Abstract
Fatty acids (FAs) are essential for cell survival, yet their overaccumulation causes lipotoxicity. To prevent lipotoxicity, cells store excess FAs as triglycerides (TGs). In cultured cells TG synthesis is activated by excess unsaturated but not saturated FAs. Here, we identify Ubxd8 as a sensor for unsaturated FAs and regulator of TG synthesis. In cultured cells depleted of FAs, Ubxd8 inhibits TG synthesis by blocking conversion of diacylglycerols (DAGs) to TGs. Excess unsaturated but not saturated FAs relieve this inhibition. As a result, unsaturated FAs are incorporated into TGs, whereas saturated FAs are incorporated into DAGs. In vitro, unsaturated but not saturated FAs alter the structure of purified recombinant Ubxd8 as monitored by changes in its thermal stability, trypsin cleavage pattern, and oligomerization. These results suggest that Ubxd8 acts as a brake that limits TG synthesis, and this brake is released when its structure is altered by exposure to unsaturated FAs.
Keywords: diacylglyceride, Insig-1, lipid droplet
In mammalian cells fatty acids (FAs) are required for the synthesis of the phospholipid components of membranes and generation of energy. However, overaccumulation of FAs is toxic. When FAs accumulate in cells, they exert at least two regulatory actions to prevent their further accumulation: (i) They inhibit their own synthesis (1–3); and (ii) they enhance the incorporation of excess FAs into triglycerides (TGs) that are stored in lipid droplets (4). In general, these regulatory functions are carried out by unsaturated but not saturated FAs (3, 5, 6). The mechanism by which cells specifically sense the level of unsaturated FAs and orchestrate their responses is not understood.
Feedback inhibition of FA synthesis is achieved at least in part by unsaturated FA-mediated inhibition of the proteolytic activation of sterol regulatory element binding protein 1 (SREBP-1) (3), a transcription factor that activates all genes necessary to synthesize FAs (7). SREBP-1 is a membrane-bound transcription factor that must be transported from the endoplasmic reticulum (ER) to the Golgi complex where it is proteolytically cleaved so that the NH2-terminal domain of the protein is able to enter the nucleus to activate its target genes (8). Transport of SREBP-1 is regulated by two ER membrane proteins: (i) Scap, an escort protein that binds SREBP-1 and carries it to the Golgi (9); and (ii) Insigs, proteins that bind to Scap and retain the Scap·SREBP-1 complex in the ER (10, 11). By blocking Scap/SREBP transport, Insigs are negative regulators of fatty acid synthesis.
Unsaturated FAs regulate FA synthesis by blocking the degradation of Insig-1, the major Insig isoform in cultured cells (12). In FA-depleted cells, Insig-1 is rapidly degraded through a process known as ER-associated degradation (ERAD) (13, 14). This degradation requires Ubxd8, a membrane-bound protein that recruits p97 to Insig-1 through its bridging interaction with both proteins (5). Recruitment of p97 is necessary for Insig-1 to be recognized and degraded by proteasomes (15). Unsaturated but not saturated FAs block the interaction between Insig-1 and Ubxd8 (5). Consequently, p97 dissociates from Insig-1, and Insig-1 is stabilized (5). The excess Insig-1 binds to Scap, blocking the proteolytic activation of SREBP-1 (5).
Our previous studies in cultured cells defined Ubxd8 as a key mediator by which unsaturated FAs inhibit their own synthesis. However, the biochemical mechanism by which unsaturated FAs influence the behavior of Ubxd8 remains a mystery. In the current study we develop in vitro assays to demonstrate that unsaturated FAs alter the structure of Ubxd8 in a way that correlates with its loss of activity. We also provide evidence that Ubxd8 is a key player in the other regulatory action of unsaturated FAs, namely the stimulation of TG synthesis and lipid droplet formation.
Results
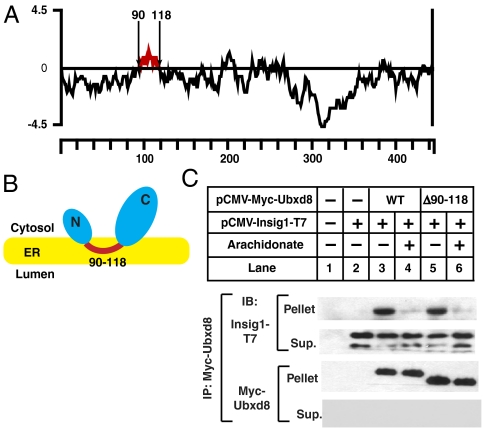
Ubxd8 is known to be an intrinsic membrane protein (16). To study the interactions of FAs with Ubxd8 we first sought to prepare a soluble form of the protein free of membrane lipids. A hydropathy plot of human Ubxd8 reveals that the protein contains a hydrophobic stretch of amino acid residues (amino acids 90–118) that may anchor the protein to membranes (Fig. 1A). To determine the membrane topology of Ubxd8, we examined the membrane orientation of the NH2- and COOH-terminal ends of the protein by transfecting cells with a plasmid encoding Ubxd8 tagged with the Myc epitope either at the NH2 (pCMV-Myc-Ubxd8) or COOH terminus (pCMV-Ubxd8-Myc) and analyzing the membrane orientation of the epitope tag by trypsin protection assay. Trypsin destroyed the Myc epitope tags at both ends of Ubxd8 in the absence or presence of Triton X-100 (Fig. S1A, Upper). Grp94, a protein localized in the ER lumen (17), was resistant to trypsin digestion in the absence of Triton X-100 but was destroyed in the presence of the detergent (Fig. S1A, Lower). These results confirmed that the membrane vesicles were sealed and impermeable to trypsin in the absence of detergent. Thus, it appears that both the NH2 and COOH ends of Ubxd8 are oriented toward the cytosol.
