Abstract
There has been considerable interest in virulence genes in the plasticity region of Helicobacter pylori, but little is known about many of these genes. JHP940, one of the virulence factors encoded by the plasticity region of H. pylori strain J99, is a proinflammatory protein that induces tumor necrosis factor-alpha and interleukin-8 secretion as well as enhanced translocation of NF-κB in cultured macrophages. Here we have characterized the structure and function of JHP940 to provide the framework for better understanding its role in inflammation by H. pylori. Our work demonstrates that JHP940 is the first example of a eukaryotic-type Ser/Thr kinase from H. pylori. We show that JHP940 is catalytically active as a protein kinase and translocates into cultured human cells. Furthermore, the kinase activity is indispensable for indirectly up-regulating phosphorylation of NF-κB p65 at Ser276. Our results, taken together, contribute significantly to understanding the molecular basis of the role of JHP940 in inflammation and subsequent pathogenesis caused by H. pylori. We propose to rename the jhp940 gene as ctkA (cell translocating kinase A).
Helicobacter pylori infection is the main cause of chronic gastritis, gastric mucosal atrophy, peptic ulcer, and some forms of gastric cancer (1). H. pylori infection up-regulates secretion of various inflammatory cytokines, including IL-1, IL-6, IL-8, and TNF-α (2). It leads to a marked recruitment and infiltration of neutrophils into the gastric mucosa, contributing directly to the pronounced inflammatory response (3). H. pylori-induced cytokine expression is regulated primarily at the transcriptional level, and several transcription factors have been implicated in H. pylori-induced transcriptional activation in gastric epithelial cells (4). Among the strongest transcriptional regulators in H. pylori-induced cytokine expression is the NF-κB family of transcription factors (5).
NF-κB is a crucial regulator of many cellular processes, such as immune responses, inflammation, and apoptosis, and is implicated in cancer development (5). NF-κB complexes are composed of homo- and heterodimers of p50 (NF-κB1), p52 (NF-κB2), p65 (RelA), c-Rel, and RelB (5). One of the prototypical, most-studied NF-κB complexes is the p50-p65 dimer. Multiple protein kinases are involved in the optimal regulation of the p50-p65 complex (5–7). Phosphorylation of the p65 subunit plays a key role in determining both the strength and duration of the NF-κB-mediated transcriptional response (6). Ser276 of p65 is phosphorylated by protein kinase A (PKA) upon triggering of the NF-κB signaling pathway by lipopolysaccharide (8) or by the mitogen- and stress-activated kinase-1 (MSK-1) after TNF-α stimulation (9). It is located at the N-terminal Rel-homology domain, which is responsible for the DNA-binding and homo- or heterodimerization abilities of p65. Phosphorylation at Ser276 disrupts the interaction between C-terminal and N-terminal regions of p65, thus unmasking an interaction site with the transcriptional coactivator CREB-binding protein (CBP)/p300 and activates transcription of the p65 target genes such as TNF-α, IL-6, and IL-8 (6, 10).
Among putative virulence genes of H. pylori, there has been considerable interest in strain-specific genes found outside of the cag pathogenicity island, especially genes in the plasticity regions (11). Nearly half of the strain-specific genes of H. pylori are located in the plasticity regions in strains 26695 and J99 (11). Because little is known about many of the genes in the plasticity regions, further studies are necessary to elucidate the roles of genes in these regions in H. pylori-associated pathogenesis. jhp940 is one of the virulence genes located in the plasticity region of H. pylori J99 (12–14). An initial systematic study showed increased prevalence of the jhp940 gene in H. pylori from gastric cancer patients (12). An in vivo study showed that JHP940 is strongly expressed in response to the interaction of H. pylori with the gerbil gastric mucosa (15, 16). Furthermore, addition of the purified recombinant JHP940 into cultured macrophages resulted in induction of TNF-α and IL-8 secretion as well as increased translocation of NF-κB (14). The induction of proinflammatory cytokines by JHP940 points to its putative role in chronic gastric inflammation and, possibly, other outcomes of H. pylori infection including gastric cancer (16). Thus, the proinflammatory gene jhp940 might play an important role during the early phases of the infection, responding to contact with the gastric mucosa and eliciting the release of inflammatory mediators (16).
The molecular details on the function of JHP940 remain elusive due to the lack of its sequence similarity with other well-characterized proteins. Interestingly, a number of human gastrointestinal bacteria such as Desulfitobacterium hafniense, Subdoligranulum variabile, Bryantella formatexigens, and Blautia hydrogenotrophica possess proteins that are highly homologous to the N-terminal 300 residues of JHP940, suggesting that this class of proteins may play an important role in these microorganisms.
