Abstract
Pex19p, a soluble cytoplasmic transport protein, is required for the traffic of the peroxisomal membrane proteins Pex3p and Pex15p from the endoplasmic reticulum (ER) to the peroxisome. We documented Pex15p traffic from the ER using a chimeric protein containing a C-terminal glycosylation acceptor peptide. Pex15Gp expressed in wild-type yeast cells is N-glycosylated and functions properly in the peroxisome. In contrast, pex19Δ-mutant cells accumulate the glycoprotein Pex15Gp in the ER. We developed a cell-free preperoxisomal vesicle-budding reaction in which Pex15Gp and Pex3p are packaged into small vesicles in the presence of cytosol, Pex19p, and ATP. Secretory vesicle budding (COPII) detected by the packaging of a SNARE protein (soluble N-ethylmaleimide-sensitive attachment protein receptor) occurs in the same incubation but does not depend on Pex19p. Conversely a dominant GTPase mutant Sar1p which inhibits COPII has no effect on Pex3p packaging. Pex15Gp and Pex3p budded vesicles sediment as low-buoyant-density membranes on a Nycodenz gradient and copurify by affinity isolation using native but not Triton X-100–treated budded vesicles. ER–peroxisome transport vesicles appear to rely on a novel budding mechanism requiring Pex19p and additional unknown factors.
Peroxisomes function in selected metabolic and biosynthetic pathways that are essential for normal development in mammals (1–3). Despite decades of work on the biosynthetic origin of the peroxisome, uncertainty remains about the source of membrane precursors. Classic morphologic work by Novikoff and Shin (4) pointed to an origin of peroxisomal membrane in the endoplasmic reticulum (ER). Subsequent work by Lazarow and Fujiki (5) and many others favored an independent origin in which peroxisomal proteins are imported from the cytoplasm into an organelle that propagates by growth and division of a preexisting organelle. However, with the development of a genetic approach to investigate peroxisome assembly in yeast, it became possible to test the biogenic dependence or independence of the organelle (6–9).
The growth and division model was challenged in genetic studies of peroxisome biogenesis in yeast where it was possible to demonstrate that the generation of new peroxisomes does not depend on a preexisting organelle (10). Two proteins, Pex3p and Pex19p, are required to initiate peroxisome formation (10, 11). Pex3p, an N-terminally anchored membrane protein, serves an unknown but essential role. Pex19p, a cytoplasmic C-terminal prenylated protein, binds to peroxisomal membrane proteins (PMPs) and may chaperone their insertion into the peroxisomal membrane (10–13). Induction of Pex19p in a pex19Δ cell results in the creation of new peroxisomes (14). Furthermore, at least two PMPs, Pex3p and Pex15p, appear to be localized to the ER in pex19Δ cells (14–16). Several other peroxisomal proteins also may originate in the ER (15). Pex15p is a C-terminal tail-anchor protein that requires the GET1, -2, and -3 gene products, which act at the ER, to assemble other C-terminal tail-anchored proteins required for secretory traffic (16). Hoepfner et al. (14) showed that newly made YFP-Pex3p migrates from the ER to the peroxisome, suggesting a novel path of intracellular traffic. They observed large ER-associated foci of YFP-Pex3p as an intermediate in this traffic path. Independently, using a photoactivable form of GFP, Kim et al. (17) showed that Pex16, a mammalian PMP, originates in the ER. However, other than Pex19p, the requirements for the formation and targeting of a membrane carrier from the ER to the peroxisome remain unknown.
We developed two independent tests of the origin and vesicular carrier mechanism of peroxisomal protein transfer from the ER. A hybrid form of Pex15p was created containing a glycosylation sequence which we used to document that Pex15p is exposed to the ER-localized N-glycosylation apparatus en route to the peroxisome. Further, we established a cell-free vesicle-budding reaction that reproduces the formation of a distinct membrane carrier responsible for transit of Pex15p and Pex3p from the ER. These ER-derived membrane carriers may fuse homotypically to create a new peroxisome or fuse with a preexisting peroxisome to sustain their growth and division.
Results
Pex15p as a Reporter of Peroxisome Traffic via the ER.
Pex15p is a C-terminal tail-anchored membrane protein that has been reported to assemble in the ER membrane (16, 18). A special insertion complex, the Get proteins, is required for ER membrane assembly, and in its absence Pex15p appears in mitochondria (16). Thus, as has been documented for Pex3p traffic, we predicted that Pex15p would assemble in the peroxisome from an ER precursor form.
We adopted a variation of an approach Schuldiner et al. (16) used to demonstrate ER insertion of tail-anchored proteins. An N-terminal protein A (pA) and GFP extension of Pex15p was tagged at the C terminus with a short opsin tag containing one potential N-glycosylation site (pA-GFP-Pex15G). This fusion gene was expressed under control of a methionine-repressible promoter in WT cells or pex19-deletion mutant cells which lack peroxisomes. Quantitative immunoblot assays using Pex15p serum showed the plasmid copy and the chromosomally encoded Pex15p were expressed at levels comparable to the endogenous levels in untransfected cells (Fig. S1). In WT cells, pA-GFP-Pex15Gp localized to discrete punctae that coincide with peroxisomes as marked by a CFP-peroxisomal target signal 1 (PTS1) (Fig. 1 A and B). In contrast, in a pex19Δ strain, pA-GFP-Pex15Gp localized to the nuclear envelope and cortical ER (Fig. 1A). The steady-state level of pA-GFP-Pex15Gp was reduced in pex19Δ vs. WT cells. Perhaps ER-localized Pex15p is subject to ER-associated degradation.