Fig. 1.
Membrane attachment of Ubxd8 is not required for FA-regulated interaction between Ubxd8 and Insig-1. (A) Hydropathy plot of Ubxd8. The residue-specific hydropathy index was calculated over a window of 18 residues by the method of Kyte and Doolitttle. The hydrophobic region of Ubxd8 between residue 90 and 118 was highlighted in red. (B) Proposed membrane topology of Ubxd8. (C) SRD-13A cells were seeded on day 0 and transfected with 0.2 μg of indicated plasmids on day 1. Following incubation for 8 h, cells were switched to medium A supplemented with 5% delipidated FCS. On day 2, cells were treated with 100 μm arachidonate in medium supplemented with 5% delipidated FCS and 10 μm MG132 for 6 h as indicated. Detergent lysates of the cells were subjected to immunoprecipitation with anti-Myc to precipitate transfected Ubxd8. Pellets (representing a 0.1 dish of cells) and supernatants (representing a 0.01 dish of cells) of the immunoprecipitation were subjected to SDS/PAGE followed by immunoblot analysis of Ubxd8 and Insig-1 with IgG-9E10 and anti-T7, respectively. Insig-1 shows two bands that represent translational products initiated with different methionine. Only the top band shown in the IP pellets as the bottom band comigrated with light chain of the antibody used for immunoprecipitation.
The results presented above suggest that Ubxd8 is inserted in the ER through a membrane localization domain (amino acid residues 90–118) that may form a hairpin loop in the membranes (Fig. 1B). If the hairpin configuration of Ubxd8 is correct, deletion of the hydrophobic sequence should alter the localization of Ubxd8 from membrane to cytosol. To test this hypothesis, we transfected cells with a plasmid encoding either wild-type or mutant Ubxd8 in which the putative membrane localization domain was deleted [Ubxd8(Δ90–118)]. As shown in Fig. S1B, All detectable wild-type Ubxd8 was found in membranes (lanes 2–4). In contrast, a significant amount of Ubxd8(Δ90–118) was found in cytosol (lanes 7–9). While wild-type Ubxd8 was integral to membranes as it was resistant to alkaline extraction (lanes 5 and 6), approximately 50% of Ubxd8(Δ90–118) were extracted by the alkaline so they were peripherally associated with membranes (lanes 10 and 11). This result suggests that amino acid residues 90–118 of Ubxd8 play an important role in anchoring the protein to membranes.
To determine whether deletion of the membrane localization domain of Ubxd8 affects its interaction with Insig-1, we performed a coimmunoprecipitation experiment in SRD-13A cells, which are mutant CHO cells auxotrophic for long-chain unsaturated FAs so that they can be easily depleted of these FAs by incubating them in medium free of FAs (18). Insig-1 was coprecipitated efficiently with both wild-type Ubxd8 and Ubxd8(Δ90–118) in cells incubated in the absence of FAs (Fig. 1C, lanes 3 and 5), and this interaction was blocked by the addition to the culture medium of arachidonate, a polyunsaturated FA (lanes 4 and 6). Similar result was observed in an experiment performed in HEK-293 cells (Fig. S1C). In cells incubated in the absence of FAs, alkaline treatment of the membranes did not affect the interaction between wild–type Ubxd8 and Insig-1 (Fig. S1D, lanes 3 and 4), but the treatment reduced the amount of Ubxd8(Δ90–118) associated with membranes and abolished its interaction with Insig-1 (Fig. S1D, lanes 5 and 6). These results suggest that deletion of the membrane localization domain in Ubxd8 did not impair unsaturated FA-regulated interaction between Ubxd8 and Insig-1, as Ubxd8(Δ90–118) peripherally associated with membranes is still able to interact with Insig-1.
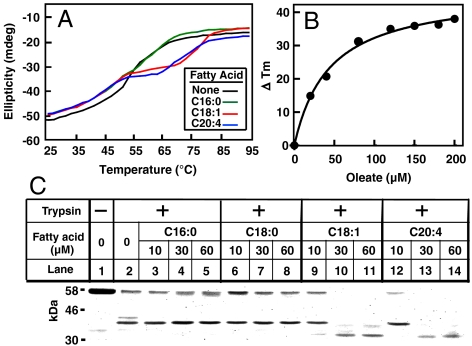
Soluble Ubxd8(Δ90–118) provided a facile tool to study the interaction of Ubxd8 with unsaturated FAs in vitro. We used the baculovirus expression system in Sf-9 insect cells to express human Ubxd8(Δ90–118) with a His6 tag at the NH2 terminus. The recombinant protein was purified to homogeneity using Ni-chromatography. To measure the interaction between FAs and Ubxd8, we first determined whether unsaturated FAs altered the thermal stability of Ubxd8. We analyzed thermal stability by incubating the protein at temperatures ranging from 25 to 95 °C and measuring circular dichroism (CD) at 222 nm. CD signals recorded at 222 nm are inversely correlated with the secondary structure content of a protein. In the absence of FAs, Ubxd8(Δ90–118) became completely denatured at approximately 70 °C (Fig. 2A, black line). Incubation with the unsaturated FAs oleate (C18:1) or arachidonate (C20:4) stabilized Ubxd8, shifting the denaturation curve to the right by approximately 10 °C (Fig. 2A, red and blue lines). In contrast to unsaturated FAs, the saturated FA palmitate (C16:0) failed to stabilize the protein (Fig. 2A, green line). In contrast to Ubxd8, the thermal stability of glutathione S-transferase was not affected by oleate (Fig. S2A). The concentration of oleate used in our studies was not high enough to form micelles because it was bellow the critical micelle concentration (> 0.5 mM) determined by a fluorometric approach (19) (Fig. S2B).