Here we have characterized the JHP940 protein both structurally and functionally to better understand the molecular mechanism of its proinflammatory action. We have determined the crystal structure of JHP940 in three forms: apo, ADP-bound, and nonhydrolyzable ATP analog-bound forms. JHP940 possesses a characteristic protein kinase fold with a conserved but distinct active site. We show that JHP940 is capable of autophosphorylating itself at a threonine residue near the N terminus, and it translocates into cultured human cells. We also show that the recombinant JHP940 leads to enhanced phosphorylation of the NF-κB p65 subunit at Ser276 in human epithelial cancer cells and demonstrate that the kinase activity of JHP940 is required for this enhanced phosphorylation. On the basis of these findings, we propose to rename the jhp940 gene as ctkA (cell translocating kinase A). Our structural and functional studies significantly contribute to our understanding of the proinflammatory mechanism caused by H. pylori infection.
Results
Overall Structure and Structural Similarity Search.
We have determined the crystal structure of C-terminally truncated CtkA (CtkAΔC; residues 1–300) by the multiwavelength anomalous diffraction method (Table S1). The apo structure has been refined to crystallographic Rwork and Rfree values of 19.4% and 23.0%, respectively, for 20–2.0 Å data without a sigma cutoff (Table S1). There are two copies of CtkA in an asymmetric unit. Two flexible loop regions (Lys14−Gly22 and Lys41−Tyr50 in chain A; Lys14−Lys24 and Pro40−Ser49 in chain B), as well as the C-terminal region containing the eight-residue affinity tag (LEHHHHHH) (13 residues in chain A; 12 residues in chain B) have no electron density and are missing from the model. The two chains in the asymmetric unit are highly similar to each other, with a rms deviation of 0.26 Å for 253 Cα atoms.
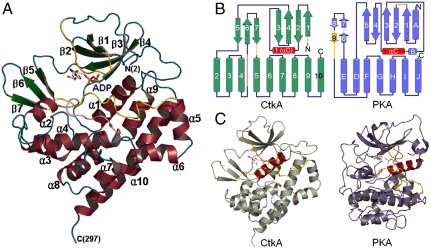
CtkAΔC contains 10 α-helices and 7 β-strands, which is folded into a characteristic protein kinase structure (Fig. 1A). It consists of two lobes of dissimilar sizes that are connected by a loop between β4 and β5. The N-terminal lobe (Pro2–Lys88) is formed by four antiparallel β-strands (β1–β4) and one prominent α-helix αC (α1 in our sequential numbering of α-helices in Fig. 1B; colored in red in Fig. 1 B and C). The αC helix is conserved among a wide range of protein kinases (17). The β-sheet in the N-terminal lobe has a strand order of 1-2-4-3, with the αC helix forming a crossover connection between β2 and β3 (Fig. 1B). The C-terminal lobe (Tyr95–Ala300) is a globular α-helical bundle with a three-stranded antiparallel β-sheet.
Fig. 1.
Overall structure of CtkA. (A) Ribbon diagram. α-Helices, β-strands, and loops are colored in wine, green, and cyan, respectively. The glycine-rich loop (Lys14–Gly22), catalytic loop (Gly152–Trp161), and activation loop (Ala175–Asp189) are colored in orange, pink, and yellow, respectively. Secondary structure elements were defined by PyMOL (http://www.pymol.org). All the structural figures were generated using PyMOL. (B) Comparison of topology diagrams of CtkA and PKA. α-Helices are shown as cylinders and β-strands as arrows. αC helix is colored in red for both proteins. (C) Overall structures of CtkA and PKA. Bound ADP molecules are shown in sticks, and αC helix is colored in red for both proteins. The glycine-rich loop (Lys14–Gly22), catalytic loop (Gly152–Trp161), and activation loop (Ala175–Asp189) of CtkA are colored in orange, pink, and yellow, respectively.
A DALI search (18) revealed that CtkA structurally resembles the catalytic domain of protein kinases, suggesting that it may function as a protein kinase. The C-terminal kinase domain of Escherichia coli multidrug tolerance Ser/Thr protein kinase HipA (19, 20) was most similar, with the highest Z score of 15.0–13.2 and a rms deviation of 3.1–3.3 Å for 202–210 Cα atom pairs despite the low sequence identity of 14% between CtkA (apo structure) and HipA.
Binding of ADP and an ATP Analog in the Active Site.
Because CtkA displayed an overall structural similarity to other protein kinases, we determined its structures in complex with either ADP or a nonhydrolyzable ATP analog (adenylyl-imidodiphosphate; AMP-PNP) to 2.5-Å and 2.9-Å resolutions, respectively (Table S1). The crystals of the ADP and AMP-PNP complexes are in the P1 space group with four chains in the asymmetric unit. The nucleotide is bound to all four chains in the asymmetric unit and is well defined by the electron density, except the terminal phosphate is disordered in one of the four AMP-PNP molecules. The rms deviations between the structures of apo, ADP-, and AMP-PNP-bound CtkA (for chain A) are less than 0.56 Å, indicating that binding of ADP or AMP-PNP induces no major structural changes (Fig. S1A). However, the nucleotide binding induces ordering in one of the two flexible loops that are disordered in the apo structure. That is, the glycine-rich loop (Lys14–Gly22) is disordered in the apo structure, whereas it becomes ordered upon binding ADP or AMP-PNP.
