1079 mutation resulted in significantly higher viral RNA loads in spleen and liver, both on day 4 and 13 after infection (Fig. 1C). Conversely, introduction of the Cl13 GP260 mutation did not augment viral loads, irrespective of the L1079 position. These experiments provided independent evidence for a key role of the L1079 mutation in determining viral loads in the organs of mice.
Genetic Determinants of LCMV-Induced CD8+ T-Cell Exhaustion and Generalized Immunosuppression.
Next, we studied whether differential virus loads and persistence correlated with the propensity of the various recombinant viruses to subvert the LCMV-specific CTL response, a process commonly referred to as exhaustion (4, 10, 11). For this, we determined the frequency of IFN-γ and TNF-α coproducing CD8+ T cells specific for nucleoprotein (NP) 396–404 (NP396) and GP33–41 (GP33) on day 22 after infection (Fig. 2 A and B). NP-specific CTLs are of particular importance for early virus control (35) and are most affected by exhaustion, whereas GP-specific cells are more resistant to this process (11). The NP396-specific response of animals infected with viruses carrying the L1079 version of ARM (A/X/X/X viruses) was significantly higher than the one of mice harboring C/X/X/X viruses, indicating that the Cl13 polymerase mutation was also the primary determinant of suppressed LCMV-NP–specific CTL responses. Notably, C/C/X/X (GP260 of Cl13) and C/A/X/X viruses (GP260 of ARM) caused similar suppression of NP396-specific CTL responses, whereas GP33-specific responses were only suppressed in C/C/X/X-infected mice (L1079 and GP260 of Cl13). Irrespective of the epitope studied, A/C/X/X-infected animals (GP260 of Cl13) exhibited a similarly intact CD8+ T-cell response as those infected with A/A/X/X viruses (GP260 of ARM), indicating that GP260 of Cl13 failed to promote CTL exhaustion in the absence of the L1079 mutation. To analyze the role of L1079 and GP260 in CD8+ T-cell exhaustion in spleen, we analyzed responses to a select set of viruses (X/X/A/A viruses) on day 13 after infection (Fig. 2 C and D). GP33-specific responses to viruses carrying the L1079 version of ARM (A/X/A/A viruses) were significantly higher than the one of mice harboring C/X/A/A viruses, indicating that the Cl13 polymerase mutation was also the primary determinant of splenic CD8+ T-cell responses to GP33 on day 13. Conversely, the introduction of the GP260 mutation did not exert obvious effects on GP33-specific responses, neither when tested in combination with L1079 (C/C/A/A vs. C/A/A/A) nor when assessed on its own (A/C/A/A vs. A/A/A/A). Analogous trends were noted for splenic NP396-specific responses, but these responses exhibited a higher degree of variability, and potential differences between groups failed to reach statistical significance. In summary, the analysis of LCMV-specific CTL exhaustion in spleen and blood revealed a very consistent role of the L1079 position, whereas the contributive effect of GP260 was more variable.
LCMV infection can also cause generalized immunosuppression, dampening the immune defense against unrelated third-party infections. To assess the viral genetic basis for immunosuppression, we infected mice with our panel of LCMV recombinants and subsequently challenged them with vesicular stomatitis virus (VSV). VSV infection elicits an early and potent neutralizing IgG response, preventing viral neuroinvasion and fatal myeloencephalitis (36, 37), and it elicits a potent CD8+ T-cell response to the dominant VSV NP-derived epitope NP52–59 (NP52). We performed VSV challenge on day 18 after LCMV infection and determined VSV-neutralizing IgG titers and NP52-specific CD8+ T-cell responses 8 d later (26 d after LCMV infection) (Fig. 2 C and D). These analyses revealed a similar hierarchy of generalized immunosuppression as observed for exhaustion of LCMV-specific CTL responses. Mice infected with C/C/X/X viruses (L1079 and GP260 of Cl13) mounted significantly lower VSV-specific IgG titers and VSV-NP52–specific CD8+ T-cell responses than animals infected with C/A/X/X, A/C/X/X, or A/A/X/X viruses. These results established a joint role of the Cl13 mutations L1079 and GP260 in suppressing humoral and cellular immune responses to an unrelated third-party antigen. Interestingly, GP260 of Cl13 and thus, enhanced DC targeting (19) failed to significantly accentuate the suppression of VSV-specific immune responses when tested in the context of the ARM polymerase (similar responses in A/C/X/X- and A/A/X/X-infected mice; P > 0.05). Conversely, VSV-neutralizing IgG responses were lower in animals infected with C/A/X/X viruses than in A/A/X/X-infected groups (P < 0.01), indicating that the polymerase position L1079 by itself enhanced the immunosuppressive capacity of LCMV independently of the GP260 mutation. Taken together, these results established the L1079 position of Cl13 as the primary determinant of peak viremia and persistence as well as of LCMV-specific CTL exhaustion and generalized immunosuppression. Additionally, the GP260 mutation, which is responsible for increased a-dystroglycan affinity and DC targeting, played an accessory role in enhancing the duration of persistence and generalized immunosuppression by those viruses that also carried the L1079 position of Cl13.
