Abstract
At least two components that modulate plant resistance against the fungal powdery mildew disease are ancient and have been conserved since the time of the monocot–dicot split (≈200 Mya). These components are the seven transmembrane domain containing MLO/MLO2 protein and the syntaxin ROR2/PEN1, which act antagonistically and have been identified in the monocot barley (Hordeum vulgare) and the dicot Arabidopsis thaliana, respectively. Additionally, syntaxin-interacting N-ethylmaleimide sensitive factor adaptor protein receptor proteins (VAMP721/722 and SNAP33/34) as well as a myrosinase (PEN2) and an ABC transporter (PEN3) contribute to antifungal resistance in both barley and/or Arabidopsis. Here, we show that these genetically defined defense components share a similar set of coexpressed genes in the two plant species, comprising a statistically significant overrepresentation of gene products involved in regulation of transcription, posttranslational modification, and signaling. Most of the coexpressed Arabidopsis genes possess a common cis-regulatory element that may dictate their coordinated expression. We exploited gene coexpression to uncover numerous components in Arabidopsis involved in antifungal defense. Together, our data provide evidence for an evolutionarily conserved regulon composed of core components and clade/species-specific innovations that functions as a module in plant innate immunity.
Keywords: (co-)regulon, glucosinolate metabolism, pathogen entry, plant defense, evolution
In nature, plants are constantly attacked by a large number of potential pathogens to most of which they exhibit robust immunity known as “nonhost” resistance (1). Preinvasive defenses at the cell periphery are part of the plant nonhost resistance machinery and represent the first layer of protection to nonadapted fungal pathogens. In the dicotyledonous model plant Arabidopsis thaliana, a number of molecular components of this defense system have been identified and their functions elucidated. One such component, the syntaxin PEN1 (At3g11820), has been shown to restrict cellular entry of the nonadapted biotrophic powdery mildew fungi Blumeria graminis f. sp. hordei (Bgh) and Erysiphe pisi, suggesting a role for N-ethylmaleimide sensitive factor adaptor protein receptor protein (SNARE)-mediated exocytosis and/or vesicle fusion events in extracellular defenses (2, 3). Biochemical studies indicated that PEN1 interacts in planta with the vesicle-associated membrane protein (VAMP) 721 (At1g04750) and VAMP722 (At2g33120), and genetic analyses revealed that these two VAMPs play a role in limiting entry of nonadapted powdery mildews (3). VAMP722 was found to preferentially form ternary SNARE complexes with PEN1 and the SNAP33 (At5g61210) SNARE protein. SNAP33 expression is pathogen-inducible, and its encoded protein accumulates beneath incipient fungal entry sites (3), suggesting that it plays a role in pathogen defense. Homozygous snap33 individuals, like vamp721/722 double mutants, are lethal (3), indicating that the secretory pathway involving these gene products is also required for viability of plant cells. Barley (Hordeum vulgare) orthologs of these proteins also interact in planta (3), suggesting that this secretory SNARE complex is ancient and may have a conserved function in both monocots and dicots.
A second genetically defined pathway contributing to extracellular defenses against nonadapted powdery mildew fungi in Arabidopsis is comprised of the atypical myrosinase PEN2 (At2g44490; refs. 4 and 5) and the ATP Binding Cassette (ABC) transporter PEN3 (At1g59870; ref. 6). It is postulated that PEN2 and PEN3 are involved in the synthesis and efflux across the plasma membrane, respectively, of toxic secondary metabolites derived from indole glucosinolates to limit pathogen invasion (4–6). In contrast to PEN1, PEN2 and PEN3 are involved in restricting the growth of a broader spectrum of pathogens, including the adapted biotrophic powdery mildews Golovinomyces orontii and G. cichoracearum, the necrotrophic fungus Plectosphaerella cucumerina, and the hemibiotrophic oomycete Phytophthora infestans. Interestingly, VAMP721/722 are also important for defense against the biotrophic oomycete Hyaloperonospora arabidopsidis (3), suggesting that SNARE-dependent secretory pathways are likewise involved in defense against a wider range of pathogens.
In Arabidopsis and barley, a monocot plant whose line separated from the dicot lineage ca. 200 Mya, loss of function mutations in seven transmembrane domain containing MLO proteins provide durable preinvasive resistance to all known isolates of powdery mildews that are normally able to colonize the two species (7, 8). In both species, this type of immunity partially depends on the respective syntaxins, PEN1 and its barley ortholog ROR2 (2, 7), suggesting that barley MLO and Arabidopsis MLO2 (At1g11310) control SNARE-mediated vesicle-associated defenses at the cell periphery. This hypothesis is further corroborated by the fact that mlo-mediated resistance in barley also requires the SNAP34 SNARE protein (9). Additionally, in Arabidopsis, mlo2-mediated resistance was shown to depend on PEN2 and PEN3 (7), indicating that MLO proteins potentially modulate a number of concurrent antifungal defenses at the cell wall.
Here, we exploited in silico analysis to identify genes that are coexpressed with these genetically defined defense components in Arabidopsis and barley. This approach revealed a similar set of coexpressed genes in the two plant species. We then took advantage of reverse genetic analysis in Arabidopsis to test the hypothesis that some of these genes might also contribute to plant immunity, which revealed several previously undescribed players in antifungal defense.