Fig. 2.
Unsaturated but not saturated FAs alters the structure of Ubxd8. (A) Thermal denaturation of Ubxd8(Δ90–118) was measured by CD at 222 nm in the absence or presence of 60 μM of indicated FAs added as stock solutions dissolved in ethanol. (B) Thermal denaturation of Ubxd8(Δ90–118) in the presence of various amounts of oleate added as stock solutions dissolved in ethanol was measured by CD at 222 nm. Tm, which was determined by the temperature at which the ellipticity reached the midpoint of the values measured at 25 and 95 °C, was recorded for each concentration of oleate. The difference in the Tm caused by the treatment with oleate (ΔTm) was plotted against oleate concentration and the points were fitted by a standard Michaelis–Menten binding curve. (C) Purified Ubxd8(Δ90–118) (7 μg) was incubated with the indicated FAs added as stock solutions dissolved in DMSO in 0.2 ml buffer A. The protein was then treated with 0.1 μg/ml of trypsin for 20 min at 25 °C, subjected to SDS/PAGE, and visualized by Coomassie blue staining.
The result shown in Fig. 2A suggests that alterations in the thermal stability of Ubxd8(Δ90–118), which can be quantified by changes in the melting temperature (Tm) of the protein, may be used to measure the interaction of unsaturated FAs with the protein. We thus examined the thermal stability of Ubxd8(Δ90–118) in the presence of various amounts of oleate. Oleate increased the Tm of Ubxd8(Δ90–118) in a concentration-dependent manner, and the effect was maximal when the concentration of oleate reached 120 μM (Fig. S2C). We then plotted the differences in the Tm as a function of oleate concentrations. Each point on the plot was nicely fitted to a single ligand-receptor complex binding curve similar to Michaelis–Menten kinetics with an apparent dissociation constant (Kd) about 40 μM (Fig. 2B).
We also characterized the interaction of unsaturated FAs with Ubxd8(Δ90–118) by subjecting the protein to partial trypsin digestion in the absence and presence of various FAs. In the absence of FAs, trypsin reduced the amount of the full length protein (58 kDa) with concomitant generation of a protected band at approximately 35 kDa (Fig. 2C, lane 2). Incubation with the saturated FAs palmitate (C16:0) or stearate (C18:0) did not change this cleavage pattern (Fig. 2C, lanes 3-8). In contrast, the cleavage pattern was altered by incubation with the unsaturated FA oleate (C18:1) or arachidonate (C20:4): The amount of full length Ubxd8(Δ90–118) and 35-kDa fragment was reduced while a new protected band at approximately 30 kDa appeared (Fig. 2C, lanes 9–14).
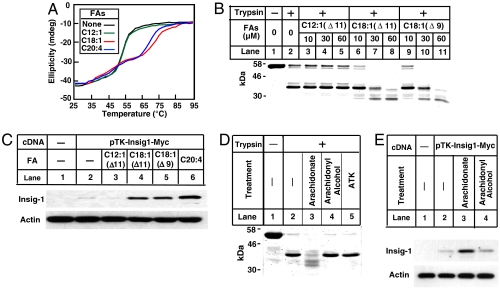
We then examined the effect of acyl chain length on the interaction of FAs with Ubxd8(Δ90–118). Unlike the long-chain unsaturated FAs oleate (C18:1) and arachidonate (C20:4), the medium chain unsaturated FA C12:1(Δ11) did not affect the thermal stability of Ubxd8(Δ90–118) (Fig. 3A). Likewise, C12:1(Δ11) failed to alter the trypsin cleavage pattern of Ubxd8(Δ90–118) (Fig. 3B, lanes 3–5). The failure of C12:1(Δ11) to influence Ubxd8(Δ90-118) is not caused by the different location of the double bond, because C18:1(Δ11) was able to change the trypsin cleavage pattern of Ubxd8(Δ90–118) as efficiently as oleate [C18:1(Δ9)] (Fig. 3B, lanes 6–11).
Fig. 3.
Specificity for the interaction between long-chain unsaturated FAs and Ubxd8(Δ90–118). (A) Thermal denaturation of Ubxd8(Δ90–118) in the presence or absence of indicated FAs was measured as described in Fig. 2A. (B and D) Trypsin cleavage of Ubxd8(Δ90–118) in the presence of indicated FAs or their acyl chain derivatives (60 μM in D) was performed as described in Fig. 2C. (C and E) SRD-13A cells were seeded on day 0 and transfected with 0.4 μg of pTK-Insig1-myc on day 1 as described in Fig. 1C. Following incubation for 8 h, cells were switched to medium A supplemented with 5% delipidated FCS. On day 2, cells were treated with 60 μm of indicated FAs or their acyl chain derivative in medium supplemented with 5% delipidated FCS for 6 h. Detergent lysates of these cells were subjected to SDS/PAGE followed by immunoblot analysis with anti-Myc IgG-9E10 (against Insig-1) and polyclonal antiactin.