Cl13 Polymerase Mutation Enhances Replication in pDCs and Increases Early Viremia.
We then aimed to differentiate between the possibilities that the L1079 position directly affected the viral replicative capacity in vivo and that increased viral loads resulted solely from subverted and thus, inefficient immune defense. For this purpose, we analyzed viremia on day 4 (i.e., before the onset of the antiviral CD8+ T-cell response). Already at this early time point, C/X/X/X viruses (L1079 of Cl13) had reached almost 10-fold higher levels of viremia than viruses carrying the ARM version of the polymerase (A/X/X/X viruses, P < 0.01 in two independent experiments) (Fig. 3A). This polymerase effect was independent of the GP260 mutation (C/C/X/X vs. C/A/X/X, P > 0.05; A/C/X/X vs. A/A/X/X, P > 0.05) and thus, was indicative for L1079-dependent differences in RNA replication by the Cl13 and ARM polymerases in vivo. Thus, we set out to directly quantify intracellular viral RNA replication in the first viral target cells in vivo. We have recently developed a platform of replication-deficient (r)LCMV vectors (38). Substitution of the LCMV envelope GP for Cre recombinase (rLCMV/Cre) (Fig. 3B) creates single-round vectors that require GP pseudotyping from producer cells for infectivity. Such vector particles infect target cells in vivo but cannot spread to neighboring cells, because they cannot produce GP, which is responsible for receptor binding and membrane fusion by infectious LCMV particles (15). When administered to transgenic GFP reporter mice (STOP-GFP mice) (39), intracellular expression of Cre recombinase mediates excision of a loxP-flanked transcriptional STOP cassette in the transgene, switching on transgenic GFP expression selectively in rLCMV/Cre-infected cells (38). We had previously shown that rLCMV vectors were virtually exclusively targeted to CD11c-positive DCs (38). Here, we analyzed this initial target cell population further (Fig. 3C); we found that the vast majority of rLCMV vector-infected DCs expressed intermediate levels of CD11c and were positive for mPDCA-1, identifying them as plasmacytoid DCs (40). We next created two versions of rLCMV/Cre vectors, expressing either the ARM (rLCMV/Cre_L1079-ARM) or Cl13 version of the L polymerase gene (rLCMV/Cre_L1079-Cl13) (Fig. 3B). Both genomes were pseudotyped with Cl13 GP from the same stably transfected producer cell (Methods) to assure identical cell tropism in vivo. We then inoculated STOP-GFP mice with either rLCMV/Cre_L1079-ARM or rLCMV/Cre_L1079-Cl13 and isolated infected (GFP-positive) pDCs by FACS sorting. Quantification by TaqMan RT-PCR and standardization for a housekeeping gene (Fig. 3D) revealed that the L gene of Cl13 (expressed by rLCMV/Cre_L1079-Cl13) resulted in ≥10-fold higher levels of viral RNA in infected pDCs than its ARM counterpart (rLCMV/Cre_L1079-ARM; P < 0.01). These results showed unequivocally that the L polymerase mutation K1079Q in Cl13 directly enhances viral RNA replication in the first viral target cell population in vivo, offering a direct explanation for higher viral loads in the early phase of infection irrespective of immunosuppression and T-cell exhaustion.