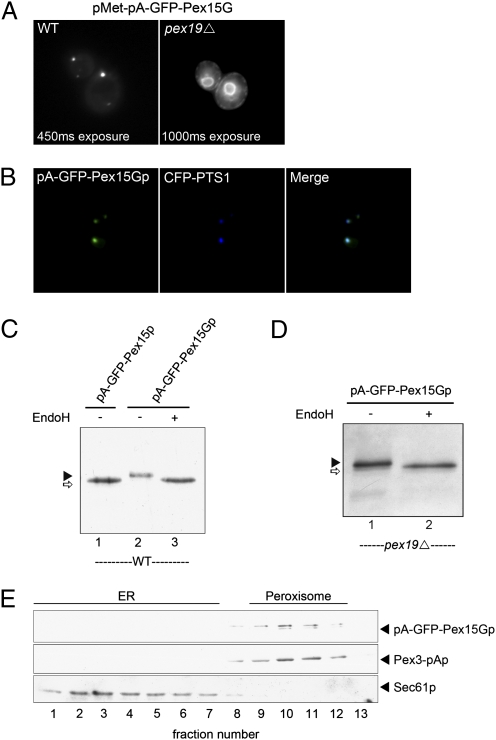
Fig. 1.
Expression of pA-GFP-Pex15Gp. (A) A fusion gene coding Pex15 with an N terminus tagged with pA and GFP and a C terminus tagged with an opsin peptide containing a predicted N-glycosylation site was cloned under control of a methionine-repressible promoter. The tagged Pex15 was expressed in WT and pex19Δ-mutant cells. GFP fluorescence was localized to punctae in WT cells and labeled the nuclear envelope and peripheral ER in pex19Δ cells. (B) Localization of pA-GFP-Pex15Gp was compared with a PMP, CFP-PTS1, in WT cells. (C) N-glycosylation of pA-GFP-Pex15Gp and pA-GFP-Pex15p (without a C-terminal glycosylation site) in WT cells was analyzed by EndoH treatment. pA-GFP-Pex15Gp displayed decreased mobility on SDS/PAGE compared with pA-GFP-Pex15p (lane1 and 2). EndoH treatment increased the mobility of pA-GFP-Pex15Gp to approximately that of pA-GFP-Pex15p (lane 3). (D) EndoH treatment of pA-GFP-Pex15Gp expressed in pex19Δ cells reduced the apparent size of the protein to that of pA-GFP-Pex15Gp. (E) pA-GFP-Pex15Gp was expressed under a methionine-repressible promoter in WT cells, and cell extracts were fractionated by centrifugation on a 10–50% step Nycodenz gradient. pA-GFP-Pex15Gp cofractionated with a genomically encoded Pex3-pAp fusion protein and separated from the ER marker Sec61p.
Elgersma et al. (18) used a Pex15-invertase fusion protein to detect ER insertion of Pex15p. Overexpressed fusion protein became N-glycosylated but remained in the ER. Our shorter opsin fragment linked to Pex15p did not interfere with Pex15 function or localization to the peroxisome (Fig. 1B). To assess the role of the ER in traffic of Pex15p, we examined the glycosylation of pA-GFP-Pex15Gp in WT and pex19Δ cells. Fig. 1C shows that pA-GFP-Pex15Gp was sensitive to endoglycosidase H (endoH), and the deglycosylated species migrated close to the same position as pA-GFP-Pex15p. Similarly, pA-GFP-Pex15Gp made in pex19Δ was sensitive to endoH (Fig. 1D).
To be certain that glycosylated Pex15Gp trafficked to the peroxisome, we fractionated membranes from WT cells expressing the hybrid protein (Fig. 1E). Glycosylated and some nonglycosylated Pex15Gp (upper and lower bands, respectively) cosedimented with peroxisomes, marked by another peroxisomal protein, Pex3-pAp, and separated from ER membranes, marked by Sec61p, on a buoyant density gradient. These results support the proposal that Pex15p passes through the ER en route to the peroxisome.
Reconstitution of Pex15Gp Budding from the ER.
Hoepfner et al. (14) showed that peroxisomes arise de novo from the ER when Pex19p expression is induced in a pex19Δ cell. They found that Pex3p accumulates in the ER in a pex19Δ strain and collects in ER punctae that migrate to peroxisomes when Pex19p expression is induced.
We sought a biochemical test of the role of Pex19p in Pex15p and Pex3p traffic from the ER. For this purpose, we created pex19Δ yeast strains harboring a methionine promoter-controlled pA-GFP-Pex15G fusion gene or a Pex-positive functional integrated form of Pex3-pA. For experiments where pA-GFP-Pex15Gp was measured, cells were grown in synthetic medium with methionine (20 mM) to reduce Pex15Gp expression, and the total Pex15p level remained the same even after Pex15Gp expression (Fig. S1). Cells were converted to spheroplasts and lysed gently under conditions in which ER membranes sediment rapidly. The gentle lysate served as a donor membrane for a vesicle-budding reaction modeled on a reconstitution that allowed us to isolate the COPII coat responsible for secretory cargo protein traffic from the ER (19–21).
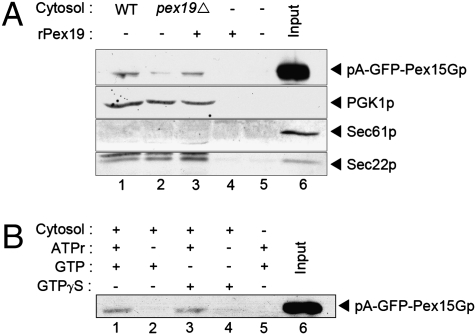
Membranes were mixed with cytosol from WT or pex19Δ-mutant cells in the presence of ATP, an ATP-regenerating system, and GTP and were incubated at 22 °C for 30 min. Donor ER membranes were centrifuged for 5 min at 20,000 × g, and the medium-speed supernatant (MSS) fraction was removed and analyzed by SDS/PAGE and immunoblot for the presence of pA-GFP-Pex15Gp. We detected Pex15Gp (ca. 1% of total) in the MSS fractions from incubations containing WT cytosol, but the signal was dramatically decreased (from 1 to <0.1%) with pex19Δ-mutant cytosol (Fig. 2A, lanes 1 and 2). Recombinant Pex19p expressed in and purified from Escherichia coli restored budding of Pex15Gp in the presence of mutant cytosol but not without cytosol (Fig. 2A, lanes 3 and 4). As controls, we detected the cytosolic protein 3-phosphoglycerate kinase (PGK1p) in the MSS fraction but not the ER-resident protein, Sec61p. In contrast, the COPII membrane cargo protein, Sec22p, was detected in the MSS fraction independent of Pex19p. Pex15Gp release into the MSS fraction was stimulated by ATP and cytosol, but GTP did not substitute for ATP, and GTPγs had no effect on the ATP-stimulated reaction (Fig. 2B).