Results
Known Components of Powdery Mildew Penetration Defenses Are Coexpressed in Arabidopsis and Barley.
To determine whether genes encoding known components of powdery mildew resistance were coexpressed in Arabidopsis, linear regression analysis was carried out on publicly available microarray data representing a wide range of conditions and developmental periods. Most of the genes tested were found to be moderately to highly coexpressed, with few exceptions (Table 1). Notably, VAMP721 was not found to be highly coexpressed with any of the other genes, consistent with results indicating a comparatively minor role in defense against nonadapted powdery mildews (3). Additionally, PEN2 was not as highly coexpressed with PEN1, SNAP33, and VAMP722 as it was with MLO2 and PEN3, consistent with results indicating that PEN1 (together with its interacting SNARE partners) and PEN2 are engaged in genetically separable pathways for defense against nonadapted pathogens (5). To determine whether these genes were also coexpressed in a monocot plant, correlation analysis was carried out on barley microarray data by using the respective barley orthologs MLO, ROR2, SNAP34, and HvVAMP721. This provided similar results to Arabidopsis, such that these genes were highly coexpressed, whereas HvVAMP721, encoding the presumptive ortholog of Arabidopsis VAMP721/722 (3), was not highly coexpressed with any of the other genes (Table 1). This finding is not unexpected, because it is likely that Arabidopsis VAMP721/722 have arisen from a recent gene duplication event and HvVAMP721 may be fulfilling the functions of both Arabidopsis VAMP genes and, hence, may not be as highly coexpressed with the other barley orthologs.
Table 1.
Known components of penetration defenses are coexpressed in Arabidopsis and barley
| Gene | MLO2 | PEN1 | SNAP33 | VAMP721 | VAMP722 | PEN2 | PEN3 | MLO | ROR2 | SNAP34 | HvVAMP721 | HvPDR8 |
| MLO2 | 1 | 0.603 | 0.677 | 0.367 | 0.580 | 0.748 | 0.780 | — | — | — | — | — |
| PEN1 | — | 1 | 0.835 | 0.445 | 0.648 | 0.545 | 0.703 | — | — | — | — | — |
| SNAP33 | — | — | 1 | 0.535 | 0.764 | 0.597 | 0.741 | — | — | — | — | — |
| VAMP721 | — | — | — | 1 | 0.466 | 0.217 | 0.352 | — | — | — | — | — |
| VAMP722 | — | — | — | — | 1 | 0.595 | 0.627 | — | — | — | — | — |
| PEN2 | — | — | — | — | — | 1 | 0.787 | — | — | — | — | — |
| PEN3 | — | — | — | — | — | — | 1 | — | — | — | — | — |
| MLO | — | — | — | — | — | — | — | 1 | 0.843 | 0.916 | 0.206 | 0.881 |
| ROR2 | — | — | — | — | — | — | — | — | 1 | 0.920 | 0.258 | 0.770 |
| SNAP34 | — | — | — | — | — | — | — | — | — | 1 | 0.299 | 0.862 |
| HvVAMP721 | — | — | — | — | — | — | — | — | — | — | 1 | 0.382 |
| HvPDR8 | — | — | — | — | — | — | — | — | — | — | — | 1 |
Distinct Arabidopsis Defense Pathways Against Nonadapted Pathogens Share Overlapping Components.
To identify further components of plant defenses against fungal pathogens, we investigated genes whose expression patterns were tightly correlated to each of the above-mentioned genes. Two hundred eighty-two genes were found to be coexpressed with the three known defense genes PEN1, SNAP33, and VAMP722 (Dataset S1), whereas 164 genes were found to be coexpressed with PEN2 and PEN3 (Dataset S1). Despite genetic evidence indicating that PEN1 and PEN2 act in separate pathways (5), there was a substantial overlap between the genes coexpressed with the PEN1/SNAP33/VAMP722- and PEN2/PEN3-associated subsets of genes (60 genes; Dataset S1). This result suggests that the two pathways may use a common set of genes in the execution of defense responses (and possibly other processes), similar to the overlap of genes that were found to be transcriptionally responsive after perception of distinct bacterial pathogen-associated molecular patterns (PAMPs) mediated by the pathogen recognition receptors EFR and FLS2 (10). In both datasets we noted a statistically significant overrepresentation of gene products involved in regulation of transcription, posttranslational modification, and signaling (Dataset S2). Protein classes related to these processes comprise receptor-like kinases, polypeptides involved in calcium signaling (e.g., calcium/calmodulin-dependent protein kinases) and transcriptional regulators.
A Common Set of Coexpressed Genes Is Identified in Monocot and Dicot Plants.
To identify defense components potentially conserved between Arabidopsis and barley, we next investigated genes that were coexpressed with MLO2, PEN1, and SNAP33 on the one hand and MLO, ROR2, and SNAP34 on the other. We chose to further study these orthologous gene sets because they represent genetically defined factors in plant defense that are highly coexpressed in both Arabidopsis and barley. A total of 107 unique genes were identified that were coexpressed with the three Arabidopsis components (Table 2 and Dataset S1). Approximately 70% of these genes were common to those found coexpressed with PEN1, SNAP33, and VAMP722 (Dataset S1) and also showed a similar overrepresentation of functional categories of the respective gene products (Dataset S2).