In the ADP- and AMP-PNP-bound structures, the ligand is deeply buried in the cleft between N- and C-terminal lobes, thus defining the potential kinase active site. The bound ligands interact with the activation loop (Ala175–Asp189, colored in yellow in Fig 1), in addition to the above-mentioned glycine-rich loop. The adenine ring of the bound ligands is surrounded by hydrophobic side chains of Phe17, Met35, Phe90, Leu97, and Phe178. The adenine ring nitrogen atoms form multiple-hydrogen bonds with Gln72, Lys88, and Phe90 (Fig. S1B). The phosphate moiety forms direct interactions with the side chains of highly conserved Lys24, Lys37, Asn160, and Asp179 (marked by blue triangles in Fig. S2), as well as the side chain and main chain nitrogen atoms of Asn21 (Fig. S1B). One or two Mg2+ ions are bound to the phosphate moiety in the AMP-PNP-bound structure, whereas one Mg2+ ion is bound to the phosphate moiety in the ADP-bound structure. The bound Mg2+ ion(s) are coordinated by the side chains of Asn160 and Asp179, and the phosphate moiety of the bound ligands (Fig. S1B). The corresponding Mg2+ ions in other protein kinases facilitate phosphotransfer by accelerating substrate association and product dissociation (17).
In all apo, ADP-bound and AMP-PNP-bound structures, the side chain of Lys37 forms a strong salt bridge with Asp179 and Glu57 (marked by a blue triangle in Fig. S2), another strictly conserved residue from the αC helix (Fig. S1C) (17). A similar mode of interaction is known to stabilize the active site structure required for the kinase activity in other protein kinases (Fig. S1C) (17). The catalytic loop of CtkA (Gly152−Trp161, colored in pink in Fig. 1 and Fig. S1C) contains the strictly conserved Asp155 (marked by a blue triangle in Fig. S2). Residues corresponding to Asp155 of CtkA are crucial in catalyzing the phosphorylation reaction by enhancing the nucleophilicity of the incoming Ser/Thr/Tyr residue of the target proteins (21). The main chain oxygen atom of Asp155 forms a hydrogen bond with the side chain of Asn160, stabilizing the conformation of the catalytic loop of CtkA (Fig. S1C).
CtkA Is Capable of Autophosphorylation as a Ser/Thr Protein Kinase.
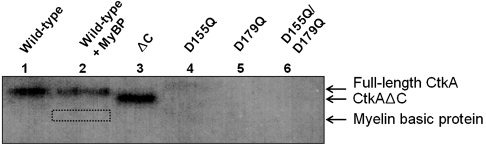
In order to test the protein kinase activity of CtkA, we performed in vitro protein kinase assays using [γ - 32P]ATP. When we employed myelin basic protein as a general Ser/Thr kinase substrate, no radio-labeled band of myelin basic protein was observed (33 kDa, indicated by a dotted box in Fig. 2, lane 2). However, we could detect a single radio-labeled band of approximately 38 kDa, which corresponds to the autophosphorylated, His-tagged, full-length CtkA (Fig. 2, lane 2). When the experiment was repeated without myelin basic protein, the same result was obtained (Fig. 2, lane 1). These results indicate that CtkA has an autophosphorylating activity. To identify the phosphorylation site(s), we compared the results of peptide mass analyses of trypsin-digested autophosphorylated CtkA. The tryptic fragment of autophosphorylated CtkA covering residues 2–14 (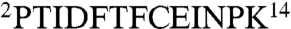 ) showed observed masses of 1,604.8 Da and 1,524.8 Da corresponding to the monophosphorylated and unphosphorylated peptides, respectively. The mass difference of 80 Da corresponds to phosphorylation at either Thr3 or Thr7, but not both. This tryptic peptide contains no tyrosine residue. Therefore, we conclude that CtkA belongs to a Ser/Thr kinase family. When the D155Q or D179Q single mutant or the D155Q/D179Q double mutant of CtkA was used in the assay, no radio-labeled 38-kDa band was present (Fig. 2, lanes 4–6). This result indicates that strictly conserved Asp155 and Asp179 are crucial for the protein kinase activity of CtkA. CtkAΔC retained the kinase activity (Fig. 2, lane 3), indicating that the C-terminal tail (His301–Arg325) of CtkA is not required for the kinase activity.
) showed observed masses of 1,604.8 Da and 1,524.8 Da corresponding to the monophosphorylated and unphosphorylated peptides, respectively. The mass difference of 80 Da corresponds to phosphorylation at either Thr3 or Thr7, but not both. This tryptic peptide contains no tyrosine residue. Therefore, we conclude that CtkA belongs to a Ser/Thr kinase family. When the D155Q or D179Q single mutant or the D155Q/D179Q double mutant of CtkA was used in the assay, no radio-labeled 38-kDa band was present (Fig. 2, lanes 4–6). This result indicates that strictly conserved Asp155 and Asp179 are crucial for the protein kinase activity of CtkA. CtkAΔC retained the kinase activity (Fig. 2, lane 3), indicating that the C-terminal tail (His301–Arg325) of CtkA is not required for the kinase activity.