Fig. 2.
Cell-free vesicle budding of pA-GFP-Pex15Gp. (A) Microsome membranes isolated from pex19Δ cells expressing pA-GFP-Pex15Gp were incubated with nucleotide WT cytosol (lane 1), pex19Δ cytosol (lane 2), pex19Δ cytosol plus Pex19 recombinant protein (rPex19, 4 μg/mL) (lane 3), rPex19 alone (4 μg/mL) (lane 4), buffer only (lane 5) or 10% load of starting microsome (lane 6). After 30 min incubation, microsomes were centrifuged, and the MSS fraction was analyzed by SDS/PAGE and immunoblot. (B) The requirements for vesicle budding of pA-GFP-Pex15Gp were assessed in incubations containing WT cytosol with ATP-regenerating system (ATPr) and 100 μM GTP (lane 1); WT cytosol with 100 μM GTP (lane 2); WT cytosol with ATPr and 100 μM GTPγS (lane 3); WT cytosol with 100 μM GTPγS (lane 4); ATPr and 100 μM GTP only (lane 5); or 10% load of starting microsome (lane 6).
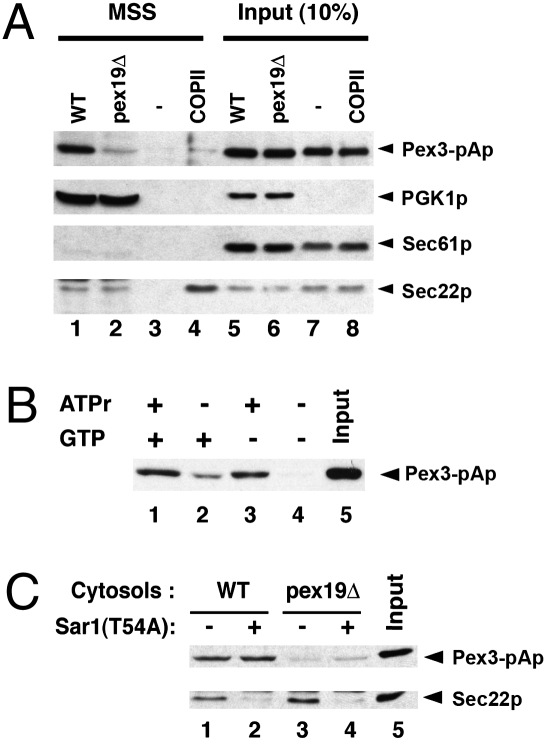
We applied the same biochemical budding reaction to evaluate the packaging of Pex3-pAp into small vesicles. As with pA-GFP-Pex15Gp, Pex3-pAp was detected in the MSS fraction (ca. 10% of total) when gently lysed cells were incubated with WT cytosol, ATP, and ATP-regenerating components, but the signal was strongly reduced with pex19Δ-mutant cytosol (from 10 to <1%), and no signal was detected with buffer alone (Fig. 3 A, lanes 1–3, and B). Additional controls showed that the COPII cargo protein, Sec22p, was packaged in reactions containing WT or pex19Δ cytosol, whereas Sec61p, the resident ER protein, was retained in the medium-speed pellet fraction. Conversely, pure COPII proteins packaged Sec22p efficiently but not Pex3-pAp (lane 4).
Fig. 3.
Cell-free packaging of Pex3-pAp. (A) Microsome membranes from pex19Δ cells expressing a chromosomally integrated form of Pex3-pA were incubated at room temperature (22 °C) for 30 min with WT cytosol (lane 1), pex19Δ cytosol (lane 2), no cytosol (lane 3), or COPII purified proteins (Sar1p, Sec23p/24p, and Sec13p/31p) only. MSS fractions were obtained by centrifugation at 15,000 rpm for 5 min in an Eppendorf refrigerated bench top centrifuge, and aliquots (17 μL) were sampled for SDS/PAGE. Lanes 5–8 represent a 10% load of total reactions. (B) Packaging of Pex3-pAp was carried in the presence of both ATPr and GTP (lane 1), GTP only (lane 2), ATPr only (lane 3), or in the absence of nucleotide (lane 4). In lane 5 there was a 10% load of starting microsome. (C) Budding reaction of Pex3-pAp was conducted in the presence of nucleotides with different cytosol fractions: WT cytosol (lane 1); WT cytosol with Sar1(T54A) (lane 2); pex19Δ cytosol (lane 3); pex19Δ cytosol with Sar1(T54A) (lane 4); or 10% load of starting microsome fraction (lane 5).
We also used selective inhibition of COPII budding to document distinct requirements for Pex3-pAp and secretory protein cargo packaging. A dominant GTPase-mutant form of Sar1p (T54A) that blocks COPII coat disassembly inhibited the packaging of Sec22p but had no effect on the incorporation of Pex3-pAp into slowly sedimenting membrane vesicles (Fig. 3C).
Pex3-pAp and pA-GFP-Pex15Gp Packaged Together into Membrane Vesicles.
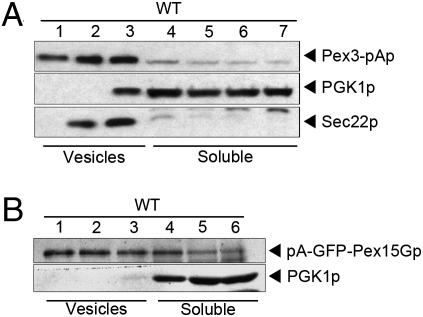
We used flotation of the MSS fraction on a Nycodenz density gradient to determine if pA-GFP-Pex15Gp and Pex3-pAp were associated with buoyant membranes. Samples were mixed with Nycodenz to a 35% final concentration and placed below less dense Nycodenz fractions. Centrifuged samples were fractionated and examined by immunoblot to detect peroxisomal proteins, cytosolic protein (PGK1p), and COPII vesicles (Sec22p). Fig. 4 A and B documents that both peroxisomal proteins associated with buoyant membranes that were slightly more buoyant than COPII vesicles and were resolved from the cytosolic marker.