Table 2.
Homologous genes are coexpressed with known defense components in Arabidopsis and barley
| Species | Coexpressed genes* | Coexpressed genes with a homolog in the reciprocal species† (%) | Common coexpressed genes‡ (%) | Best hit,§ (%) |
| Arabidopsis | 107 | 85 (79) | 47 (55) | 22 (47) |
| Barley | 356 | 272 (76) | 94 (35) | 22 (23) |
*Number of genes coexpressed with known defense genes in each species (see text) at r ≥ 0.60.
†Number of coexpressed genes (percentage of total) showing homology to genes in the reciprocal species at an e value ≤ 1 x 10−20 based on BLASTX (barley versus Arabidopsis) and TBLASTN (Arabidopsis versus barley) analysis.
‡Number of coexpressed genes (percentage of those with homologs) common to both Arabidopsis and barley at an e value ≤ 1 × 10−20; please note that the difference in numbers between Arabidopsis and barley results from the fact that the Arabidopsis genome is fully sequenced (unambiguous gene models), whereas in several cases multiple barley unigenes (assembled EST contigs) represented on the Barley1 Affymetrix gene chip relate to a single Arabidopsis gene. Additional factors such as the presence of co-orthologs in one of the species also contribute to the differences.
§Number (and percentage) of common coexpressed genes that represent the best hit in the reciprocal BLAST analysis.
In silico analysis indicated that most of the genes show elevated transcript levels in response to biotic stresses, as well as fungal (chitin) and bacterial (elf18) MAMPs (Fig. S1A). Consistently, many of the genes had lower transcript abundance in an fls2 mutant background (Fig. S1B). Additionally, many were highly expressed in response to the translation inhibitor cycloheximide, suggesting that their transcription may be under negative control of short-lived regulatory polypeptides. Analysis of gene expression during plant development indicated that numerous coexpressed genes were strongly expressed during the reproductive stage and during senescence (Fig. S1C).
In barley, a total of 356 unique EST assemblies (unigenes; corresponding to 389 probe sets of the Affymetrix Barley1 gene chip; Dataset S3) were found to be coexpressed with MLO, ROR2, and SNAP34 (Table 2 and Dataset S4). Analysis of the protein categories encoded by the respective unigenes indicated an overrepresentation of proteins involved in biotic stress, signaling, regulation of transcription and hormone metabolism (Dataset S2). Reciprocal BLAST analysis of the coexpressed barley and Arabidopsis genes indicated that ≈55% (Arabidopsis versus barley) and 35% (barley versus Arabidopsis) of the coexpressed query genes showed homology to coexpressed genes in the respective other species (e value ≤ 1 × 10−20; Table 2 and Dataset S4), suggesting that the three genes are part of a conserved set of genes that are commonly expressed in both monocot and dicot plants. Interestingly, a unigene encoding a protein with high sequence relatedness to PEN3/PDR8 (e value = 0.0; designated HvPDR8) was coexpressed with MLO, ROR2, and SNAP34 (Table 1). Because PEN2 is a recent acquisition of Arabidopsis and glucosinolate biosynthesis is a clade-specific innovation limited to the Capparales (7), this result suggests that although the extrusion of toxic metabolites via a PEN3-like ABC transporter appears conserved in both Arabidopsis and barley, the structure of the extruded phytochemicals likely has been subject to evolutionary diversification. Of note, another ABC transporter has been recently identified in wheat that confers broad-spectrum resistance against rust and powdery mildew fungi (11).
A unique cis-Acting Element Mediates Expression in Older Leaf Tissue and Is Responsive to Wounding and Pathogens.
The 500-bp 5′ upstream regions of genes that were coexpressed with Arabidopsis MLO2, PEN1, and SNAP33 were analyzed to identify known cis-acting regulatory motifs. A number of elements implicated in abiotic stress and pathogen responsiveness were overrepresented in these genes (Dataset S5), consistent with a potential role in pathogen defense. Additionally, a considerable number of light-responsive motifs were also overrepresented, suggesting that the expression of these genes may be in part regulated by light or photoperiod. The 500-bp 5′ upstream regions of the same Arabidopsis genes were then analyzed by MEME and MAST (http://meme.nbcr.net) to search for novel cis-acting elements. A number of motifs were identified that were overrepresented in the gene list, one of which was identified by MEME in all of the genes, suggesting that this motif is a determinant of common transcriptional control. The best match of this cis-acting element identified by MAST (Fig. 1A) showed similarity to a previously described element (TL1) involved in induction of a set of secretion-related defense-associated genes whose expression depends on the transcriptional regulator NPR1 (12). Notably, none of the 13 genes used to identify the TL1 element was found in any of our lists of coexpressed genes, indicating that despite their sequence similarity the two cis-regulatory elements likely control distinct gene sets.
Fig. 1.