Fig. 2.
In vitro protein kinase assay of wild-type CtkA and its mutant variants. Lane 1, full-length wild-type CtkA; lane 2, full-length wild-type CtkA plus myelin basic protein (MyBP); lanes 3–6, CtkAΔC, D155Q, D179Q, D155Q/D179Q mutants, respectively.
CtkA Activation Loop Exhibits an Active Conformation Without Phosphorylation.
The activation segment of eukaryotic protein kinases is composed of the Mg2+-binding loop, the activation loop, the P + 1 loop, and a β-strand connecting the Mg2+-binding loop and the activation loop (Fig. S3A, Right) (22). The activation segment sequences of eukaryotic protein kinases contain conserved features (Fig. S3B). The activation segment of most protein kinases undergoes large conformational changes when they switch between inactive and active states (17, 22). It is well established that activation of many eukaryotic-type protein kinases requires phosphorylation(s) at the activation loop for the substrate binding and a proper orientation of the conserved DFG motif of the Mg2+-binding loop for coordinating Mg2+ ions (22).
Despite the overall structural similarities to the eukaryotic protein kinases, the structure and amino acid sequence of the CtkA activation segment differ significantly from those of other eukaryotic protein kinases. The activation segment of CtkA (Ala175–Asp189, colored in yellow in Fig. S3A, Left) consists of a single loop lacking the β-strand between the Mg2+-binding loop and the activation loop. It shows a more extended conformation, lacking the P + 1 loop within the winding activation segments of other eukaryotic protein kinases (Fig. S3A, Right). The potential phosphorylation site (Ser182) in the activation segment of CtkA shows no sign of phosphorylation in the electron density, and it was not phosphorylated by autophosphorylation. Interestingly, however, the DxG motif of CtkA (boxed in orange in Figs. S2 and S3B, x stands for any amino acid) coordinates a Mg2+ ion in the active site, thus resembling the DFG motif of activated PKA (Fig. S4 A and B).
In the structure of activated PKA (23), the conformation of the activation loop is stabilized by multiple-hydrogen bonds between phosphorylated Thr197 and the side chains of neighboring His87, Arg165, and Lys189 (Fig. S4D). The APE motif (boxed in blue in Fig. S3B), which marks the end of the activation segment, is conserved among many eukaryotic protein kinases. In the PKA structure, the APE motif maintains the active conformation of the activation segment by forming a salt bridge between Glu208 of the APE motif and a conserved Arg280 (22). CtkA lacks such an APE motif. Instead, the activation loop of CtkA is stretched out by helix α5 (colored in yellow in Fig. 1A and Fig. S3A) and the interaction between the side chains of Tyr185 and Arg278 stabilizes the active conformation of this loop at its C-terminal end (Fig. S4C).
Many eukaryotic protein kinases possess the conserved RD motif in the catalytic loop (boxed in green in Fig. S3B). The conserved Arg residue of the RD motif makes a direct interaction with a main chain oxygen atom adjacent to the conserved DFG motif of the activation segment (Fig. S4B), stabilizing the orientation of the DFG motif for the proper phospho-transfer reaction (22). The corresponding sequence of the RD motif is inverted as DR (Asp155–Arg156) in the catalytic loop of CtkA and its close homologs in the human gastrointestinal tract bacteria (boxed in green in Figs. S2 and S3B). In the CtkA structure, Arg156 of the DR motif interacts with the main chain oxygen atoms of Phe178 and Asp179 and stabilizes the conformation of the DxG motif for Mg2+ binding (Fig. S4A).
CtkA Translocates into Human Cancer Cells.
It has been reported that treatment of cultured human cells with the purified recombinant CtkA resulted in induction of TNF-α and IL-8 secretion as well as increased translocation of NF-κB (14). Therefore, we examined using flow cytometry if recombinant CtkA translocates into HeLa cells. Treatment of HeLa cells with recombinant EGFP-fused full-length CtkA for 2 h significantly increased cellular fluorescence in a dose-dependent manner, whereas treatment with recombinant EGFP alone showed no sign of cellular fluorescence (Fig. S5). Treatment with recombinant EGFP-fused CtkAΔC gave a similar result with that of the EGFP-fused full-length CtkA, indicating that the C-terminal tail (residues 301–325) of CtkA is not required for translocation into HeLa cells.