Discussion
This study in the LCMV model shows that enhanced intracellular replication—a property lent by the L1079 mutation in the Cl13 polymerase—is a primary determinant of viral chronicity and exhaustion of the virus-specific T-cell response as well as generalized immunosuppression. The abundance of antigen is known to determine the rate of T-cell exhaustion (14, 41). Here, we report that the L1079 mutation increased intracellular viral RNA levels under conditions of single-round vector infection in vivo (rLCMV/Cre) (38) and thus, in the absence of spreading infectivity or T-cell exhaustion. This indicates that enhanced replication with a resulting increase in viral loads is the cause rather than the consequence of viral persistence, T-cell exhaustion, and generalized immunosuppression.
We acknowledge that DCs not only are initial targets of LCMV infection (38), spreading the infection to other cell types throughout the body (42), but also represent the main cell type priming LCMV-specific CTLs in vivo (43), presumably after virus-induced phenotypic conversion of pDCs into CD11c high-expressing classical DCs (44). Higher levels of intracellular viral RNA owing to the L1079 mutation may, thus, exert diverse effects on DC homeostasis and may have direct immunomodulatory influence on the T-cell response (21). Such potential L1079 effects on DC homeostasis are, however, insufficient to explain the wide-ranging effects of this mutation, because we have recently reported that rLCMV vectors equipped with the Cl13 version of the LCMV polymerase trigger highly functional and protective CTL immunity (38). Still, the contributive effects of GP260 on long-term persistence of Cl13 are not a result of early viral replication kinetics. Similar viremia in C/C/X/X and C/A/X/X viruses up to day 7 of infection contrasts with accelerated clearance of C/A/X/X viruses. The impact of GP260 on long-term persistence may, thus, reflect immunomodulatory effects owing to DC targeting (19, 21). Alternatively and not mutually exclusively, delayed clearance of C/C/X/X viruses may reflect alterations in viral dissemination to specific cell types and tissue microcompartments, where immune-mediated clearance is more difficult to achieve. This possibility is supported by the observation that Cl13 GP-expressing viruses disseminate efficiently to the splenic white pulp, whereas their ARM GP-carrying counterparts are mostly found in the red pulp (20). Additional studies will be needed to investigate these possibilities.
Given the limited structural information on RNA-dependent RNA polymerases of arenaviruses (45, 46), the molecular impact of the L1079 mutation on the LCMV L protein structure, interactions, and supramolecular complex formation remains elusive. The 1,079 position does not map to any of the known catalytic core domains in the L protein (47). Still, several not mutually exclusive possibilities remain as to how L1079 affects the Cl13 immunobiology. Differential resistance of ARM and Cl13 replication to IFN has been reported (48) and may be related to the L1079 mutation. Also, polymerase processivity may be affected directly by influencing interactions of the L protein with cellular cofactors of viral replicase and transcriptase complexes (31, 49). Differential availability of specific cellular cofactors and cell type-specific cofactor isoforms could, therefore, explain the differential impact of L1079 on viral replication in different cell types (50). Of note, L-protein mutations other than K1079Q can also enhance persistence of ARM (19, 42), spreading the infection to other cell types throughout the body. In line with this observation, a comparison of the GP260 and L1079 positions in several commonly used LCMV strains (Table S1) suggests that other strains such as Docile establish persistence, despite sharing the K1079 residue of the L protein with ARM.
Systemic persistent infections like HIV and HCV infections represent important global health problems. It is, therefore, of paramount importance that we understand better how viruses establish persistence, how they subvert the antiviral T-cell defense (51, 52), and which parameters of infection facilitate generalized immunosuppression. Using reverse genetic techniques with LCMV, we show here that a single-point mutation enhancing viral replication in the initial target cell population in vivo can dictate all of the above three parameters. Importantly, viral growth assessments in standard immortalized cell lines fail to detect these differences (34), despite wide-ranging effects on the viral immunobiology. The mechanisms governing persistence or clearance of hepatitis C and B virus infection are still poorly defined (27–29). Our present findings in a widely used model of systemic persistent infection suggest that isolate-specific differences in replicative capacity can profoundly influence the viral immunobiology, including the resulting antiviral defense of the host. Hence, such differences should be considered anew as potential strain- and isolate-specific determinants of hepatitis virus chronicity as well as a key pathogenetic determinant in other persistent viral infections.