Fig. 4.
Pex3-pAp and pA-GFP-Pex15Gp are packaged into buoyant membrane. (A) The MSS fraction from a cell-free incubation of Pex3-pAp microsomes was evaluated using a density gradient flotation assay. Membrane-bound Sec22p and Pex3-pAp centrifuged near the top of a Nycodenz gradient (fractions 1–3), whereas soluble protein (PGK1p) remained in the densest Nycodenz fractions (fractions 4–7). (B) The MSS fraction from a cell-free incubation of pA-GFP-Pex15Gp microsomes was evaluated by density gradient flotation. Membrane-bound Sec22p and pA-GFP-Pex15Gp centrifuged near the top of a Nycodenz gradient (fractions 1–3), whereas soluble protein (PGK1p) remained in the dense Nycodenz phase (fractions 4–6).
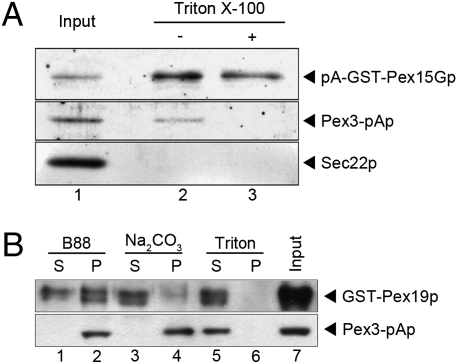
Two further experiments were conducted to examine the membrane integration of Pex15Gp and Pex3p. The pex19Δ strain harboring an integrated Pex3-pA was transformed with a centromere (CEN) plasmid carrying a pA-GST-Pex15G construct under control of the methionine promoter. Cells grown in methionine-minimal medium were converted to spheroplasts, and membranes prepared for the budding reaction were incubated with WT cytosol and nucleotide. The centrifuged MSS fraction was incubated with glutathione-agarose beads in the absence or presence of 1% Triton X-100 to capture exposed pA-GST-Pex15Gp. pA-GST-Pex15Gp, Pex3-pAp, and Sec22p were present in the starting fraction, but after extensive washing with buffer only Pex15Gp and Pex3p were retained on the glutathione beads (Fig. 5A, lanes 1 and 2). However, in the presence of detergent, only Pex15Gp was recovered on beads (Fig. 5A, lanes 2 and 3). We conclude that some Pex15Gp and Pex3p are packaged together in a common membrane vesicle, although the possibility of separate vesicles for a fraction of each protein remains possible.
Fig. 5.
pA-GFP-Pex15Gp and Pex3-pAp co-isolate in budded vesicles. (A) A fivefold scaled-up vesicle-budding incubation was carried out with microsomes prepared from pex19Δ cells coexpressing pA-GST-Pex15Gp (CEN plasmid) and Pex3-pAp (chromosomally integrated). The resulting MSS fraction (lane 1) was incubated with glutathione-agarose beads in buffer with (lane 3) or without (lane 2) Triton X-100. Sec22p is a COPII vesicle cargo protein. (B) The MSS fraction was prepared from a budding reaction using membranes from the Pex3-pAp strain mixed with pex19Δ cytosol and purified GST-Pex19p. The MSS fraction was subjected to high-speed centrifugation to obtain vesicles enriched in an HSP. The HSP was resuspended with B88 buffer (lanes 1 and 2), 0.1 M Na2CO3 (lanes 3 and 4), and 1% Triton X-100 (lanes 5 and 6). The resuspended HSP was centrifuged again at 55,000 rpm in TLA100.3 for 30 min and separated into pellet (P) and supernatant (S). Lane 7, 10% load of the starting microsome fraction.
In another experiment, we examined the membrane association of Pex3-pAp in vesicles in the MSS fraction after a budding reaction. A standard incubation of pex19Δ membranes was conducted with pex19Δ cytosol and purified GST-Pex19p (to provide a tag for Pex19p detection). We found that GST-Pex19p promoted Pex3-pAp budding as efficiently as Pex19p produced by proteolytic cleavage of the GST hybrid protein. The MSS fraction was centrifuged at high speed to collect small vesicles in a pellet which was resuspended in buffer (B88), 0.1 M Na2CO3 (to extract peripheral membrane proteins), or 1% Triton X-100 (to solubilize membranes). After a repeat high-speed centrifugation, the supernatant and pellet fractions were resolved on SDS PAGE and examined by immunoblot for the pA and GST-tagged peroxisomal proteins. GST-Pex19p was membrane associated, possibly by direct interaction with Pex3p (22), but was readily removed by carbonate or detergent. In contrast, Pex3-pAp was retained in membranes except in the presence of detergent. We conclude that Pex3p is packaged as an integral membrane protein, along with Pex15Gp, into buoyant vesicles in a budding reaction that requires Pex19p and other cytosolic proteins.
Other Cytoplasmic Peroxisomal Proteins Are Not Required for Pex3p Budding.
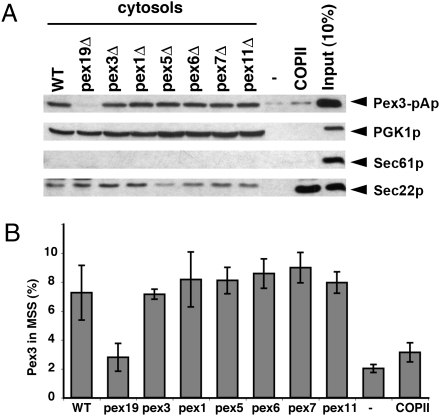
We observed that Pex19p is not sufficient to package pA-GFP-Pex15Gp into budded vesicles (Fig. 2A, lanes 3 and 4). The other known cytoplasmic, Pex proteins (Pex1p, Pex5p, Pex6p, Pex7p, and Pex11p) are required for protein import into the organelle or for organelle dynamics but not for the formation of the organelle per se (23–25). Nonetheless, we tested their role in the budding reaction using cytosol prepared from strains harboring deletions in each of these genes and from pex3Δ cells and compared the activity of these fractions with the standard pex19Δ cytosol in a Pex3-pAp budding reaction. As seen before, pex19Δ cytosol was defective in Pex3-pAp budding but not in Sec22p budding (Fig. 6). Conversely, pure COPII proteins packaged Sec22p but not Pex3-pAp. Cytosols from the other pex-mutant cells showed no defect in Pex3-pAp packaging. The additional soluble proteins required to produce Pex3-pAp budded vesicles remain unknown.