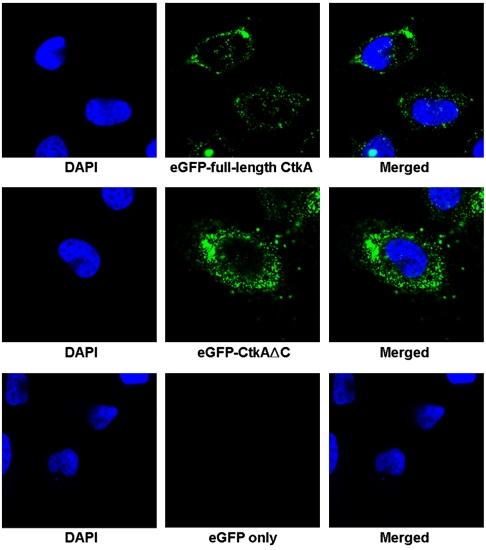
We have also confirmed the cell-translocating activity of CtkA using confocal microscopy. A strong fluorescence signal was observed from HeLa cells after treatment for 2 h with either EGFP-fused full-length CtkA or EGFP-fused CtkAΔC, whereas no fluorescence was observed from the cells treated with EGFP alone (Fig. 3). The instability of the recombinant EGFP-fused CtkA prevented us from monitoring the translocation upon prolonged incubation longer than 2 h. These results establish that CtkA possesses a cell-translocating activity, and the C-terminal tail (residues 301–325) is not required for translocation into HeLa cells.
Fig. 3.
Confocal microscopy analysis of HeLa cells treated for 2 h with EGFP-fused full-length CtkA (Top), with EGFP-fused CtkAΔC (Middle), and with EGFP only (Bottom). DAPI (4′-6-Diamidino-2-phenylindole) stain (blue) indicates the position of nuclei. Strong green fluorescence signals clearly show intracellular translocation of the EGFP-fused CtkA or EGFP-fused CtkAΔC proteins into the cells.
CtkA Enhances Phosphorylation of NF-κB in a Kinase Activity-Dependent Manner.
To characterize the cellular target(s) of the CtkA kinase activity, a microarray-based kinase substrate screening experiment was performed (Kinexus Custom Kinase Substrate Profiling service at www.kinexus.ca). Briefly, it utilizes an antibody microarray of 270 specific phospho antibodies to screen for increased phosphorylation in the protein components of cultured human gastric epithelial cancer cell (AGS cell) lysates after treatment of the cells with the recombinant full-length CtkA protein. The most significant result was a 2.5-fold increase in phosphorylation at Ser276 of the p65 subunit of NF-κB transcription factor.
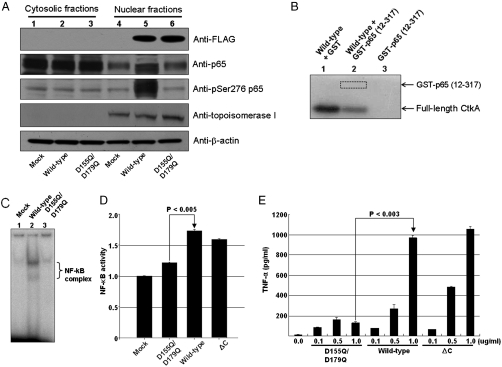
Because the recombinant CtkA protein in culture media was unstable under the growth condition, we transiently expressed the full-length wild-type ctkA gene or the full-length D155Q/D179Q double mutant gene in AGS cells. Then we performed a Western blot analysis of the cytosolic and nuclear fractions of AGS cells 48 h after transfection with p3XFLAG-CMV-10 plasmid (Sigma) containing either the full-length wild-type ctkA gene or the full-length D155Q/D179Q double mutant gene. This experiment additionally confirmed significantly enhanced phosphorylation of p65 Ser276 by wild-type CtkA (lane 5 in Fig. 4A). In contrast, expression of the catalytically inactive D155Q/D179Q double mutant CtkA did not affect phosphorylation of p65 Ser276 (lane 6 in Fig. 4A). This result suggests that the kinase activity of CtkA is required for enhanced phosphorylation of the NF-κB p65 subunit at Ser276. When we examined cellular location of the transiently expressed CtkA after 0, 12, 24, and 48 h of transfection, the transiently expressed CtkA translocated from cytosol into nucleus in a time-dependent manner (Fig. S6).
Fig. 4.
Protein kinase activity of CtkA and NF-κB activation. (A) Enhanced phosphorylation of NF-κB p65 at Ser276. Lanes 1–3 correspond to Western blot results of the cytosolic fractions of the AGS cells transfected with the mock and the plasmid p3XFLAG-CMV-10 containing either the full-length wild-type ctkA gene or the full-length D155Q/D179Q mutant ctkA gene, respectively. Lanes 4–6 correspond to the nuclear fractions of the cells transfected with mock, full-length wild-type, and full-length D155Q/D179Q ctkA genes, respectively. (B) In vitro protein kinase assay with recombinant p65. Lane 1, full-length wild-type CtkA plus GST; lane 2, full-length wild-type CtkA plus GST-p65 (12–317); lane 3, GST-p65 (12–317). (C) Electrophoretic mobility shift assay using an NF-κB binding DNA probe. (D) Monitoring of NF-κB activation by SEAP reporter assay. Student’s t test was used to analyze the data. P values less than 0.05 are considered statistically significant. (E) Estimation of levels of TNF-α in culture supernatants after stimulation of the cultured Thp1 cells with recombinant CtkA and variants at different concentrations.