Methods
Mice and Animal Experiments.
C57BL/6 and STOP-GFP mice (39) were bred under SPF conditions at the Institut für Labortierkunde, University of Zurich, Switzerland. Animal experiments were performed at the Universities of Zurich and Geneva with permission by the respective Cantonal Veterinary Office and in accordance with the Swiss law for animal protection.
Viruses, Virus Titration, and Assessment of Virus-Neutralizing Antibodies.
LCMV Armstrong ARM5.3b (ARM), LCMV Cl13, and VSV have been described (30). The 16 recombinant LCM viruses were engineered by standard reverse genetic techniques as reported (30) and were grown on BHK-21 cells. LCMV infectivity was determined in a standard immunofocus assay (53). rLCMV/Cre_L1079-Cl13 and rLCMV/Cre_L1079-ARM vectors were generated as described (38), with the sole modification that 293T cells stably expressing LCMV Cl13 GP were used as producer cells. VSV-neutralizing IgG was determined in a plaque reduction assay (54).
Enumeration of Virus-Specific CD8+ T Cells.
Mouse peripheral blood mononuclear cells were incubated with either the H-2Db–restricted NP396-404 peptide of LCMV or the H-2Kb–restricted N52-59 peptide of VSV at a concentration of 10−6 M for 5–6 h, stained for the surface markers CD8 and B220 as well as the intracellular cytokines IFN-γ and TNF-α, and were enumerated on a FACS Calibur (BD). Values shown in the figures represent the percentage of cytokine-positive cells within CD8+B220– lymphocytes.
Determination of Intracellular S Segment RNA in rLCMV Vector-Infected pDCs.
For purification of GFP-expressing (rLCMV/Cre-infected) pDCs from STOP-GFP reporter mice, DCs were enriched by first incubating them with CD11c-biotin–labeled antibody followed by cell separation with antibiotin MACS beads (Miltenyi Biotech). Subsequently, the cells were sorted on a FACS Aria by gating on the population of live CD11c-intermediate mPDCA-1+ lymphocytes and frozen in RNAprotect Cell Reagent (Qiagen). Total cellular RNA was extracted using the RNA Plus Mini kit (Qiagen) and was subject to linear preamplification by the WT-Ovation FFPE System kit (NuGen). LCMV RNA was determined by TaqMan PCR as previously described (55) and normalized by the housekeeping gene EF1-α to obtain arbitrary units.
Statistical Analysis.
For comparison of multiple groups, we performed one-way ANOVA with Bonferroni's posttest. Two groups were compared by two-tailed Student t test. The analysis was carried out using Graphpad Prism software (vs. 4.0b). P values of P < 0.05 were considered statistically significant (*), and P < 0.01 was considered highly significant (**). P > 0.05 was considered not statistically significant (ns).
Supplementary Material
Acknowledgments
A.B. is a European Molecular Biology Organization long-term fellow and was supported by a PhD fellowship of the Boehringer Ingelheim Fonds, postdoctoral scholarships of the Roche Research Foundation, and the Schweizerische Stiftung für Medizinisch-Biologische Stipendien foundation of the Swiss National Science Foundation. A.N.H. was a fellow of GRK1121 of the German Research Foundation. S.J. is a recipient of an Australian National Health and Medical Research Council postdoctoral training fellowship. M.L. is a Lichtenberg fellow funded by the Volkswagen Foundation. D.D.P. was supported by Grant 3100A0-104067/1 of the Swiss National Science Foundation, the European Community seventh framework program (European Virus Archive Grant 228292), and the Prix Scientifique of the Fondation Leenaards, and he holds a stipendiary professorship from the Swiss National Science Foundation (PP00A–114913/1).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011998107/-/DCSupplemental.