To test the relevance of these in vitro observations to the in vivo function of Ubxd8, we examined the ability of various fatty acids to block the function of Ubxd8 in degrading Insig-1. In agreement with our in vitro observations, the long-chain unsaturated FAs arachidonate, oleate, and C18:1(Δ11) increased the amount of Insig-1, whereas C12:1(Δ11) had no such effect (Fig. 3C).
To further define the specificity of the interaction between FAs and Ubxd8, we tested the ability of arachidonyl alcohol and arachidonyl trifluoromethyl ketone (ATK) to influence trypsin cleavage of Ubxd8(Δ90–118). Neither of these compounds changed the trypsin cleavage pattern of Ubxd8(Δ90–118) (Fig. 3D). In agreement with this in vitro result, arachidonate but not arachidonyl alcohol stabilized Insig-1 in cultured cells (Fig. 3E). We were unable to determine the in vivo effect of ATK because the compound was toxic to cells. These results suggest that in addition to acyl chains, the carboxyl groups of unsaturated FAs also determine their specificity to bind Ubxd8.
To further define the influence of FAs on the structure of Ubxd8, we performed blue native polyacrylamide gel electrophoresis (PAGE), a technique that allows detection of protein complexes in their native state (10). In the absence of FAs, Ubxd8(Δ90–118) migrated as a band at approximately 100 kDa (Fig. 4A, lane 1). Incubation with C12:1(Δ11) or stearate (C18:0) did not change the migration of the protein (Fig. 4A, lanes 2–3). In the presence of oleate (C18:1) or arachidonate (C20:4), Ubxd8(Δ90-118) migrated as a broad band at approximately 700–1,000 kDa (Fig. 4A, lanes 4 and 5). The calculated molecular mass for Ubxd8(Δ90–118) is 58 kDa. Thus, the result shown in Fig. 4A suggests that the protein forms a dimer in the absence of FAs and polymerizes into oligomers that contain at least 12 Ubxd8(Δ90–118) molecules when treated with unsaturated FAs. This result was confirmed by gel filtration analysis. In the absence of oleate, Ubxd8(Δ90–118) emerged from the column as a 100-kDa species (Fig. 4B, blue line). In the presence of oleate, the protein eluted as broad species of approximately 700 kDa (Fig. 4B, red line). Oleate did not affect the filtration of molecular weight marker proteins, indicating that the effect of oleate on Ubxd8(Δ90–118) was specific to this protein. Oleate stimulated oligomerization of Ubxd8(Δ90–118) regardless of whether it is added to the reaction in stock solution dissolved in dimethyl sulfoxide (DMSO), ethanol, or water (for oleate sodium salt) (Fig. S2D).
Fig. 4.
Long-chain unsaturated FAs induce oligomerization of Ubxd8. (A) Ubxd8(Δ90–118) (0.7 μg) was incubated with 100 μM of indicated FAs added as stock solutions dissolved in ethanol, subjected to blue native PAGE, and visualized with Coomassie blue staining. Molecular weights for protein standards are indicated. (B) Ubxd8(Δ90–118) (0.4 mg) incubated with or without 100 μM Na oleate was applied to FPLC using a Superose 6 size exclusion column. Absorbance at 280 nm was monitored continuously to identify position of elution of Ubxd8(Δ90–118). Standard molecular weight markers were chromatographed on the same column under identical buffer conditions and eluted at the positions shown by arrows. (C) Oligomerization of Ubxd8(Δ90–118) in the presence of indicated FAs and their CoA derivatives was determined by blue native PAGE as described in A.
FAs are activated by Co-enzyme A (CoA) before they are incorporated into various lipids. We thus examine the effect of acyl-CoA on oligomerization of Ubxd8(Δ90–118). As shown in Fig. 4C, only oleate but not oleoyl-CoA, stearate, or stearoyl-CoA stimulated oligomerization of Ubxd8(Δ90–118). This result agrees with our earlier observation that the carboxyl groups in unsaturated FAs are important for their interaction with Ubxd8.
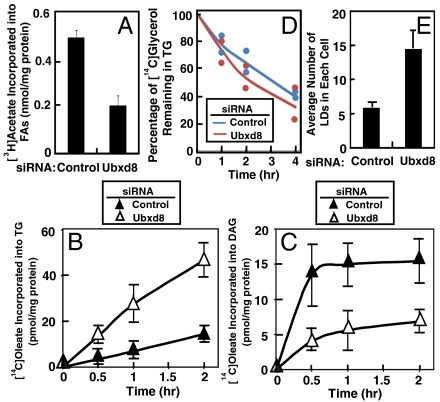
Our previous study indicated that Ubxd8 promotes proteolytic activation of SREBP-1 by facilitating degradation of Insig-1 in FA-depleted cells (5). Thus, Ubxd8 is expected to activate FA synthesis in these cells. To test this hypothesis, we transfected SV589 cells with siRNA targeting Ubxd8 or a control siRNA targeting GFP. SV589 cells are an immortalized line of human fibroblasts (20) efficiently transfected by siRNAs. Following incubation in medium supplemented with delipidated FCS to deplete FAs from the cells, the cells were labeled with [3H]acetate, and its incorporation into FAs was determined by thin layer chromatography (TLC). As shown in Fig. 5A, knockdown of Ubxd8 inhibited synthesis of FAs by approximately threefold in FA-depleted cells.
Fig. 5.