We further tested whether CtkA directly phosphorylates Ser276 of the p65 subunit by monitoring phosphorylation of the GST-fused recombinant p65 subunit (residues 12–317) after treatment with the wild-type CtkA in the presence of [γ - 32P]ATP. GST-fused recombinant p65 subunit (residues 12–317) was not phosphorylated (lane 2 in Fig. 4B), whereas autophosphorylation of CtkA was evident (lanes 1 and 2 in Fig. 4B). Therefore, CtkA likely enhances phosphorylation at Ser276 of NF-κB p65 indirectly via phosphorylation of up-stream element(s) of NF-κB.
Phosphorylation at Ser276 of the NF-κB p65 subunit is known to activate the transcriptional activity of NF-κB (7). As described above, we have discovered that CtkA increases phosphorylation of p65 Ser276. Thus we checked if the activation of NF-κB is dependent on the kinase activity of CtkA. Our electrophoretic mobility shift assay using an NF-κB binding DNA probe showed that cellular expression of wild-type CtkA stimulated activation of NF-κB (lane 2 in Fig. 4C), whereas expression of the kinase-inactive D155Q/D179Q double mutant did not (lane 3 in Fig. 4C). Activation of NF-κB via cellular expression of CtkA implies an interaction(s) between CtkA and a cellular component(s) involved in the NF-κB signaling pathway. We also analyzed the NF-κB activity after transfection of AGS cells with the ctkA gene; we performed a secreted alkaline phosphatase (SEAP) assay using a reporter plasmid carrying the NF-κB promoter (Clontech). A 70% increase in the NF-κB activity was observed 24 h after transfection with the full-length wild-type ctkA gene (Fig. 4D), whereas transfection with the full-length D155Q/D179Q double mutant gene resulted in a much less significant increase (Fig. 4D). Therefore, we conclude that the kinase activity of CtkA is important for NF-κB activation through enhanced phosphorylation of the p65 subunit at Ser276.
We have further tested whether the kinase activity of CtkA is directly involved in the reported increase in proinflammatory cytokine secretion from macrophages (14). We monitored the secretion of TNF-α from cultured Thp1 cells using enzyme-linked immunosorbent assay, after treatment with the purified recombinant CtkA and its variants. Induction of TNF-α secretion increased in a dose-dependent manner upon treatment with wild-type full-length CtkA and CtkAΔC (Fig. 4E). In contrast, cells treated with the kinase-inactive D155Q/D179Q double mutant did not show such a dose-dependent increase (Fig. 4E). This result indicates that the kinase activity of CtkA is crucial for CtkA-mediated proinflammatory cytokine secretion.
Discussion
This study establishes that CtkA, a previously uncharacterized proinflammatory protein JHP940 from H. pylori J99, is a unique eukaryotic-type Ser/Thr protein kinase that is capable of translocating into cultured human cells to up-regulate NF-κB in a kinase-dependent manner. It is an unusual example of a Ser/Thr kinase from bacteria that can translocate into human cells by itself. CtkA was reported to enhance nuclear translocation of NF-κB in cultured macrophage cells (14). In the light of our work, the reported result can be interpreted not only as enhanced translocation but also as the enhancement of the NF-κB transcriptional activity through phosphorylation of p65 Ser276. The activation of NF-κB leads to an enhanced recruitment and infiltration of neutrophils into the gastric mucosa, contributing to a pronounced inflammatory response that is a hallmark of H. pylori infection (24). Phosphorylation of the NF-κB p65 subunit at Ser276 enhances the recruitment of the coactivator CBP/p300 (10), leading to an increased transcriptional activation of the p65 target genes such as TNF-α, IL-6, and IL-8 (6).
It is tempting to speculate that the kinase activity of CtkA plays an important role in the induction of host inflammatory responses during H. pylori infection, although further studies are needed to elucidate a detailed event(s) leading to enhanced Ser276 phosphorylation of the p65 subunit. Given the genetic variability of clinical H. pylori strains, the presence of the ctkA gene might confer selective advantages to some of the strains to colonize the gastric mucus of different host genotypes and therefore become more invasive (16).
Our results establish that CtkA is a unique H. pylori virulence factor that has the eukaryotic-type protein kinase fold, is catalytically active as Ser/Thr kinase, and is capable of translocating into human cells. However, we do not rule out the possibility that CtkA phosphorylates a small molecule(s). Recent studies have shown that eukaryotic-type protein kinases play major roles in growth, virulence, and persistence within a number of pathogenic bacteria such as Pseudomonas aeruginosa, Mycobacterium tuberculosis, Yersinia pseudotuberculosis, and Staphylococcus aureus (25). In P. aeruginosa, for example, threonine phosphorylation by the PpkA kinase regulates assembly of the Type VI secretion system that influences virulence in clinical isolates (26). Among the 11 eukaryotic-type protein kinases expressed by M. tuberculosis (PknA–PknL) (27), PknG was reported to be secreted by M. tuberculosis and to play an essential role in signaling inside the host cells (28).