References
- 1.Traub E. The epidemiology of lymphocytic choriomeningitis in white mice. J Exp Med. 1936;64:183–200. doi: 10.1084/jem.64.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zinkernagel RM. Lymphocytic choriomeningitis virus and immunology. Curr Top Microbiol Immunol. 2002;263:1–5. doi: 10.1007/978-3-642-56055-2_1. [DOI] [PubMed] [Google Scholar]
- 3.Pfau CJ, Valenti JK, Jacobson S, Pevear DC. Cytotoxic T cells are induced in mice infected with lymphocytic choriomeningitis virus strains of markedly different pathogenicities. Infect Immun. 1982;36:598–602. doi: 10.1128/iai.36.2.598-602.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 6.Blackburn SD, et al. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat Immunol. 2009;10:29–37. doi: 10.1038/ni.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tinoco R, Alcalde V, Yang Y, Sauer K, Zuniga EI. Cell-intrinsic transforming growth factor-beta signaling mediates virus-specific CD8+ T cell deletion and viral persistence in vivo. Immunity. 2009;31:145–157. doi: 10.1016/j.immuni.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brooks DG, et al. Interleukin-10 determines viral clearance or persistence in vivo. Nat Med. 2006;12:1301–1309. doi: 10.1038/nm1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ejrnaes M, et al. Resolution of a chronic viral infection after interleukin-10 receptor blockade. J Exp Med. 2006;203:2461–2472. doi: 10.1084/jem.20061462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moskophidis D, Lechner F, Pircher H, Zinkernagel RM. Virus persistence in acutely infected immunocompetent mice by exhaustion of antiviral cytotoxic effector T cells. Nature. 1993;362:758–761. doi: 10.1038/362758a0. [DOI] [PubMed] [Google Scholar]
- 11.Zajac AJ, et al. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, et al. Visualizing antigen-specific and infected cells in situ predicts outcomes in early viral infection. Science. 2009;323:1726–1729. doi: 10.1126/science.1168676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blattman JN, Wherry EJ, Ha SJ, van der Most RG, Ahmed R. Impact of epitope escape on PD-1 expression and CD8 T-cell exhaustion during chronic infection. J Virol. 2009;83:4386–4394. doi: 10.1128/JVI.02524-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mueller SN, Ahmed R. High antigen levels are the cause of T cell exhaustion during chronic viral infection. Proc Natl Acad Sci USA. 2009;106:8623–8628. doi: 10.1073/pnas.0809818106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchmeier MJ. Arenaviruses: Protein structure and function. Curr Top Microb Immunol. 2002;262:159–173. doi: 10.1007/978-3-642-56029-3_7. [DOI] [PubMed] [Google Scholar]
- 16.Perez M, Craven RC, de la Torre JC. The small RING finger protein Z drives arenavirus budding: Implications for antiviral strategies. Proc Natl Acad Sci USA. 2003;100:12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee KJ, Perez M, Pinschewer DD, de la Torre JC. Identification of the lymphocytic choriomeningitis virus (LCMV) proteins required to rescue LCMV RNA analogs into LCMV-like particles. J Virol. 2002;76:6393–6397. doi: 10.1128/JVI.76.12.6393-6397.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee KJ, Novella IS, Teng MN, Oldstone MB, de La Torre JC. NP and L proteins of lymphocytic choriomeningitis virus (LCMV) are sufficient for efficient transcription and replication of LCMV genomic RNA analogs. J Virol. 2000;74:3470–3477. doi: 10.1128/jvi.74.8.3470-3477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sevilla N, et al. Immunosuppression and resultant viral persistence by specific viral targeting of dendritic cells. J Exp Med. 2000;192:1249–1260. doi: 10.1084/jem.192.9.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smelt SC, et al. Differences in affinity of binding of lymphocytic choriomeningitis virus strains to the cellular receptor alpha-dystroglycan correlate with viral tropism and disease kinetics. J Virol. 2001;75:448–457. doi: 10.1128/JVI.75.1.448-457.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sevilla N, McGavern DB, Teng C, Kunz S, Oldstone MB. Viral targeting of hematopoietic progenitors and inhibition of DC maturation as a dual strategy for immune subversion. J Clin Invest. 