Ubxd8 promotes FA synthesis and inhibits TG synthesis in FA-depleted cells. (A) SV589 cells were set up at 1 × 105 cells/60 mm dish on day 0 and transfected with 400 pmol of siRNA targeting Ubxd8 or GFP as a control on day 1. After incubation for 48 h on day 3, they were switched to medium supplemented with 10% delipidated FCS. Following incubation for 16 h on day 4, cells were labeled with 2 μCi/ml [3H]acetate for 4 h. The amount of [3H]acetate incorporated into FAs was determined as described in Material and Methods. Data are reported as mean ± S.E. from three independent experiments. (B and C) SV-589 cells were seeded, transfected with indicated siRNA, and incubated as described in A for the first 3 days. On day 4, cells were labeled with 1 μM [14C]oleate for the indicated amount of time. Lipid extracts of the cells were subjected to TLC analysis. The activity of lipid synthesis was determined by the radioactivity found in TG (B) or DAG (C) normalized by the amount of cellular protein. Data are reported as mean ± S.E. from three independent experiments. (D) SV-589 cells were seeded, transfected with indicated siRNA, and incubated as described in Fig. 5A for the first 3 days. On day 4, cells were pulse labeled with 10 μM [14C]glycerol for 4 h. They were then chased in medium supplemented with delipidated FCS and 100 μM unlabeled glycerol for the indicated amount of time. The amount of radiolabeled glycerol found in TG was determined. Value shown are relative to the amount of [14C]glycerol found in TG at time 0, which is set at 100%. Results from two independent experiments were presented. (E) SV589 cells were seeded, transfected with indicated siRNA, and incubated as described in A for the first 3 days. On day 4, cells were treated with 10 μM oleate for 6 h, fixed and stained with oil red O, and subjected to immunofluorescent microscopy analysis. Average number of lipid droplets (LDs) in each cell was calculated by counting lipid droplets in images that contain at least 10 cells. Data are reported as mean ± S.E. from four different sets of images.
The results shown above suggest that Ubxd8 senses long-chain unsaturated FAs to control FA synthesis by regulating Insig-1 degradation. We then tested whether Ubxd8 mediates the other regulatory action of unsaturated FAs—namely, activation of TG synthesis. The synthesis of TGs begins with attachment of FAs to glycerol-3-phosphate, leading to the generation of diacylglycerols (DAGs), which can be converted to TGs or phospholipids (21). Diacylglycerol acyltransferases (DGATs) catalyze the final step in TG synthesis by adding the third FA molecule to DAGs (21). To determine whether Ubxd8 regulates TG synthesis, we first examined the effect of Ubxd8 on TG synthesis in cells depleted of FAs. We transfected SV589 cells with siRNA targeting Ubxd8 or GFP as a control and cultured the cells in medium depleted of FAs. The cells were then radiolabeled with a low concentration of [14C]oleate, and its incorporation into TGs and DAGs was determined by TLC. The amount of [14C]oleate incorporated into TGs in cells treated with the siRNA targeting Ubxd8 was increased by approximately threefold (Fig. 5B), while that found in DAGs was decreased by approximately threefold (Fig. 5C). This result suggests that either DGATs are activated or the TG lipases that hydrolyze TGs to DAGs are inhibited when Ubxd8 is depleted. To exclude the possibility that depletion of Ubxd8 inhibits TG lipase activity, we performed a pulse-chase experiment in which SV589 cells transfected with the siRNA targeting Ubxd8 or a control siRNA were cultured in delipidated FCS. The cells were pulsed labeled with [14C]glycerol and chased in medium containing excessive nonradiolabeled glycerol. The rate of disappearance of [14C]glycerol from TGs in cells transfected with the siRNA targeting Ubxd8 was not slower than that in control cells (Fig. 5D). Thus, Ubxd8 inhibits the incorporation of oleate into TGs by reducing TG synthesis rather than enhancing TG hydrolysis in cells depleted of FAs. In agreement with this result, SV589 cells transfected with the siRNA targeting Ubxd8 contained more lipid droplets than those transfected with a control siRNA when they were cultured in medium supplemented with delipidated FCS and a small amount of oleate (Fig. 5E). Representative images of lipid droplets in these cells were shown in Fig. S3.
The results shown in Fig. 5 suggest that in cells depleted of FAs, Ubxd8 inhibits TG synthesis at the last step by blocking incorporation of FAs into DAGs to produce TGs. This observation was not unique to SV589 cells; similar results were obtained in HEK-293 cells (see below). Inasmuch as long-chain unsaturated FAs stimulate polymerization of Ubxd8, we hypothesized that these FAs may activate TG synthesis by inhibiting Ubxd8. To determine whether unsaturated FAs activate TG synthesis, we incubated HEK-293 cells with a tracer amount of [14C]oleate (10 μM) in the absence or presence of a 10-fold excess of unlabeled oleate. Even though the specific radioactivity of [14C]oleate was reduced by dilution with unlabeled oleate, the amount of radioactivity found in TGs rose markedly in cells treated with nonradiolabeled oleate (Fig. 6A). Thus, oleate is not only a substrate but also an activator of TG synthesis.
Fig. 6.