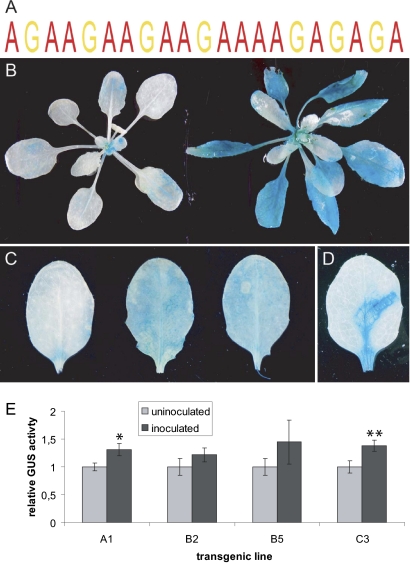
Gene expression governed by a unique cis element in Arabidopsis is light- and pathogen-responsive. Histochemical analysis of transgenic Arabidopsis plants expressing a promoter-GUS construct containing a tandem repeat of a unique cis element (see text). (A) Overrepresented cis element identified by MEME and MAST analysis. (B) Four-week-old T3 plants soil-grown under short (Left) and long (Right) days. (C) Unchallenged, G. orontii, and E. pisi-inoculated (left to right) rosette leaves of 4-wk-old short-day-grown T3 plants at 3 d after inoculation. (D) Wounded (forceps mark) rosette leaf of 4-wk-old short-day-grown T3 plant. Similar expression patterns were observed in four independent transgenic lines. (E) Results of quantitative GUS assay on multiple T2 plants (4–9 per line) of four different transgenic lines upon inoculation with Bgh (at 3 d after inoculation). Values of each line were normalized to the uninoculated control (set as 1); error bars represent the SEM. Asterisks indicate a significant difference from the uninoculated control (*P ≤ 0.05; **P ≤ 0.01; Student's t test). Similar results were obtained in two independent experiments.
Analysis of transgenic A. thaliana plants expressing the β-glucuronidase (GUS) reporter gene driven by a synthetic promoter containing four tandem repeats of this element indicated that the majority of GUS expression was in older rosette leaves, particularly under long day conditions (Fig. 1B), suggestive of a role for this cis element in regulating gene expression during senescence or light stress. Expression was also observed in the cotyledons, floral organs, and in the stele of the root and in the root cap (Fig. S2), which is consistent with the observed expression pattern of the coexpressed genes (Fig. S1C). Inoculation of these plants with both adapted and nonadapted powdery mildews indicated that gene expression governed by this element may also be responsive to fungal pathogens (Fig. 1 C and E). Additionally, GUS was strongly expressed in wounded leaf tissue (Fig. 1D). Interestingly, reporter gene expression did not respond to exogenous salicylic acid (SA) or methyl jasmonate application, suggesting that either the cognate transcription machinery acts upstream or independent of these known defense signaling metabolites.
Identification of Novel Genes Required for Defense Against Diverse Fungal Pathogens.
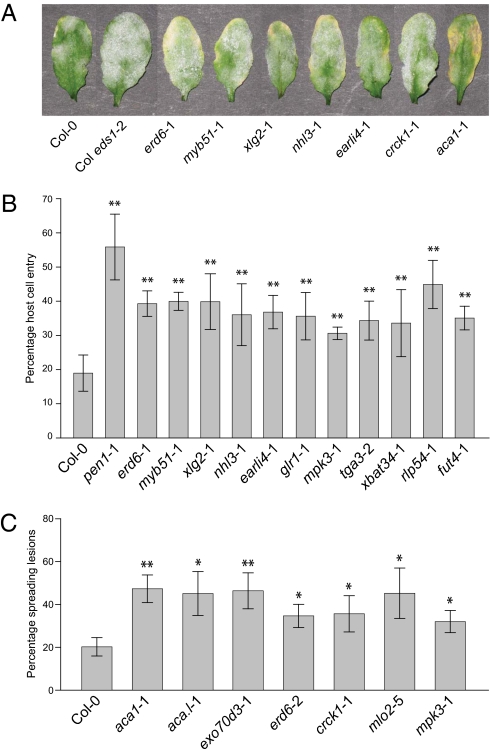
We selected homozygous Arabidopsis insertion/mutant lines for a number of genes that were commonly coexpressed with each of the query genes. The target genes for subsequent functional analysis were chosen, in part, based on a presumed function in pathogen defense and, in part, based on the availability of suitable knock-out lines at the stock centers. The 88 confirmed mutant lines, mostly predicted to represent null mutants, represent 58 Arabidopsis genes, of which 31 (53%) were also found in the list of barley coexpressed genes (Dataset S6). With the known exception of snap33, none of the mutant lines tested was lethal and only an insertion in CYP83B1 showed significant growth defects, as has been reported (13). Challenge of the mutant lines with the adapted powdery mildew pathogen, G. orontii revealed in most cases wild-type–like disease phenotypes (Dataset S6). However, a few lines exhibited enhanced disease symptoms similar to eds1 (Fig. 2A and Fig. S3C), whereas one (aca1-1) exhibited chlorotic necrosis similar to that observed in pen3 mutants in response to infection by G. cichoracearum (6). Infection-induced chlorosis in combination with enhanced fungal sporulation was observed in case of erd6-1 and myb51-1 (Fig. 2A). Phenotypes of most mutants were confirmed by a second independent mutant allele (Fig. S3C and Dataset S6).
Fig. 2.
