Abstract
The activity of P-type plasma membrane H+-ATPases is modulated by H+ and cations, with K+ and Ca2+ being of physiological relevance. Using X-ray crystallography, we have located the binding site for Rb+ as a K+ congener, and for Tb3+ and Ho3+ as Ca2+ congeners. Rb+ is found coordinated by a conserved aspartate residue in the phosphorylation domain. A single Tb3+ ion is identified positioned in the nucleotide-binding domain in close vicinity to the bound nucleotide. Ho3+ ions are coordinated at two distinct sites within the H+-ATPase: One site is at the interface of the nucleotide-binding and phosphorylation domains, and the other is in the transmembrane domain toward the extracellular side. The identified binding sites are suggested to represent binding pockets for regulatory cations and a H+ binding site for protons leaving the pump molecule. This implicates Ho3+ as a novel chemical tool for identification of proton binding sites.
Keywords: allosteric regulation, membrane protein crystallography, proton transport
Members of the large family of P-type ATPases operate as pumps that transport cations across biological membranes. The transport process includes discernible catalytic intermediates termed E1, E1P, E2P, and E2, with P denoting a phosphorylated intermediate (1). Structures of the Na+, K+-ATPase (2, 3), the sarcoplasmic reticulum (SR) Ca2+-ATPase (4, 5), and the plasma membrane (PM) H+-ATPase (6) show that different members of the P-type ATPase family exhibit the same three-dimensional fold with four discrete domains involved in the basic cation transporting machinery: a nucleotide-binding domain (N), a phosphorylation domain (P), an actuator domain (A), and a transmembrane domain (M) (2, 4, 6). The three cytosolic domains are responsible for ATP hydrolysis, and the associated domain movements are coupled to cation translocation through the opening and closing of transport pathways that leads to the cation binding site(s) residing in the central part of the M domain (5, 7–9).
The plasma membrane (PM) H+-ATPase is a central constituent of all plant and fungal cells where its ejection of protons from the cell generates the essential electrochemical gradient across the plasma membrane. The molecular mechanism of proton transport by the ATPase involves a conserved proton acceptor/donor (Asp684) in the membranous part of the protein (10). Proton inlet and exit pathways leading to and from this central proton binding site have been suggested based on structural data (6, 11). Besides protons, alkali cations, and K+ in particular, increase ATP hydrolytic activity of the PM H+-ATPase protein (12–14). As the membrane potential approaches the reversal potential, the accumulation of the phosphorylated intermediate is likely to occur. Under these conditions, K+ might serve as an uncoupling factor by introducing a safety valve in the pump (15). Further, direct reversible inactivation of the PM H+-ATPase by increased cytosolic Ca2+ has been proposed to be a key step in closing of the stomatal pore in plants (13). No structural data of the proposed K+ and Ca2+ binding sites on the PM H+-ATPase has so far been presented.
Results
The Delipidated PM H+-ATPase Is Folded in a Catalytically Competent State.
The E1 protein crystals used for localization of the ion-binding sites were produced in the absence of exogenously added lipids and likely represent the integral membrane protein in a delipidated form (6). The PM H+-ATPase construct used for crystallization is a truncated form of the protein, which lacks the C-terminal part of its autoinhibitory domains and is consequently constitutively activated. After purification, the protein was fully functional following the addition of minor amounts of exogenous lipid (100–200 lipid molecules per protein molecule) (Fig. S1). This indicates that the crystallized PM H+-ATPase represents a correctly folded and functional ATPase and therefore is suitable as a tool for identification of binding pockets by soaking and/or cocrystallization techniques.
Crystallographic Identification of Rb+, Tb3+ and Ho3+ Sites in the PM H+-ATPase.
We initially tried to obtain direct structural information of Ca2+ and K+ binding to the PM H+-ATPase by crystal soaking and cocrystallization with Ca2+ ions and K+ ions. However, the low anomalous signal of K+ and Ca2+ did not allow for structural identification of these cations in the crystallographic data obtained. We therefore turned to Ca2+ and K+ congeners with a higher anomalous signal.
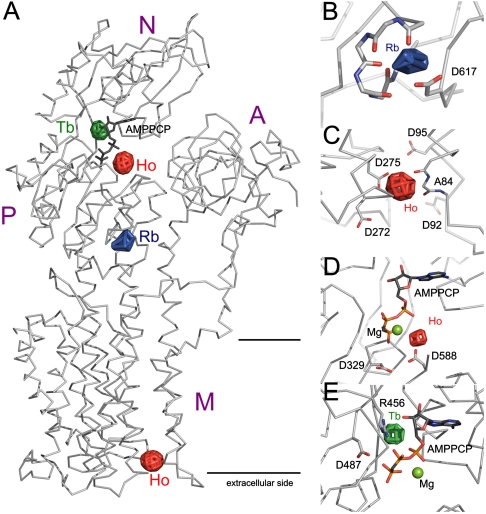
Rb+ is a congener of K+, and a Rb+ binding site was identified by replacing K+ with Rb+ in the crystallization setup. The published structure of the E1-AMPPCP form refined at 3.6-Å resolution was used as model, and anomalous difference Fourier maps were calculated based on data from 40 to 7.5 Å resolution and model phases (Table S1). Two peaks at 6.4 and 5.3 sigma (background maximum of 4.0) were identified at two sites related by noncrystallographic symmetry in the asymmetric unit containing two protein monomers. The single Rb+ ion peak observed on each protein monomer was localized to a conserved ion-binding pocket in the PM H+-ATPase (Fig. 1 A and B). The Rb+/K+ ion is positioned next to four backbone carbonyl groups (residues 598 to 601) and the side chain oxygen atoms from Asp617, previously demonstrated by mutagenesis to be necessary for any K+-mediated effects on the PM H+-ATPase (15). Rb+ was the only anomalous scatter at the applied wavelength, and the anomalous difference Fourier density therefore allows us to unambiguously assign the peak densities as Rb+ ions.