2004;113:737–745. doi: 10.1172/JCI20243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patterson S, Rae A, Hockey N, Gilmour J, Gotch F. Plasmacytoid dendritic cells are highly susceptible to human immunodeficiency virus type 1 infection and release infectious virus. J Virol. 2001;75:6710–6713. doi: 10.1128/JVI.75.14.6710-6713.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Donaghy H, Gazzard B, Gotch F, Patterson S. Dysfunction and infection of freshly isolated blood myeloid and plasmacytoid dendritic cells in patients infected with HIV-1. Blood. 2003;101:4505–4511. doi: 10.1182/blood-2002-10-3189. [DOI] [PubMed] [Google Scholar]
- 24.Feldman S, et al. Decreased interferon-alpha production in HIV-infected patients correlates with numerical and functional deficiencies in circulating type 2 dendritic cell precursors. Clin Immunol. 2001;101:201–210. doi: 10.1006/clim.2001.5111. [DOI] [PubMed] [Google Scholar]
- 25.Kanto T, et al. Reduced numbers and impaired ability of myeloid and plasmacytoid dendritic cells to polarize T helper cells in chronic hepatitis C virus infection. J Infect Dis. 2004;190:1919–1926. doi: 10.1086/425425. [DOI] [PubMed] [Google Scholar]
- 26.Ulsenheimer A, et al. Plasmacytoid dendritic cells in acute and chronic hepatitis C virus infection. Hepatology. 2005;41:643–651. doi: 10.1002/hep.20592. [DOI] [PubMed] [Google Scholar]
- 27.Thimme R, et al. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rehermann B, Nascimbeni M. Immunology of hepatitis B virus and hepatitis C virus infection. Nat Rev Immunol. 2005;5:215–229. doi: 10.1038/nri1573. [DOI] [PubMed] [Google Scholar]
- 29.Lechner F, et al. Analysis of successful immune responses in persons infected with hepatitis C virus. J Exp Med. 2000;191:1499–1512. doi: 10.1084/jem.191.9.1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flatz L, Bergthaler A, de la Torre JC, Pinschewer DD. Recovery of an arenavirus entirely from RNA polymerase I/II-driven cDNA. Proc Natl Acad Sci USA. 2006;103:4663–4668. doi: 10.1073/pnas.0600652103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qanungo KR, Shaji D, Mathur M, Banerjee AK. Two RNA polymerase complexes from vesicular stomatitis virus-infected cells that carry out transcription and replication of genome RNA. Proc Natl Acad Sci USA. 2004;101:5952–5957. doi: 10.1073/pnas.0401449101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed R, et al. Genetic analysis of in vivo-selected viral variants causing chronic infection: Importance of mutation in the L RNA segment of lymphocytic choriomeningitis virus. J Virol. 1988;62:3301–3308. doi: 10.1128/jvi.62.9.3301-3308.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinschewer DD, Perez M, de la Torre JC. Dual role of the lymphocytic choriomeningitis virus intergenic region in transcription termination and virus propagation. J Virol. 2005;79:4519–4526. doi: 10.1128/JVI.79.7.4519-4526.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matloubian M, Kolhekar SR, Somasundaram T, Ahmed R. Molecular determinants of macrophage tropism and viral persistence: Importance of single amino acid changes in the polymerase and glycoprotein of lymphocytic choriomeningitis virus. J Virol. 1993;67:7340–7349. doi: 10.1128/jvi.67.12.7340-7349.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schildknecht A, Welti S, Geuking MB, Hangartner L, van den Broek M. Absence of CTL responses to early viral antigens facilitates viral persistence. J Immunol. 2008;180:3113–3121. doi: 10.4049/jimmunol.180.5.3113. [DOI] [PubMed] [Google Scholar]
- 36.Bründler MA, et al. Immunity to viruses in B cell-deficient mice: Influence of antibodies on virus persistence and on T cell memory. Eur J Immunol. 1996;26:2257–2262. doi: 10.1002/eji.1830260943. [DOI] [PubMed] [Google Scholar]
- 37.Kalinke U, et al. Monovalent single-chain Fv fragments and bivalent miniantibodies bound to vesicular stomatitis virus protect against lethal infection. Eur J Immunol. 1996;26:2801–2806. doi: 10.1002/eji.1830261202. [DOI] [PubMed] [Google Scholar]