Fig. 6.
Pex3-pAp packaging is dependent on Pex19p but not on other cytoplasmic peroxisomal proteins. Microsomes were incubated with cytosol prepared from WT, pex19Δ, pex3Δ, pex1Δ, pex5Δ, pex6Δ, pex7Δ, pex11Δ, without cytosol, and with COPII recombinant proteins only. MSS fractions were obtained from each reaction and detected as described in Materials and Methods. Pex3-pAp in the MSS fraction was examined by SDS/PAGE and immunoblot (A) and quantified (B) by Typhoon and Image Quant Software (Molecular Dynamics). n = 2.
Discussion
The biogenic origin of peroxisomes has been a controversial subject for decades. In recent years, most investigators have focused on the ER as a source of membrane and at least some of the integral membrane proteins that end up in the peroxisome. Most of the evidence in support of a role for the ER comes from visualization of peroxisomal precursors in the ER membrane followed kinetically by movement to the peroxisome (14, 15). Although it is clear that Pex3p and Pex15p accumulate in the ER in a pex19Δ yeast mutant (14, 15), the possibility that this path is a minor one taken when no peroxisome exists for these proteins to enter could not be ruled out. Thus it has been difficult to test the role of the ER as an essential station in this pathway.
We have developed two lines of evidence concerning the role of the ER in peroxisome biogenesis. Using an opsin tag, we introduced a single potential N-glycosylation site at the C terminus of Pex15 to create a functional fusion protein. We find this protein accumulates as a glycoprotein in WT and pex19Δ-mutant cells. Fluorescence localization and cell fractionation of PEX-positive cells confirms that the glycoprotein is in bona fide peroxisomes. Thus, even without an interruption of normal traffic in the pex19 mutant, Pex15Gp shows the mark of an ER intermediate in peroxisome assembly.
Pex19p has been found in molecular complexes with newly synthesized PMPs (12, 26). This finding was interpreted as a role for Pex19p in chaperoning the import of membrane proteins directly from the cytoplasm into the peroxisome. What role could Pex19p play if PMPs are first assembled in the ER? To examine the mechanism of peroxisomal protein transit from the ER, we developed a cell-free vesicle-budding reaction in which two different PMPs are incorporated into a slowly sedimenting vesicle fraction dependent on Pex19p, ATP, and additional cytosolic factors. Our evidence demonstrates that this budding process is independent of the COPII machinery responsible for secretory cargo packaging at the ER.
Pex3p is the one of the most crucial PMPs required for peroxisome formation. No peroxisomal structures are detected in pex3-mutant cells (10). New peroxisomes form when Pex3p synthesis is induced, and Pex19p is required for this de novo process (10, 14). Our cell-free budding reaction recapitulates the first step of this pathway. Indeed, the importance of Pex3p in peroxisome biogenesis may be reflected in the efficiency with which it is packaged in our cell-free budding reaction in relation to Pex15p (10% vs. 1% of total). Pex19p contains a dedicated domain for Pex3p interaction and a common domain for the other PMPs (13), perhaps explaining why Pex3p has a higher packaging efficiency than Pex15p. Another possibility is that pA-GFP in some way sterically hinders the preperoxisomal packaging of Pex15p. Preperoxisomal vesicle biogenesis may begin with the recruitment of Pex19p to the membrane peroxisomal targeting signal (mPTS) on an ER-integrated form of Pex3p. Likewise, Pex15p and other PMPs may display mPTS on the ER that also are bound by Pex19p. In contrast, resident ER proteins or biosynthetic membrane proteins destined for transport through the secretory pathway would lack such mPTS sequences. Thus, Pex19p may constitute an adaptor-like protein to initiate the sorting of PMPs from other membrane proteins in the ER.
Our recombinant Pex19p was made in E. coli; thus it lacks the C-terminal prenylation group attached to the native protein in yeast (11, 27). Nonetheless, the recombinant proteins, as well as a GST-Pex19p chimera, were functional in our cell-free budding reaction. However, activity depended on additional heat-sensitive cytosolic factors contained in the WT or pex19Δ-mutant cell lysates. The unidentified factors, probably proteins, do not include known peroxisomal cytoplasmic proteins, COPII, or COPI. One or more of the additional proteins or possibly a membrane protein may confer ATP dependence on this budding reaction. Neither Pex3p nor Pex19p is predicted to contain an ATP-binding domain (10–12). The unknown factors could bind ER-associated Pex19p and convey Pex19p-bound cargo into a peroxisomal transport vesicle.
Peroxisomes are not essential for yeast cell growth on glucose medium. Thus, the genetic screens for pex mutants were conducted initially by mutagenesis and then through a survey of the entire set of viable deletion-mutant yeast strains (6, 8, 28). Despite this exhaustive search, no additional candidate genes encoding cytosolic proteins as important as Pex19p have emerged. The unidentified genes may be essential for yeast cell viability (e.g., if preperoxisome vesicle budding requires proteins also engaged in an essential vesicle traffic event), or they may encode redundant functions (29). Additional genetic screens that incorporate these possibilities may turn up such candidates. However, our cell-free reaction provides an alternative to allow the purification of functional forms of the missing components.
Materials and Methods
Yeast Strains and Media.