Mutants with insertions in unique coexpressed genes exhibit enhanced disease symptoms to a range of pathogens with diverse lifestyles. (A) Representative macroscopic phenotypes of Col-0 and selected insertion lines 10 d after inoculation with G. orontii. (B) Quantitative analysis of host cell entry of E. pisi on selected lines determined at 3 d after inoculation. Results represent mean ± SD of at least five leaves per genotype. (C) Quantification of disease symptoms 6 days after inoculation with B. cinerea (Materials and Methods). Results represent mean ± SD of three independent samples per genotype. Comparable results were obtained in at least three independent experiments. For all data, asterisks indicate a significant difference from Col-0 (*P ≤ 0.05; **P ≤ 0.01; Student's t test).
The mutant lines from the coexpressed genes were then assayed for their pre- and postinvasive defense against the nonadapted powdery mildews Bgh and E. pisi. Consistent with previous reports (5, 14, 15), eds1, npr1, and pad4 plants, all known to be defective in SA-dependent defense responses, showed significant increases in pre- (Fig. S3A) as well as postpenetration growth of E. pisi compared with wild type (Fig. S3B). Although not in the list of coexpressed genes, we added a mutant defective in the isochorismate synthase SID2, responsible for the majority of pathogen-induced SA production, as a control in our experiments. sid2 plants exhibited wild-type–like symptoms, suggesting that EDS1, NPR1, and PAD4 act independently of SA accumulation in defense against nonadapted powdery mildews. A number of the insertion lines (in most cases supported by two independent alleles) in the newly identified genes exhibited enhanced penetration of E. pisi but not Bgh (Fig. 2B, Fig. S3D, and Dataset S6). None of the insertion lines assayed exhibited significant secondary hyphal growth compared with the wild type, suggesting that these genes have a specific role in preinvasion defense.
Both Arabidopsis mlo2 and barley mlo plants exhibit enhanced disease symptoms toward necrotrophic and hemibiotrophic pathogens (7, 16, 17), and a number of the coexpressed genes also have a known role in defense against these classes of pathogens (Dataset S6). Therefore, we tested a subset of the insertion lines for their responses to the necrotrophic fungi Alternaria brassicicola and Botrytis cinerea. A number of the lines showed enhanced disease symptoms in response to B. cinerea, but none of the lines showed consistently altered responses to A. brassicicola on the basis of independent mutant alleles (Fig. 2C, Fig. S3E, and Dataset S6).
Unique Components of the PEN2-Mediated Defense Pathway.
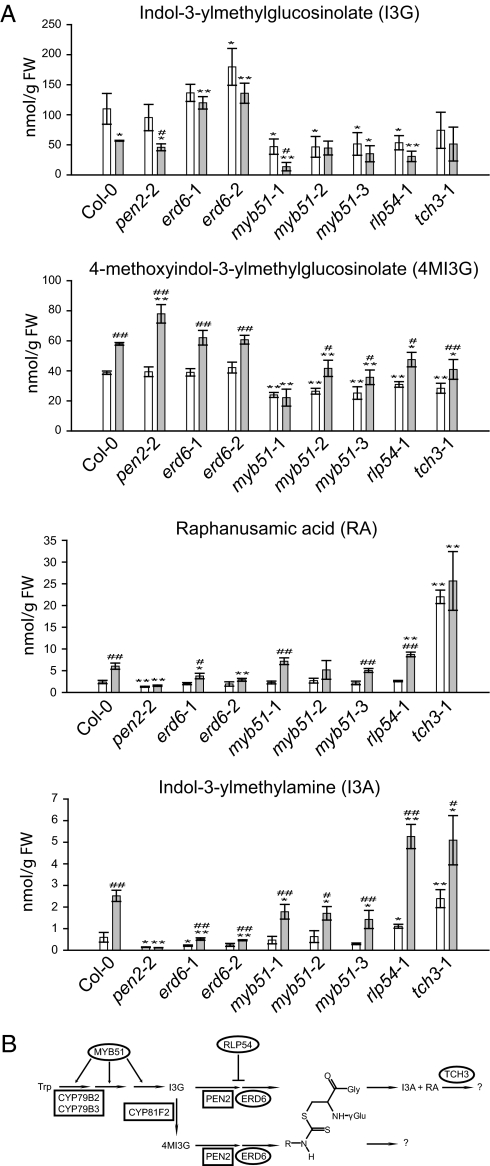
We next investigated a number of insertion lines for the accumulation of secondary metabolites, including indole glucosinolates and their derivatives that play a role in PEN2/PEN3-mediated defense (4, 18). Several cytochrome P450 enzyme-encoding genes were coexpressed with PEN2 and PEN3 (Dataset S1), including CYP83B1 (SUR2), which is required for indole glucosinolate biosynthesis (13). Other genes include MPK3 and MKK9, which were recently shown to regulate biosynthesis of the indolic phytoalexin camalexin (19, 20). Consistent with a role for the R2R3-MYB transcription factor MYB51 in regulating indole glucosinolate biosynthetic genes (21), the respective insertion lines exhibited enhanced fungal entry in conjunction with globally reduced levels of all metabolites tested (Figs. 2 and 3A and Fig. S3). These findings suggest that MYB51 is involved in regulating genes critical for glucosinolate metabolism that also impact antifungal defense (Fig. 3B). Reminiscent of pen2 mutants (4), insertion lines in the putative sucrose transporter ERD6 exhibited reduced levels of the indole glucosinolate hydrolysis products indol-3-ylmethylamine (I3A) and raphanusamic acid (RA; Fig. 3A). However, unlike in pen2 or wild-type plants, the pathogen-induced indol-3-ylmethylglucosinolate (I3G), but not 4-methoxyindol-3-ylmethyglucosinolate (4MI3G) level, is specifically elevated in erd6 plants, suggesting a function of ERD6 in the transport of the PEN2 substrate precursor I3G. An insertion in RLP54 (encoding a receptor-like kinase) exhibited increased I3A levels coupled with reduced I3G, suggesting that its gene product may be involved in negatively regulating a proposed branch pathway for pathogen-inducible I3G turnover (Fig. 3B). Additionally, an insertion in TCH3 (encoding a calmodulin-like protein) exhibited constitutively increased levels of the presumed glucosinolate hydrolysis products I3A and RA (Fig. 3A), suggesting that this gene may be involved in regulating the further turnover of these compounds independent of plant defense (Fig. 3B).