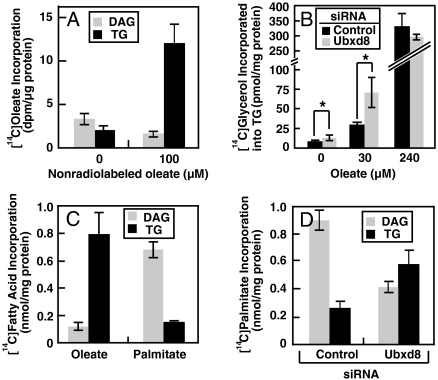
Long-chain unsaturated but not saturated FAs relieve the inhibition of TG synthesis imposed by Ubxd8. (A) HEK-293 cells were set up at 4 × 105 cells/60 mm dish on day 0. On day 2 they were switched to medium supplemented with 10% delipidated FCS. After incubation for 16 h on day 3, the cells were labeled with 10 μM [14C]oleate in the absence or presence of 100 μM nonradiolabeled oleate for 4 h. The amount of [14C]oleate incorporated into TGs and DAGs was determined. (B) HEK-293 cells were set up at 1.5 × 105 cells/60 mm dish on day 0. They were transfected with the indicated siRNA and incubated as described in Fig. 5A for the first three days. On day 4, these cells were labeled with 10 μM [14C]glycerol in the presence of indicated concentrations of nonradiolabeled oleate for 4 h. The amount of [14C]glycerol incorporated into TGs was determined. *, p < 0.05. (C) HEK-293 cells were set up and incubated as described in A for the first 2 days. On day 3 the cells were labeled with 10 μM [14C]oleate in the presence of 100 μM nonradiolabeled oleate (oleate) or 10 μM [14C]palmitate in the presence of 100 μM nonradiolabeled palmitate (palmitate) for 4 h. The amount of radio-labeled FAs incorporated into TGs and DAGs was determined. (D) HEK-293 cells were set up, transfected with indicated siRNA, and incubated as described in B for the first 3 days. On day 4, these cells were labeled with 10 μM [14C]palmitate in the presence of 100 μM nonradiolabeled palmitate for 4 h. The amount of [14C]palmitate incorporated into TGs and DAGs was determined. (A–D) Data are reported as mean ± S.E. from three independent experiments.
If unsaturated FAs activate TG synthesis by inhibiting Ubxd8, then knockdown of Ubxd8 is not expected to further enhance TG synthesis in cells treated with excess unsaturated FAs. To test this hypothesis, we transfected HEK-293 cells with siRNA targeting Ubxd8 or a control siRNA and incubated the cells with various amounts of oleate. The cells were then labeled with [14C]glycerol, and its incorporation into TG was measured. Knockdown of Ubxd8 activated TG synthesis only in cells incubated in the absence of excess oleate (Fig. S4A), such as those treated with no oleate or 30 μM of oleate (Fig. 6B). In cells treated with 240 μM oleate, TG synthesis was markedly elevated and knockdown of Ubxd8 no longer had any stimulatory effect (Fig. 6B).
Unsaturated FAs activate TG synthesis and stabilize Insig-1 through Ubxd8. To determine whether stabilization of Insig-1 is responsible for activation of TG synthesis in cells exposed to excess unsaturated FAs, we analyzed TG synthesis in SRD-14 cells, a line of mutant CHO-7 cells deficient in Insig-1 (12). Oleate stimulated TG synthesis in SRD-14 cells as efficiently as that in parental CHO-7 cells (Fig. S4B). Thus, Ubxd8 appears to regulate TG synthesis through proteins other than Insig-1.
Unlike long-chain unsaturated FAs, saturated FAs do not bind Ubxd8. If Ubxd8 is the regulator of TG synthesis, saturated FAs should not stimulate TG synthesis in the same manner as unsaturated FAs. To test this hypothesis, we incubated HEK-293 cells in medium supplemented with delipidated FCS. The cells were then treated with oleate or palmitate, and their incorporation into DAGs and TGs was determined. We observed a striking difference in the labeling pattern. Whereas oleate was incorporated efficiently into TGs, palmitate accumulated in DAGs (Fig. 6C). To determine whether the failure of palmitate to be incorporated into TGs is caused by Ubxd8-mediated inhibition in TG synthesis, we knocked down the expression of Ubxd8 by RNAi and performed an experiment similar to that shown in Fig. 6C. As shown in Fig. 6D, knockdown of Ubxd8 reduced the amount of [14C]palmitate incorporated into DAGs and increased that incorporated into TGs. We observed a similar result when [14C]glycerol was used as the radiolabeled tracer to perform the experiment shown in Fig. 6D (Fig. S4C). Treatment with oleate, which inactivates Ubxd8, also increased the amount of palmitate incorporated into TGs and decreased that incorporated into DAGs (Fig. S4D).
Discussion
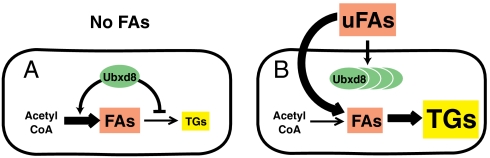
The current study reveals that Ubxd8 functions as a sensor for long-chain unsaturated FAs in mammalian cells. A working model that summarizes the regulatory role of Ubxd8 is shown in Fig. 7. We earlier showed that Ubxd8 facilitates the degradation of Insig-1 in cells deprived of FAs, thereby activating FA synthesis by promoting proteolytic processing of SREBP-1 (5). In the current study we show that Ubxd8 blocks TG synthesis by limiting the conversion of DAGs to TGs in these cells as well. Thus, in cells deprived of FAs, the concerted regulatory actions of Ubxd8 make FAs available for incorporation into phospholipids by limiting their diversion into TGs. When long-chain unsaturated FAs are supplied externally, these FAs change the structure of Ubxd8, promoting its polymerization and inhibiting its activity. Consequently, Insig-1 is stabilized and FA synthesis decreases. TG synthesis increases so that excess exogenous FAs are stored as TGs in lipid droplets.
Fig. 7.