Protein A was integrated at the C-terminal end of the Pex3 locus by a PCR-based one-step transformation procedure (30). Primers for C-terminal protein A integration were 5′-ATACAGCAACTTTGGCGTCTCCAGCTCGTTTTCCTTCAAGCCT and 3′-CGCTATATATATATATATTCTGGTGTGAGTGTCAGTACTTATTCA. The PCR product was introduced into pex19Δ cells from the Euroscarf collection (Mat α; his3Δ; leu2 Δ; lys2 Δ; ura3 Δ; pex19::kanMX4) to obtain NYY9 (Mat α; leu2 Δ; lys2 Δ; ura3 Δ; pex19::kanMX4 Pex3-pA::His3), which was used for microsome preparation. Pex15G was cloned from yeast genomic DNA with an additional opsin tag sequence (ATGATGTCTAGAATGAATGGTACTGAAGGTCCAAATTTTTATGTTCCATTTTCTAATAAAACAGTTGAT) at the 3′ end and was inserted downstream of pA and GFP in a MET25 vector (31). For cytosol preparation, all the deletion-mutant cells were from the Euroscarf deletion collection. Cells were grown at 30 °C in yeast-peptone-dextrose (YPD) medium (1% yeast extract, 2% peptone, 2% glucose) or synthetic medium (2% glucose, Difco yeast nitrogen base without amino acid, CSM-Ura from Sunrise Science with 20 mg/L of methionine) for the preparation of microsomal and cytosolic fractions. For oleic acid selection, cultures were grown at 30 °C on oleate plates containing 0.67% yeast nitrogen base, 0.25% oleic acid, 0.25% Tween 40, 2% agar, with the supplement of amino acids.
EndoH Treatment.
pA-GFP-Pex15p or pA-GFP-Pex15Gp was expressed under a methionine-repressible promoter in WT cells and pex19Δ cells, and 5 mL of cell culture (OD600 = 0.8) was centrifuged at 4,000 × g for 5 min. The cell pellet was resuspended in 1 mL of 20 mM Tris·HCl buffer. Total proteins were extracted by grinding in liquid nitrogen and heated at 95 °C with 1% SDS for 5 min. The lysate was centrifuged at 15,000 rpm in a bench-top ultracentrifuge (Eppendorf 5417C with rotor F45-30-11), and the supernatant fraction was collected. Aliquots (1–20 μg protein) were mixed with 1 μL 10× denaturing buffer (New England Biolabs), and water was added to a volume of 10 μL. The mixture was heated at 95 °C for 10 min. Once the mixture cooled, 5 μL (5,000 units) of EndoH (New England Biolabs) and 2 μL 10× G5 reaction buffer (New England Biolabs) were added, and the volume was adjusted to 20 μL with water. The reaction was incubated at 37 °C for 1 h. Samples (15 μL) were heated with SDS-loading buffer and sampled for SDS/PAGE and immunoblot analysis.
Subcellular Fractionation of Peroxisome.
pA-GFP-Pex15G was expressed under a methionine-repressible promoter in WT cells, and 5 mL of cell culture (OD600 = 0.8) was centrifuged at 4,000 × g for 5 min. The cell pellet was resuspended in 1 mL of lyticase buffer (0.7 M sorbitol, 0.75× yeast extract Peptone, 0.5% glucose, 10 mM Hepes/NaOH, pH 7.4) and digested with lyticase (6,000 units/mL) at 30 °C for 30 min. Cells were centrifuged at 4,000 × g for 10 min at 4 °C and resuspended in 1 mL of B88 [20 mM Hepes-KOH (pH 6.8), 0.25 M Sorbitol, 0.15 M KOAc, 1 mM MgOAc2] with protease inhibitor mixture (Roche). The suspension was homogenized with 20 strokes in a Potter-Elvehjem homogenizer. The lysate was centrifuged at 4,000 × g for 10 min. The postnuclear supernatant (300 μL) was loaded on the top of a Nycodenz step gradient ranging from 10 to 50% (200 μL: 50%, 40%, 30%, 20%; 150 μL: 10%) and centrifugated for 2 h at 55,000 rpm at 4 °C in a Beckman TLS-55 ultracentrifuge. The top 150 μL was removed, and 13 fractions (60 μL) were collected. An aliquot (15 μL) of each fraction was heated at 65 °C with SDS loading buffer for 10 min and sampled for SDS/PAGE and immunoblot analysis.
Protein Purification.
GST-Pex19 was prepared as an N-terminal GST fusion protein from pGEX-2T (GE Healthcare) in E. coli BL21 (DE3). The GST-tagged protein was purified by a method described previously (32, 33) with slight modification. GST-Pex19 expression was induced by isopropyl-β-d-thio-galactoside (0.1 mM) at 30 °C for 3 h. GST was cleaved from Pex19 by thrombin cleavage in 20 mM Tris·HCl (pH 8) buffer with 2.5 mM CaCl2 at 22 °C for 2 h. Sar1, Sec13/31, and Sec23/24 were prepared as described (20). A dominant-negative form of Sar1 (T54A) (34) was purified from E. coli expressing GST-fusion proteins. Thrombin was used to cleave the GST from Sar1p, which was purified further by MonoQ column.
Cell-Free Vesicle Budding Reaction.