Fig. 3.
Mutants with insertions in coexpressed genes exhibit perturbed levels of glucosinolates and glucosinolate metabolites. (A) Accumulation of indicated selected secondary metabolites (nmol/g of fresh tissue weight) in indicated Arabidopsis genotypes without (open bars) or 16 h after inoculation with E. pisi conidiospores (filled bars). Results represent mean ± SD of three independent plants per genotype. Asterisks indicate a significant difference from respective Col-0 sample (*P ≤ 0.05; **P ≤ 0.01), and hash marks indicate a significant difference between inoculated and uninoculated plants of particular genotypes (#P ≤ 0.05; ##P ≤ 0.01; Student's t test). Comparable results were obtained in at least three independent experiments. (B) Schematic diagram of indole glucosinolate biosynthesis, showing locations of genes previously identified and unique genes identified in this study. Biosynthetic enzymes are highlighted by boxes; proteins with other activities by ovals. Diagram adapted from ref. 4.
Discussion
We have shown here on the basis of gene expression meta analysis that a common set of genes is coexpressed with genetically defined core components of antifungal defense in the monocot barley and the dicot Arabidopsis (Table 2). An explicit implication of our data is that MLO proteins play a direct role in regulating plant defense rather than being only indirectly involved in fungal pathogenesis. The identification of a cis-element common to the 5′ upstream regions of many of the coexpressed genes suggests that their transcriptional regulation is under coordinated control in Arabidopsis, which is the hallmark of genetic regulons. We noted a statistically significant overrepresentation of genes encoding receptor-like kinases, components involved in calcium signaling and transcriptional regulators in the coexpressed Arabidopsis genes (Datasets S1 and S2). Together with the knowledge that genes encoding proteins involved in secondary metabolite biosynthesis (PEN2, cytochrome P450s) and exocytosis/extrusion (SNAREs, exocyst subunits, ABC transporters) are present in the gene list, we hypothesize that the regulon encodes a functional unit comprising all components necessary for one subtype of plant defense, including membrane-resident pattern recognition receptors, signaling proteins, transcriptional regulators, and defense execution factors devoted to the biosynthesis and delivery of antimicrobial cargo. Our findings suggest that the plant defense machinery is possibly substructured in individual modules that might be selectively activated upon particular stress cues (e.g., fungal versus bacterial infection). This hypothesis is consistent with the fact that genes known to be exclusively crucial for defense against bacterial intruders, e.g., those encoding the pattern recognition receptor FLS2, its coreceptor BAK1 (22), and the SNARE protein SYP132 (23), are not part of the regulon described here. However, the proposed modules appear to be interconnected because many of the genes of the present regulon show elevated transcript levels also in response to bacterial MAMPs (Fig. S1A) and had lower transcript abundance in an fls2 mutant background (Fig. S1B). Furthermore, at least some components of the proposed defense modules (BAK1, CYP83B1, MLO2, SNAP33, VAMP721/722) have additional functions during plant development (3, 7, 8, 13), indicating shared protein functions in different biological processes.
Results of reciprocal BLAST searches suggest that at least a subset of the homologous coexpressed gene pairs in barley and Arabidopsis represent orthologs (Dataset S4). These findings support the assumption that the core group of genes identified in this study (≈50 genes that are shared between barley and Arabidopsis; Table 2) comprises an ancient regulon that is evolutionarily conserved in monocot and dicot plants. In addition, a number of innovations have shaped the regulons in a species- or clade-specific manner. Intriguingly, these species/clade-specific innovations (such as PEN2, encoding an atypical myrosinase restricted to the Capparales; ref. 7), must have been integrated in the transcriptional core network within short evolutionary time frames. It remains a future challenge to disentangle the transcriptional regulators that control expression of the regulon members.
We have demonstrated that exploiting gene coexpression is a powerful data mining tool with predictive power for identifying further components of a given physiological process and a metabolic pathway. It is unlikely that many of the genes found here as required for preinvasive resistance to nonadapted powdery mildews would have been identified in a forward genetic screen because the respective mutant lines exhibited relatively subtle phenotypes. Coexpression might therefore be a very sensitive and unbiased approach for the identification of novel gene function(s). In Escherichia coli, yeast, and animals, coexpression analysis is an established tool to predict functional networks (24, 25), and sophisticated software tools have been especially developed for this purpose (26). Although this approach has also been suggested as a tool for the identification of linked gene functions in plants (27), few studies have addressed this method experimentally in this taxonomic group of organisms (28–31). In the interim, several Web-based online resources based on microarray data have become available to study gene coexpression networks in Arabidopsis and other plant species (http://www.cressexpress.org, http://atted.jp, http://genecat.mpg.de, and http://bar.utoronto.ca). These tools pave the way for a widespread employment of coexpression analysis for the discovery of novel plant gene function(s).