Model for Ubxd8-mediated cellular responses to exogenous unsaturated FAs. (A) Cells incubated in the absence of exogenous unsaturated FAs. In these cells Ubxd8 promotes synthesis of FAs and inhibits incorporation of FAs into TGs. These two regulatory actions allow cells to maintain enough FAs for their survival. (B) Cells incubated in the presence of exogenous unsaturated fatty acids (uFAs). In these cells excessive unsaturated FAs inactivate Ubxd8 by promoting its polymerization. In the absence of active Ubxd8, synthesis of endogenous FAs is inhibited and excessive FAs are channeled into TGs. These two regulatory actions prevent toxic overaccumulation of FAs.
The strongest piece of evidence that Ubxd8 is a sensor for long-chain unsaturated FAs comes from the observations that these FAs alter the structure of purified Ubxd8(Δ90–118). Using various techniques, we show that unsaturated FAs alter the thermal stability, trypsin cleavage pattern, and oligomerization state of the protein. This effect is specific to long-chain unsaturated FAs because saturated FAs, medium chain unsaturated FAs, or long-chain unsaturated alcohols do not alter the structure of the protein. A major limitation in the current study is that we were unable to perform these in vitro assays on full length Ubxd8. This is because detergents, which are required to solubilize the full length protein, interfered with our assays. However, our in vitro characterization performed with Ubxd8(Δ90–118) might still be a good indication of the full length protein for the following reasons: (i) Deletion of the membrane localization domain in Ubxd8 did not affect FA-regulated interaction between Insig-1 and Ubxd8 in cultured cells, suggesting that the membrane localization domain is not required for Ubxd8 to sense unsaturated FAs; (ii) the specificity of FAs and their analogues to alter the structure of Ubxd8(Δ90–118) in vitro matches their specificity to stabilize Insig-1 inside cells; and (iii) the ability of oleate and palmitate to alter the structure of Ubxd8(Δ90–118) in vitro matches their ability to stimulate TG synthesis inside cells.
The current study indicates that Ubxd8-facilitated degradation of Insig-1 is required to activate FA synthesis in cells depleted of FAs. However, Insig-1 is not required for unsaturated FA-stimulated TG synthesis, an event also regulated by Ubxd8. Ubxd8 inhibits TG synthesis by blocking the conversion of DAGs to TGs, suggesting that Ubxd8 may regulate a DGAT activity. Further study will be required to determine which proteins are targeted by Ubxd8 to regulate TG synthesis.
The current findings may have relevance to insulin resistance, the primary risk factor for the development of type 2 diabetes. In obese individuals, elevated levels of plasma FAs lead to enhanced FA uptake into nonadipose tissues, resulting in insulin resistance in muscle and liver (22). Recent evidence suggests that accumulation of DAGs is responsible for the development of insulin resistance (23). The effect of DAGs on insulin resistance was most extensively studied in skeletal muscle cells (6, 23). Overaccumulation of DAGs in these cells is known to be caused by exposure of cells to excessive saturated FAs (6). Our current finding that Ubxd8 is inactivated by unsaturated but not saturated FAs may help to explain this observation. Because of their inability to inactivate Ubxd8, saturated FAs are preferentially incorporated into DAGs. Thus, a pharmacologic agent that mimics unsaturated FAs in inactivating Ubxd8 might enhance the conversion of DAGs to TGs and thus relieving insulin resistance.
Materials and Methods
CD Spectroscopy.
The CD spectra of Ubxd8(Δ90–118) were recorded by Aviv model 62DS CD spectrometer (Aviv Associates Inc.). Ellipticity was measured in 0.5 ml of buffer A containing 2.5 μM of the protein at 222 nm using a 2-mm cell. Thermal denaturation was determined by CD recorded as a temperature scan with temperature ranging from 25 to 95 °C.
Blue Native PAGE.
Ubxd8(Δ90–118) (0.7 μg) was incubated with 100 μM of indicated FAs in buffer A at room temperature for 5 min (final volume, 18 μl). After receiving 2 μl of a 10x loading buffer (5 mM Bis-Tris, pH 7.0, 60% glycerol, 0.5 μg/ml Coomassie G250, and 10 mg/ml 6-aminohexanoic acid), the samples were subjected to 4–12% blue native gel electrophoresis for 2 h at room temperature as previously described (24).
Gel Filtration Analysis.
Ubxd8(Δ90–118) (0.4 mg) incubated with or without 100 μM Na Oleate (final volume, 0.4 ml) was applied to FPLC using a Superose 6 size exclusion column(Amersham) preequilibrated with buffer A supplemented with or without 100 μM Na Oleate at a flow rate of 0.4 ml/ min at 4 °C. Absorbance at 280 nm was monitored continuously to identify position of elution of the protein.
Supplementary Material
Acknowledgments.
We thank Drs. Michael S. Brown and Joseph L. Goldstein for their continued support and advice; Lisa Beatty, Angela Carroll, Shomanike Head, and Ijeoma Onwuneme for invaluable help with tissue culture; and Saada Abdalla for excellent technical assistance. This work was supported by grants from the National Institutes of Health (HL-20948) and Perot Family Foundation. J.Y. is supported by American Heart Association National Scientific Development Grant 0630029N.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1011859107/-/DCSupplemental.