- 38.Flatz L, et al. Development of replication-defective lymphocytic choriomeningitis virus vectors for the induction of potent CD8+ T cell immunity. Nat Med. 2010;16:339–345. doi: 10.1038/nm.2104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- 40.Krug A, et al. TLR9-dependent recognition of MCMV by IPC and DC generates coordinated cytokine responses that activate antiviral NK cell function. Immunity. 2004;21:107–119. doi: 10.1016/j.immuni.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 41.Wherry EJ, Blattman JN, Murali-Krishna K, van der Most R, Ahmed R. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J Virol. 2003;77:4911–4927. doi: 10.1128/JVI.77.8.4911-4927.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bergthaler A, Merkler D, Horvath E, Bestmann L, Pinschewer DD. Contributions of the lymphocytic choriomeningitis virus glycoprotein and polymerase to strain-specific differences in murine liver pathogenicity. J Gen Virol. 2007;88:592–603. doi: 10.1099/vir.0.82428-0. [DOI] [PubMed] [Google Scholar]
- 43.Probst HC, van den Broek M. Priming of CTLs by lymphocytic choriomeningitis virus depends on dendritic cells. J Immunol. 2005;174:3920–3924. doi: 10.4049/jimmunol.174.7.3920. [DOI] [PubMed] [Google Scholar]
- 44.Zuniga EI, McGavern DB, Pruneda-Paz JL, Teng C, Oldstone MB. Bone marrow plasmacytoid dendritic cells can differentiate into myeloid dendritic cells upon virus infection. Nat Immunol. 2004;5:1227–1234. doi: 10.1038/ni1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vieth S, Torda AE, Asper M, Schmitz H, Günther S. Sequence analysis of L RNA of Lassa virus. Virology. 2004;318:153–168. doi: 10.1016/j.virol.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 46.Morin B, et al. The N-terminal domain of the arenavirus L protein is an RNA endonuclease essential in mRNA transcription. PLoS Pathog. 2010;6:e1001038. doi: 10.1371/journal.ppat.1001038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sánchez AB, de la Torre JC. Genetic and biochemical evidence for an oligomeric structure of the functional L polymerase of the prototypic arenavirus lymphocytic choriomeningitis virus. J Virol. 2005;79:7262–7268. doi: 10.1128/JVI.79.11.7262-7268.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moskophidis D, et al. Resistance of lymphocytic choriomeningitis virus to alpha/beta interferon and to gamma interferon. J Virol. 1994;68:1951–1955. doi: 10.1128/jvi.68.3.1951-1955.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pinschewer DD, Perez M, de la Torre JC. Role of the virus nucleoprotein in the regulation of lymphocytic choriomeningitis virus transcription and RNA replication. J Virol. 2003;77:3882–3887. doi: 10.1128/JVI.77.6.3882-3887.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gromeier M, Bossert B, Arita M, Nomoto A, Wimmer E. Dual stem loops within the poliovirus internal ribosomal entry site control neurovirulence. J Virol. 1999;73:958–964. doi: 10.1128/jvi.73.2.958-964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wedemeyer H, et al. Impaired effector function of hepatitis C virus-specific CD8+ T cells in chronic hepatitis C virus infection. J Immunol. 2002;169:3447–3458. doi: 10.4049/jimmunol.169.6.3447. [DOI] [PubMed] [Google Scholar]
- 52.Kostense S, et al. Persistent numbers of tetramer+ CD8(+) T cells, but loss of interferon-gamma+ HIV-specific T cells during progression to AIDS. Blood. 2002;99:2505–2511. doi: 10.1182/blood.v99.7.2505. [DOI] [PubMed] [Google Scholar]
- 53.Battegay M, et al. Quantification of lymphocytic choriomeningitis virus with an immunological focus assay in 24- or 96-well plates. J Virol Methods. 1991;33:191–198. doi: 10.1016/0166-0934(91)90018-u. [DOI] [PubMed] [Google Scholar]
- 54.Pinschewer DD, et al. Kinetics of protective antibodies are determined by the viral surface antigen. J Clin Invest. 2004;114:988–993. doi: 10.1172/JCI22374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pinschewer DD, et al. Innate and adaptive immune control of genetically engineered live-attenuated arenavirus vaccine prototypes. Int Immunol. 2010;22:749–756. doi: 10.1093/intimm/dxq061. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.