Methods
Bioinformatic Analyses.
To investigate coexpression, correlation/regression analysis was carried out by using publicly available microarray data sources. For the Arabidopsis data analysis, version 2.0 of the online tool CressExpress (http://www.cressexpress.org) (31) was used with a Kolmogorov–Smirnov quality-control statistic (D) value of 0.15 or greater to filter for potential outlier chips as described (32). For each of the query genes tested, the P values observed for those genes classified as coexpressed with an r value cutoff of 0.6 were ≤1 × 10−40. Arabidopsis query genes were represented by the following Affymetrix ATH1 probe ID numbers: 262455_at (MLO2), 258786_at (PEN1), 247571_at (SNAP33), 264595_at (VAMP721), 245167_s_at (VAMP722), 267392_at (PEN2), and 262899_at (PEN3). For barley, microarray data were extracted from the Plant Expression Database (http://www.plexdb.org), and correlation analysis was carried out on RMA-normalized data by using the statistical package R. A total of 300 randomly chosen hybridizations were analyzed. Barley genes were represented by the following contigs on the Barley1 microarray: Contig6351_at (MLO; u35_16561 in assembly 35 of the HarvEST database; http://harvest.ucr.edu/), Contig22370_at (ROR2; u35_5407), Contig12026_at (SNAP34; u35_4293), Contig4900_at (HvVAMP721; u35_1946) and Contig8122_at (HvPDR8; u35_17048). Genes were sorted according to their r value (Pearson's correlation coefficient) and a cutoff of r ≥ 0.6 (empirically determined P < 0.05) was used to infer correlated gene expression to the respective query genes (via calculating the p value as the probability of identifying an r value greater than the observed r value, i.e., P = p(r) ≥ robserved). Details of the Arabidopsis and barley microarray datasets used for coexpression analyses are shown in Dataset S7. Gene sets were manually collated by using the Microsoft Excel software and curated by using The Bio-Array Resource for Arabidopsis Functional Genomics (http://bar.utoronto.ca). The reciprocal BLAST analysis was performed on the basis of HarvEST barley EST unigenes (assembly 35), identified by BLASTN analysis using the Barley1 contigs as a query (Dataset S3), and the TAIR10 Arabidopsis genome release. Analysis of overrepresented gene processes was carried out by using the online MapMan tool (http://mapman.gabipd.org/web/guest/home). Genevestigator (https://www.genevestigator.com/gv/index.jsp) was used for metaanalysis of gene expression.
Plant Material and Genetic/Biochemical Analyses.
To analyze the expression of the previously undescribed cis-acting element, a construct containing GUS under the control of four tandem repeats of the 21-mer identified (Fig. 1A) and a minimal 35S promoter was generated as described (33). For analysis in A. thaliana, the entire fragment was ligated into the binary vector pCAMBIA3300 and this construct was introduced into wild-type plants (Col-0) via Agrobacterium tumefaciens-mediated transformation. Histochemical and quantitative β-glucoronidase reporter assays of either T2 or T3 transgenic lines were carried out as described (33).
T-DNA insertional mutant lines were obtained from the Nottingham Arabidopsis Stock Center (NASC) from the John Innes Center SM line, GABI-Kat, SALK, and Syngenta collections. Plants were grown at 20–23 °C in growth chambers under ≈150 μmol m−2·s−1 light intensity. For all pathogen and GUS assays, plants were grown under a 12-h light cycle, unless otherwise indicated.
Inoculations with G. orontii (anonymous isolate), Bgh (K1), and E. pisi (Birmingham isolate) were carried out on 3- to 4-wk old plants as described (5). Droplet inoculation with B. cinerea (strain iMi 169558) was carried out using 1 × 106 spores per mL, and disease severity was quantified as the percentage of leaves exhibiting spreading lesions. Infection assays with A. brassicicola (isolate MUCL 20297) were carried out as described (7). For biochemical analyses, A. thaliana wild-type, and T-DNA insertional mutant lines were analyzed by HPLC with or without inoculation with E. pisi as described (4).
Supplementary Material
Acknowledgments
We thank Staffan Persson for valuable advice on coexpression and gene networks, Volker Lipka for discussions, and Renier van der Hoorn for his assistance with B. cinerea infections. M.H. was partially supported by a postdoctoral fellowship from the Alexander von Humboldt Foundation. Work in the R.P. laboratory is funded by Deutsche Forschungsgemeinschaft Grants SFB670 and PA861/6-1 and the Max Planck Society.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1003619107/-/DCSupplemental.