References
- 1.Ntambi JM. Dietary regulation of stearoyl-CoA desaturase 1 gene expression in mouse liver. J Biol Chem. 1992;267:10925–10930. [PubMed] [Google Scholar]
- 2.Ou J, et al. Unsaturated fatty acids inhibit transcription of the sterol regulatory element-binding protein-1c (SREBP-1c) gene by antagonizing ligand-dependent activation of the LXR. Proc Natl Acad Sci USA. 2001;98:6027–6032. doi: 10.1073/pnas.111138698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hannah VC, Ou J, Luong A, Goldstein JL, Brown MS. Unsaturated fatty acids down-regulate SREBP isoforms 1a and 1c by two mechanisms in HEK-293 cells. J Biol Chem. 2001;276:4365–4372. doi: 10.1074/jbc.M007273200. [DOI] [PubMed] [Google Scholar]
- 4.Farese RV, Walther TC. Lipid droplets finally get a little R-E-S-P-E-C-T. Cell. 2009;139:855–860. doi: 10.1016/j.cell.2009.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee JN, Zhang X, Feramisco JD, Gong Y, Ye J. Unsaturated fatty acids inhibit proteasomal degradation of insig-1 at a post-ubiquitination step. J Biol Chem. 2008;283:33772–33783. doi: 10.1074/jbc.M806108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coll T, et al. Oleate reverses palmitate-induced insulin resistance and inflammation in skeletal muscle cells. J Biol Chem. 2008;283:11107–11116. doi: 10.1074/jbc.M708700200. [DOI] [PubMed] [Google Scholar]
- 7.Horton JD, et al. Combined analysis of oligonucleotide microarray data from transgenic and knockout mice identifies direct SREBP target genes. Proc Natl Acad Sci USA. 2003;100:12027–12032. doi: 10.1073/pnas.1534923100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc Natl Acad Sci USA. 1999;96:11041–11048. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeBose-Boyd RA, et al. Transport-dependent proteolysis of SREBP: Relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 1999;99:703–712. doi: 10.1016/s0092-8674(00)81668-2. [DOI] [PubMed] [Google Scholar]
- 10.Yang T, et al. Crucial step in cholesterol homeostasis: Sterols promote binding of Scap to Insig-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 2002;110:489–500. doi: 10.1016/s0092-8674(02)00872-3. [DOI] [PubMed] [Google Scholar]
- 11.Yabe D, Brown MS, Goldstein JL. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc Natl Acad Sci USA. 2002;99:12753–12758. doi: 10.1073/pnas.162488899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sever N, Lee PCW, Song BL, Rawson RB, DeBose-Boyd RA. Isolation of mutant cells lacking Insig-1 through selection with SR-12813, an agent that stimulates degradation of 3-hydroxy-3-methylglutaryl-coenzyme A reductase. J Biol Chem. 2004;279:43136–43147. doi: 10.1074/jbc.M406406200. [DOI] [PubMed] [Google Scholar]
- 13.Meusser B, Hirsch C, Jarosch E, Sommer T. ERAD: The long road to destruction. Nat Cell Biol. 2005;7:766–772. doi: 10.1038/ncb0805-766. [DOI] [PubMed] [Google Scholar]
- 14.Vembar S, Brodsky JL. One step at a time: endoplasmic reticulum-associated degradation. Nat Rev Mol Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ikeda Y, et al. Regulated endoplasmic reticulum-associated degradation of a polytopic protein: p97 recruits proteasomes to Insig-1 before extraction from membranes. J Biol Chem. 2009;284:34889–34900. doi: 10.1074/jbc.M109.044875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zehmer JK, et al. Targeting sequences of UBXD8 and AAM-B reveal that the ER has a direct role in the emergence and regression of lipid droplets. J Cell Sci. 2009;122:3694–3702. doi: 10.1242/jcs.054700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Munro S, Pelham HRB. A C-terminal signal prevents secretion of luminal ER proteins. Cell. 1987;48:899–907. doi: 10.1016/0092-8674(87)90086-9. [DOI] [PubMed] [Google Scholar]
- 18.Rawson RB, DeBose-Boyd R, Goldstein JL, Brown MS. Failure to cleave sterol regulatory element-binding proteins (SREBPs) causes cholesterol auxotrophy in Chinese hamster ovary cells with genetic absence of SREBP cleavage-activating protein. J Biol Chem. 1999;274:28549–28556. doi: 10.1074/jbc.274.40.28549. [DOI] [PubMed] [Google Scholar]
- 19.Ananthapadmanabhan KP, Goddard ED, Turro NJ, Kuo PL. Fluorescence probes for critical micelle concentration. Langmuir. 1985;1:352–355. doi: 10.1021/la00063a015. [DOI] [PubMed] [Google Scholar]
- 20.Yamamoto T, et al. The human LDL receptor: A cysteine-rich protein with multiple Alu sequences in its mRNA. Cell. 1984;39:27–38. doi: 10.1016/0092-8674(84)90188-0. [DOI] [PubMed] [Google Scholar]
- 21.Yen CLE, Stone SJ, Koliwad S, Harris C, Farese RV., Jr DGAT enzymes and triacylglycerol biosynthesis. J Lipid Res. 2008;49:2283–2301. doi: 10.1194/jlr.R800018-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unger RH. Lipotoxic diseases. Annu Rev Med. 2002;53:319–336. doi: 10.1146/annurev.med.53.082901.104057. [DOI] [PubMed] [Google Scholar]
- 23.Erion DM, Shulman GI. Diacylglycerol-mediated insulin resistance. Nat Med. 2010;16:400–402. doi: 10.1038/nm0410-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wittig I, Braun HP, Schagger H. Blue native PAGE. Nat Protoc. 2006;1:418–428. doi: 10.1038/nprot.2006.62. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.