For the preparation of concentrated cytosol, yeast was grown in 1 L YPD at 30 °C, and cells were harvested at ∼1 OD600/mL, washed in cold water, and resuspended in 2 mL of B88 with protease inhibitor mixture (Roche). A concentrated suspension was dropped into a mortar filled with liquid nitrogen to form small beads which were ground to a fine powder with a pestle. The frozen lysate was transferred into a 50-mL tube, thawed on ice, and centrifuged at 3,000 rpm for 5 min, then at 15,000 rpm for 10 min (Beckman SS34), and finally at 34,000 rpm for 1 h (Beckman SW55Ti). The supernatant was collected carefully to avoid a lipid phase at the meniscus. Microsomal membranes for use in vesicle-budding reactions were prepared from NYY9 cells as described (20). The conditions of incubation for peroxisomal vesicle budding were similar to those used to form COPII vesicles (20). Aliquots (15 μg protein) of the microsome fraction were washed twice in 2.5 M urea in B88, rinsed twice in B88, and the washed/rinsed microsome fraction was centrifugated at 15,000 rpm in an Eppendorf refrigerated bench-top centrifuge. The final pellet was resuspended in 800 μL of B88. An aliquot (100 μL) of the microsomal suspension was mixed with cytosol (final concentration 4 mg protein/mL) in the presence of ATP (1 mM) with an ATP regeneration system (ATPr) (35) and GTP (0.1 mM) in a final volume of 250 μL. In one experiment, a dominant-negative form of Sar1 (T54A) was added at the concentration of 10 μg/mL. Purified COPII proteins were added at a concentration of 10 μg/mL Sar1p, 20 μg/mL Sec23p/24p complex, and 20 μg/mL Sec13p/31p complex. The reaction mixture was incubated at 22 °C for 30 min. The MSS fraction was obtained after centrifugation at 20,000 × g for 5min. In some experiments, the MSS fraction was analyzed further by high-speed centrifugation at 55,000 rpm for 30 min at 4 °C in a Beckman TLA100 ultracentrifuge or by equilibrium density flotation (20) at 55,000 rpm for 2 h at 4 °C in a Beckman TLS-55 ultracentrifuge. Briefly, MSS fractions (150 μL) were mixed with 70% Nycodenz (300 μL) in B88 (final Nycodenz concentration ∼35%) and placed at the bottom of a TLS-55 tube. Three solutions of different density (200 μL each of 35%, 25%, and 10% Nycodenz in B88) were layered on top. Tubes were centrifuged at 55,000 rpm for 2 h (Beckman TLS-55). Fractions (150 μL) were collected from the top, mixed with SDS loading buffer, and heated at 55 °C for 10 min, followed by SDS/PAGE and immunoblot analysis. Antibodies used in this study were rabbit peroxidase anti-peroxidase (Jackson ImmunoResearch Laboratories, Inc.) for the detection of Pex3-pAp, pA-GFP-Pex15p, pA-GFP-Pex15Gp, and pA-GST-Pex15Gp; anti-Sec22p, anti-Sec61p, anti-GST (Santa Cruz), and anti-Pgk1p (Molecular Probes). ECL plus Western blotting detection reagent (GE Healthcare) was used for detection. An alkaline sodium carbonate fractionation assay was performed as follows: Microsomal membranes were incubated with GST-Pex19p (20 μg/mL) with pex19 cytosol (4 mg/mL). The MSS fraction (150 μL) was collected after the incubation, followed by high-speed centrifugation as described above. The high-speed pellet (HSP) fraction was resuspended in 500 μL of B88 in three tubes. After a repeat centrifugation, the HSP was resuspended in 150 μL B88, 0.1 M Na2CO3, or 1% Triton X-100 and incubated on ice for 30 min; then soluble and integral membrane proteins were separated by high-speed centrifugation (Beckman TLA100) at 55,000 rpm for 30 min.
Glutathione Bead Co-Isolation.
A fivefold vesicle budding was carried out with microsomes prepared from pex19Δ cells coexpressing pA-GST-Pex15G (CEN plasmid) and Pex3-pA (chromosomally integrated). The resulting MSS fraction (1 mL) was incubated with 50 μL glutathione-agarose beads in B88 with or without 1% Triton X-100 overnight at 4 °C. Beads were settled by gravity, and the supernatant was removed. The beads were rinsed three times with 1.5 mL of B88 with or without 1% Triton X-100. The settled beads were heated to 95 °C with SDS loading buffer for 5 min and sampled for SDS/PAGE and immunoblot analysis.
Supplementary Material
Acknowledgments
We thank Jonathan Weissman (Howard Hughes Medical Institute, Department of Cellular and Molecular Pharmacology, University of California, San Francisco, and California Institute for Quantitative Biosciences, San Francisco) for the opsin tag constructs and Suresh Subramani (Section of Molecular Biology, Division of Biological Sciences, University of California at San Diego, La Jolla, CA) for the Pex15 antibody. We also thank Lazar Dimitrov for insightful discussions. S.K.L. is supported by a Human Frontier Science Program Long-term Fellowship. R.S. is an investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013397107/-/DCSupplemental.
References
- 1.Kunau WH. Peroxisome biogenesis: From yeast to man. Curr Opin Microbiol. 1998;1:232–237. doi: 10.1016/s1369-5274(98)80016-7. [DOI] [PubMed] [Google Scholar]
- 2.Steinberg SJ, et al. Peroxisome biogenesis disorders. Biochim Biophys Acta. 2006;1763:1733–1748. doi: 10.1016/j.bbamcr.2006.09.010. [DOI] [PubMed] [Google Scholar]
- 3.Brosius U, Gärtner J. Cellular and molecular aspects of Zellweger syndrome and other peroxisome biogenesis disorders. Cell Mol Life Sci. 2002;59:1058–1069. doi: 10.1007/s00018-002-8486-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Novikoff AB, Shin WY. The endoplasmic reticulum in the Golgi zone and its relations to microbodies, Golgi apparatus, and autophagic vacuoles in rat liver cells. J Microsc (Paris) 1964;3:187–206. [Google Scholar]
- 5.Lazarow PB, Fujiki Y. Biogenesis of peroxisomes. Annu Rev Cell Biol. 1985;1:489–530. doi: 10.1146/annurev.cb.01.110185.002421. [DOI] [PubMed] [Google Scholar]
- 6.Erdmann R, Veenhuis M, Mertens D, Kunau WH. Isolation of peroxisome-deficient mutants of Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1989;86:5419–5423. doi: 10.1073/pnas.86.14.5419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tsukamoto T, Yokota S, Fujiki Y. Isolation and characterization of Chinese hamster ovary cell mutants defective in assembly of peroxisomes. J Cell Biol. 1990;110:651–660. doi: 10.1083/jcb.110.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elgersma Y, van den Berg M, Tabak HF, Distel B. An efficient positive selection procedure for the isolation of peroxisomal import and peroxisome assembly mutants of Saccharomyces cerevisiae. Genetics. 1993;135:731–740. doi: 10.1093/genetics/135.3.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gould SJ, Valle D. Peroxisome biogenesis disorders: Genetics and cell biology. Trends Genet. 2000;16:340–345. doi: 10.1016/s0168-9525(00)02056-4. [DOI] [PubMed] [Google Scholar]