Interestingly, the amino acid sequence of the CtkA kinase domain (residues 1–300) is conserved among a number of human gastrointestinal bacteria (Fig. S2). The human gastrointestinal tract is a very complex environment that is populated by diverse species of microorganisms (29). The gastrointestinal microfloras have the potential to exert both pro- and anti-inflammatory responses (29). Although it is not known whether the CtkA-like kinases in other human gastrointestinal bacteria modulate inflammatory responses of the host, they are likely to play important biological roles.
As mentioned above, the activation loop of H. pylori CtkA adopts an active conformation without autophosphorylation. It is well established that the activation loops of a number of eukaryotic protein kinases adopt an active conformation without phosphorylation (22). These kinases possess regulatory domains or recruit a regulatory protein to control their kinase activity (22). CtkA may recruit binding partner(s) for the regulation of its kinase activity within host cells, because it lacks a regulatory domain. Further studies are required to understand the regulatory mechanism of controlling the CtkA kinase activity.
In this study, we have demonstrated that CtkA can enter cultured human cells. Similar incorporation of the H. pylori virulence factor TNF-α inducing protein-α (TIP-α) into the gastric epithelial cells was reported (30). Because CtkA as well as TIP-α induces NF-κB activation in a cag pathogenicity island-independent manner, the mechanism of CtkA penetration into host cells is expected to be different from that of CagA, which is delivered into gastric epithelial cells via the type IV secretion system. In summary, our structural and functional characterization of CtkA from H. pylori J99 shows that it is a unique eukaryotic-type Ser/Thr kinase that translocates into human cells to up-regulate the NF-κB activity in a kinase-dependent manner.
Materials and Methods
Detailed methods are provided in SI Materials and Methods. Briefly, the ctkA gene was subcloned into pET-21a, expressed in E. coli Rosetta2(DE3)pLysS cells, purified, and crystallized. Details of X-ray structure determination as well as in vitro kinase assay, cell culture, transfection, Western blotting, cellular translocation analysis by a fluorescence-activated cell sorter and a confocal microscope, electrophoretic mobility shift assay, SEAP reporter assay, and enzyme-linked immunosorbent assay are provided in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We are grateful to the staffs at Pohang Light Source beamlines 4A and 6C for assistance with synchrotron data collection. We thank Prof. Chin Ha Chung (Seoul National University, Seoul, Korea) for providing the p65 gene and Dr. Hye-Jin Yoon for assisting the refinement calculation. This work is funded by Korea Ministry of Education, Science, and Technology, National Research Foundation of Korea, Innovative Drug Research Center for Metabolic and Inflammatory Disease (Grant R11-2007-107-00000-0), Basic Science Outstanding Scholars Program, World-Class University Program, and Korea Healthcare Technology R&D Project, Ministry of Health, Welfare and Family Affairs, Republic of Korea (Grant A092006 to S.W.S.) and National Creative Research Initiative Grant of National Research Foundation of Korea (2010-0000733 to S.K.L.). The funding agencies had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
Data deposition: Atomic coordinates of the refined structures have been deposited in the Protein Data Bank, http://www.pdb.org [PDB ID codes 3AKJ (Apo), 3AKK (ADP-complex), and 3AKL (AMP-PNP-complex)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010153107/-/DCSupplemental.
References
- 1.Shanks AM, El-Omar EM. Helicobacter pylori infection, host genetics and gastric cancer. J Dig Dis. 2009;10:157–164. doi: 10.1111/j.1751-2980.2009.00380.x. [DOI] [PubMed] [Google Scholar]
- 2.Yamaoka Y, et al. Induction of various cytokines and development of severe mucosal inflammation by cagA gene positive Helicobacter pylori strains. Gut. 1997;41:442–451. doi: 10.1136/gut.41.4.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida N, et al. Mechanisms involved in Helicobacter pylori-induced inflammation. Gastroenterologia. 1993;105:1431–1440. doi: 10.1016/0016-5085(93)90148-6. [DOI] [PubMed] [Google Scholar]
- 4.Keates S, Hitti YS, Upton M, Kelly CP. Helicobacter pylori infection activates NF-κB in gastric epithelial cells. Gastroenterologia. 1997;113:1099–1109. doi: 10.1053/gast.1997.v113.pm9322504. [DOI] [PubMed] [Google Scholar]
- 5.Ghosh S, Karin M. Missing pieces in the NF-κB puzzle. Cell. 2002;109:S81–S96. doi: 10.1016/s0092-8674(02)00703-1. [DOI] [PubMed] [Google Scholar]
- 6.Chen LF, Greene WC. Shaping the nuclear action of NF-kappaB. Nat Rev Mol Cell Biol. 2004;5:392–401. doi: 10.1038/nrm1368. [DOI] [PubMed] [Google Scholar]