References
- 1.Heath MC. Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol. 2000;3:315–319. doi: 10.1016/s1369-5266(00)00087-x. [DOI] [PubMed] [Google Scholar]
- 2.Collins NC, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425:973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 3.Kwon C, et al. Co-option of a default secretory pathway for plant immune responses. Nature. 2008;451:835–840. doi: 10.1038/nature06545. [DOI] [PubMed] [Google Scholar]
- 4.Bednarek P, et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323:101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- 5.Lipka V, et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310:1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 6.Stein M, et al. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18:731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Consonni C, et al. Conserved requirement for a plant host cell protein in powdery mildew pathogenesis. Nat Genet. 2006;38:716–720. doi: 10.1038/ng1806. [DOI] [PubMed] [Google Scholar]
- 8.Büschges R, et al. The barley Mlo gene: A novel control element of plant pathogen resistance. Cell. 1997;88:695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 9.Douchkov D, Nowara D, Zierold U, Schweizer P. A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Mol Plant Microbe Interact. 2005;18:755–761. doi: 10.1094/MPMI-18-0755. [DOI] [PubMed] [Google Scholar]
- 10.Zipfel C, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- 11.Krattinger SG, et al. A putative ABC transporter confers durable resistance to multiple fungal pathogens in wheat. Science. 2009;323:1360–1363. doi: 10.1126/science.1166453. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Weaver ND, Kesarwani M, Dong X. Induction of protein secretory pathway is required for systemic acquired resistance. Science. 2005;308:1036–1040. doi: 10.1126/science.1108791. [DOI] [PubMed] [Google Scholar]
- 13.Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R. CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell. 2001;13:101–111. doi: 10.1105/tpc.13.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yun BW, et al. Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non-host resistance in Arabidopsis against wheat powdery mildew. Plant J. 2003;34:768–777. doi: 10.1046/j.1365-313x.2003.01773.x. [DOI] [PubMed] [Google Scholar]
- 15.Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Somerville S. Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J. 2004;40:633–646. doi: 10.1111/j.1365-313X.2004.02236.x. [DOI] [PubMed] [Google Scholar]
- 16.Jarosch B, Kogel KH, Schaffrath U. The ambivalence of the barley Mlo locus: Mutations conferring resistance against powdery mildew (Blumeria graminis f. sp. hordei) enhance susceptibility to the rice blast fungus Magnaporthe grisea. Mol Plant Microbe Interact. 1999;12:508–514. [Google Scholar]
- 17.Kumar J, Hückelhoven R, Beckhove U, Nagarajan S, Kogel KH. A compromised Mlo pathway affects the response of barley to the necrotrophic fungus Bipolaris sorokiniana (teleomorph: Cochliobolus sativus) and its toxins. Phytopathology. 2001;91:127–133. doi: 10.1094/PHYTO.2001.91.2.127. [DOI] [PubMed] [Google Scholar]
- 18.Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323:95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren D, et al. A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc Natl Acad Sci USA. 2008;105:5638–5643. doi: 10.1073/pnas.0711301105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu J, et al. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J Biol Chem. 2008;283:26996–27006. doi: 10.1074/jbc.M801392200. [DOI] [PubMed] [Google Scholar]
- 21.Gigolashvili T, et al. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- 22.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 23.Kalde M, Nühse TS, Findlay K, Peck SC. The syntaxin SYP132 contributes to plant resistance against bacteria and secretion of pathogenesis-related protein 1. Proc Natl Acad Sci USA. 2007;104:11850–11855. doi: 10.1073/pnas.0701083104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bergmann S, Ihmels J, Barkai N. Similarities and differences in genome-wide expression data of six organisms. PLoS Biol. 2004;2:E9. doi: 10.1371/journal.pbio.0020009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stuart JM, Segal E, Koller D, Kim SK. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. doi: 10.1126/science.1087447. [DOI] [PubMed] [Google Scholar]
- 26.Luo F, et al. Modular organization of protein interaction networks. Bioinformatics. 2007;23:207–214. doi: 10.1093/bioinformatics/btl562. [DOI] [PubMed] [Google Scholar]
- 27.Aoki K, Ogata Y, Shibata D. Approaches for extracting practical information from gene co-expression networks in plant biology. Plant Cell Physiol. 2007;48:381–390. doi: 10.1093/pcp/pcm013. [DOI] [PubMed] [Google Scholar]
- 28.Hirai MY, et al. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc Natl Acad Sci USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Persson S, Wei HR, Milne J, Page GP, Somerville CR. Identification of genes required for cellulose synthesis by regression analysis of public microarray data sets. Proc Natl Acad Sci USA. 2005;102:8633–8638. doi: 10.1073/pnas.0503392102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wei HR, et al. Transcriptional coordination of the metabolic network in Arabidopsis. Plant Physiol. 2006;142:762–774. doi: 10.1104/pp.106.080358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srinivasasainagendra V, Page GP, Mehta T, Coulibaly I, Loraine AE. CressExpress: A tool for large-scale mining of expression data from Arabidopsis. Plant Physiol. 2008;147:1004–1016. doi: 10.1104/pp.107.115535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ehlting J, Provart NJ, Werck-Reichhart D. Functional annotation of the Arabidopsis P450 superfamily based on large-scale co-expression analysis. Biochem Soc Trans. 2006;34:1192–1198. doi: 10.1042/BST0341192. [DOI] [PubMed] [Google Scholar]
- 33.Rushton PJ, Reinstädler A, Lipka V, Lippok B, Somssich IE. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell. 2002;14:749–762. doi: 10.1105/tpc.010412. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.