- 10.Höhfeld J, Veenhuis M, Kunau WH. PAS3, a Saccharomyces cerevisiae gene encoding a peroxisomal integral membrane protein essential for peroxisome biogenesis. J Cell Biol. 1991;114:1167–1178. doi: 10.1083/jcb.114.6.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Götte K, et al. Pex19p, a farnesylated protein essential for peroxisome biogenesis. Mol Cell Biol. 1998;18:616–628. doi: 10.1128/mcb.18.1.616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sacksteder KA, et al. PEX19 binds multiple peroxisomal membrane proteins, is predominantly cytoplasmic, and is required for peroxisome membrane synthesis. J Cell Biol. 2000;148:931–944. doi: 10.1083/jcb.148.5.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Y, Morrell JC, Jones JM, Gould SJ. PEX3 functions as a PEX19 docking factor in the import of class I peroxisomal membrane proteins. J Cell Biol. 2004;164:863–875. doi: 10.1083/jcb.200311131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoepfner D, Schildknegt D, Braakman I, Philippsen P, Tabak HF. Contribution of the endoplasmic reticulum to peroxisome formation. Cell. 2005;122:85–95. doi: 10.1016/j.cell.2005.04.025. [DOI] [PubMed] [Google Scholar]
- 15.van der Zand A, Braakman I, Tabak HF. Peroxisomal membrane proteins insert into the endoplasmic reticulum. Mol Biol Cell. 2010;21:2057–2065. doi: 10.1091/mbc.E10-02-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schuldiner M, et al. The GET complex mediates insertion of tail-anchored proteins into the ER membrane. Cell. 2008;134:634–645. doi: 10.1016/j.cell.2008.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim PK, Mullen RT, Schumann U, Lippincott-Schwartz J. The origin and maintenance of mammalian peroxisomes involves a de novo PEX16-dependent pathway from the ER. J Cell Biol. 2006;173:521–532. doi: 10.1083/jcb.200601036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elgersma Y, et al. Overexpression of Pex15p, a phosphorylated peroxisomal integral membrane protein required for peroxisome assembly in S.cerevisiae, causes proliferation of the endoplasmic reticulum membrane. EMBO J. 1997;16:7326–7341. doi: 10.1093/emboj/16.24.7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuoka K, et al. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–275. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- 20.Barlowe C, et al. COPII: A membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- 21.Rexach MF, Schekman RW. Distinct biochemical requirements for the budding, targeting, and fusion of ER-derived transport vesicles. J Cell Biol. 1991;114:219–229. doi: 10.1083/jcb.114.2.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matsuzono Y, Matsuzaki T, Fujiki Y. Functional domain mapping of peroxin Pex19p: Interaction with Pex3p is essential for function and translocation. J Cell Sci. 2006;119:3539–3550. doi: 10.1242/jcs.03100. [DOI] [PubMed] [Google Scholar]
- 23.Titorenko VI, Rachubinski RA. Peroxisomal membrane fusion requires two AAA family ATPases, Pex1p and Pex6p. J Cell Biol. 2000;150:881–886. doi: 10.1083/jcb.150.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li X, Gould SJ. PEX11 promotes peroxisome division independently of peroxisome metabolism. J Cell Biol. 2002;156:643–651. doi: 10.1083/jcb.200112028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meinecke M, et al. The peroxisomal importomer constitutes a large and highly dynamic pore. Nat Cell Biol. 2010;12:273–277. doi: 10.1038/ncb2027. [DOI] [PubMed] [Google Scholar]
- 26.Jones JM, Morrell JC, Gould SJ. PEX19 is a predominantly cytosolic chaperone and import receptor for class 1 peroxisomal membrane proteins. J Cell Biol. 2004;164:57–67. doi: 10.1083/jcb.200304111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vastiau IM, et al. Farnesylation of Pex19p is not essential for peroxisome biogenesis in yeast and mammalian cells. Cell Mol Life Sci. 2006;63:1686–1699. doi: 10.1007/s00018-006-6110-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lockshon D, Surface LE, Kerr EO, Kaeberlein M, Kennedy BK. The sensitivity of yeast mutants to oleic acid implicates the peroxisome and other processes in membrane function. Genetics. 2007;175:77–91. doi: 10.1534/genetics.106.064428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perry RJ, Mast FD, Rachubinski RA. Endoplasmic reticulum-associated secretory proteins Sec20p, Sec39p, and Dsl1p are involved in peroxisome biogenesis. Eukaryot Cell. 2009;8:830–843. doi: 10.1128/EC.00024-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kragler F, Lametschwandtner G, Christmann J, Hartig A, Harada JJ. Identification and analysis of the plant peroxisomal targeting signal 1 receptor NtPEX5. Proc Natl Acad Sci USA. 1998;95:13336–13341. doi: 10.1073/pnas.95.22.13336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mumberg D, Müller R, Funk M. Regulatable promoters of Saccharomyces cerevisiae: Comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 1994;22:5767–5768. doi: 10.1093/nar/22.25.5767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang CW, Hamamoto S, Orci L, Schekman R. Exomer: A coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J Cell Biol. 2006;174:973–983. doi: 10.1083/jcb.200605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang F, et al. Development of antibody-based assays for omega-conotoxin MVIIA. J Biochem Biophys Methods. 2006;67:49–56. doi: 10.1016/j.jbbm.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Saito Y, Kimura K, Oka T, Nakano A. Activities of mutant Sar1 proteins in guanine nucleotide binding, GTP hydrolysis, and cell-free transport from the endoplasmic reticulum to the Golgi apparatus. J Biochem. 1998;124:816–823. doi: 10.1093/oxfordjournals.jbchem.a022185. [DOI] [PubMed] [Google Scholar]
- 35.Baker D, Hicke L, Rexach M, Schleyer M, Schekman R. Reconstitution of SEC gene product-dependent intercompartmental protein transport. Cell. 1988;54:335–344. doi: 10.1016/0092-8674(88)90196-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.