- 7.Viatour P, Merville MP, Bours V, Chariot A. Phosphorylation of NF-kappaB and IkappaB proteins: Implications in cancer and inflammation. Trends Biochem Sci. 2005;30:43–52. doi: 10.1016/j.tibs.2004.11.009. [DOI] [PubMed] [Google Scholar]
- 8.Zhong H, SuYang H, Erdjument-Bromage H, Tempst P, Ghosh S. The transcriptional activity of NF-kappaB is regulated by the IkappaB-associated PKAc subunit through a cyclic AMP-independent mechanism. Cell. 1997;89:413–424. doi: 10.1016/s0092-8674(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 9.Vermeulen L, De Wilde G, Van Damme P, Vanden Berghe W, Haegeman G. Transcriptional activation of the NF-kappaB p65 subunit by mitogen- and stress-activated protein kinase-1 (MSK1) EMBO J. 2003;22:1313–1324. doi: 10.1093/emboj/cdg139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhong H, Voll RE, Ghosh S. Phosphorylation of NF-kappa B p65 by PKA stimulates transcriptional activity by promoting a novel bivalent interaction with the coactivator CBP/p300. Mol Cell. 1998;1:661–671. doi: 10.1016/s1097-2765(00)80066-0. [DOI] [PubMed] [Google Scholar]
- 11.Yamaoka Y. Roles of the plasticity regions of Helicobacter pylori in gastroduodenal pathogenesis. J Med Microbiol. 2008;57:545–553. doi: 10.1099/jmm.0.2008/000570-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Occhialini A, et al. Distribution of open reading frames of plasticity region of strain J99 in Helicobacter pylori strains isolated from gastric carcinoma and gastritis patients in Costa Rica. Infect Immun. 2000;68:6240–6249. doi: 10.1128/iai.68.11.6240-6249.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Santos A, et al. New pathogenicity marker found in the plasticity region of the Helicobacter pylori genome. J Clin Microbiol. 2003;41:1651–1655. doi: 10.1128/JCM.41.4.1651-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rizwan M, Alvi A, Ahmed N. Novel protein antigen (JHP940) from the genomic plasticity region of Helicobacter pylori induces tumor necrosis factor alpha and interleukin-8 secretion by human macrophages. J Bacteriol. 2008;190:1146–1151. doi: 10.1128/JB.01309-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham JE, Peek RM, Jr, Krishna U, Cover TL. Global analysis of Helicobacter pylori gene expression in human gastric mucosa. Gastroenterologia. 2002;123:1637–1648. doi: 10.1053/gast.2002.36589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romo-Gonzalez C, et al. Differences in genome content among Helicobacter pylori isolates from patients with gastritis, duodenal ulcer, or gastric cancer reveal novel disease-associated genes. Infect Immun. 2009;77:2201–2211. doi: 10.1128/IAI.01284-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 18.Holm L, Rosenström P. Dali server: Conservation mapping in 3D. Nucleic Acids Res. 2010;38(Suppl):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schumacher MA, et al. Molecular mechanisms of HipA-mediated multidrug tolerance and its neutralization by HipB. Science. 2009;323:396–401. doi: 10.1126/science.1163806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evdokimov A, et al. New kinase regulation mechanism found in HipBA: A bacterial persistence switch. Acta Crystallogr D. 2009;65:875–879. doi: 10.1107/S0907444909018800. [DOI] [PubMed] [Google Scholar]
- 21.Adams JA. Kinetic and catalytic mechanisms of protein kinases. Chem Rev. 2001;101:2271–2290. doi: 10.1021/cr000230w. [DOI] [PubMed] [Google Scholar]
- 22.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 23.Knighton DR, et al. Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science. 1991;253:407–414. doi: 10.1126/science.1862342. [DOI] [PubMed] [Google Scholar]
- 24.Shimada T, Terano AJ. Chemokine expression in Helicobacter pylori-infected gastric mucosa. J Gastroenterol. 1998;33:613–617. doi: 10.1007/s005350050146. [DOI] [PubMed] [Google Scholar]
- 25.Ohlsen K, Donat S. The impact of serine/threonine phosphorylation in Staphylococcus aureus. Int J Med Microbiol. 2010;300:137–141. doi: 10.1016/j.ijmm.2009.08.016. [DOI] [PubMed] [Google Scholar]
- 26.Mougous JD, Gifford CA, Ramsdell TL, Mekalanos JJ. Threonine phosphorylation post-translationally regulates protein secretion in Pseudomonas aeruginosa. Nat Cell Biol. 2007;9:797–803. doi: 10.1038/ncb1605. [DOI] [PubMed] [Google Scholar]
- 27.Alber T. Signaling mechanisms of the Mycobacterium tuberculosis receptor Ser/Thr protein kinases. Curr Opin Struct Biol. 2009;19:650–657. doi: 10.1016/j.sbi.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walburger A, et al. Protein kinase G from pathogenic mycobacteria promotes survival within macrophages. Science. 2004;304:1800–1804. doi: 10.1126/science.1099384. [DOI] [PubMed] [Google Scholar]
- 29.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suganuma M, et al. TNF-alpha-inducing protein, a carcinogenic factor secreted from H. pylori enters gastric cancer cells. Int J Cancer. 2008;123:117–122. doi: 10.1002/ijc.23484. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.