Fig. 1.
Location of the identified cation binding sites in the PM H+-ATPase. (A) Ribbon representation of the PM H+-ATPase is shown in light gray with the nucleotide AMPPCP shown as sticks (dark gray) and with overlay of the anomalous difference density maps shown as green mesh (Tb3+), red mesh (Ho3+), and blue mesh (Rb+). The positions of the different domains are indicated. For B–E, the color code of the anomalous difference density maps is as in A. (B) Close-up of the cytoplasmic Rb+ site with an indication of the residues thought to contribute back bone carbonyl groups (598–601) and side-chain oxygen atom(s) (Asp617). (C) Four acidic residues (Asp92, Asp95, Asp272, and Asp275) are located in the vicinity of the extracellular Ho3+ binding site. (D) The Ho3+ ion identified at the interface between the N and P domains is positioned adjacent to the nucleotide (stick representation) and the well-conserved Asp329 and Asp588 residues. (E) The Tb3+ ion binds at a site distinct from that of Ho3+ close to the nucleotide (stick representation) and the conserved residues Arg456 and Asp487.
Lanthanide ions have been widely used as spectroscopic and crystallographic probes for Ca2+ binding sites (reviewed in ref. 16). We grew PM H+-ATPase crystals and soaked them with Tb3+. The Tb3+-ion was easily identified in both anomalous and isomorphous difference maps and gave rise to peaks at 6.8 and 6.2 sigma related by noncrystallographic symmetry. The Tb3+ peak is located at the N domain, just above the hinge connecting the N domain to the P domain, in the binding pocket of ATP at a position next to the ribose and base of the nucleotide adjacent to the phosphate group of the bound nucleotide (Fig. 1E). Only a single negatively charged residue, the highly conserved Asp487, is in close proximity and is therefore a prime candidate for being involved in the direct binding of the Tb3+ ion. The positively charged residue Arg456 also resides in the vicinity and could be involved in the cation coordination of Tb3+-ion through a network of water molecules.
Likewise, we used crystals soaked with Ho3+ to collect data to 6-Å resolution at 1.2782-Å wavelength where there is strong anomalous signal from Ho3+ (Table S1). Despite similar chemical properties, Tb3+ and Ho3+ do not bind at same locations in the PM H+-ATPase. The Ho3+ signal gave rise to four clear anomalous peaks related two and two by noncrystallographic symmetry. One site is located at the interface between the N and P domains, approximately at 10-Å distance from the Tb3+ site, and is coordinated by well-conserved residues Asp588 and Asp329 (Fig. 1D). The second Ho3+ binding position is located at the extracellular side of the transmembrane domain in a groove between transmembrane segments M1, M2, M4, and M6 (Fig. 1C). It is coordinated by Asp275 (situated in M4), Asp95 (M2), and the backbone oxygen of Ala84 (M1). Asp92 and Asp272 are situated in the vicinity of the binding site and could contribute to the local electrostatic environment. Asp92 and Asp95 are almost completely conserved (respectively, 35 and 39 of 39 H+-ATPase sequences) in the family of PM H+-ATPases.
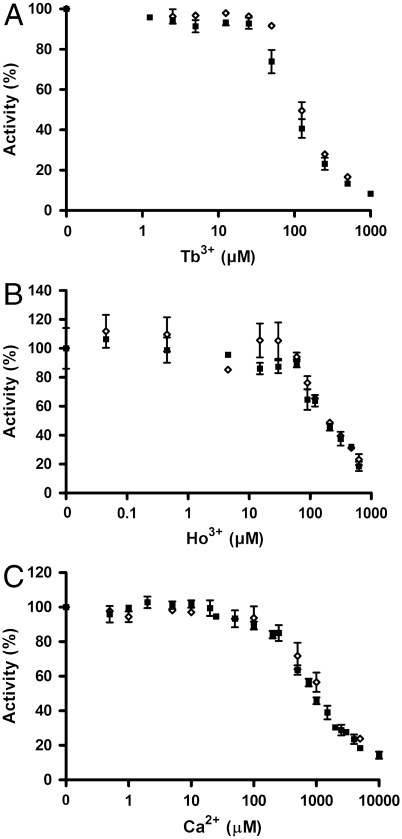
Testing the biochemical effect of Tb3+ and Ho3+ ions on the purified PM H+-ATPase, both ions were found to inhibit the catalytic turnover as well as the accumulation of the phosphorylated intermediate (EP). The apparent Ki of Tb3+ was found to be approximately 100 μM (Fig. 2A and Fig. S2), and the apparent Ki of Ho3+ was approximately 150 μM (Fig. 2B and Fig. S2).
Fig. 2.
Ca2+, Tb3+ and Ho3+ ions inhibit the activity of the purified PM H+-ATPase independently of an intact autoinhibitory domain. ATP hydrolytic activity of the C-terminal truncated PM H+-ATPase (aha2Δ73) and the full-length PM H+-ATPase (AHA2) was measured with varying concentrations of: (A) Tb3+ ions, (B) Ho3+ ions, and (C) Ca2+ ions. The activity without the addition of ions were set to 100% in A, B, and C. ▪, aha2Δ73; ◊, AHA2. All experiments are represented as ± S.D.
A Single Point Mutation of Arg456 Decreases the Apparent Affinity for Tb3+ and Ca2+.
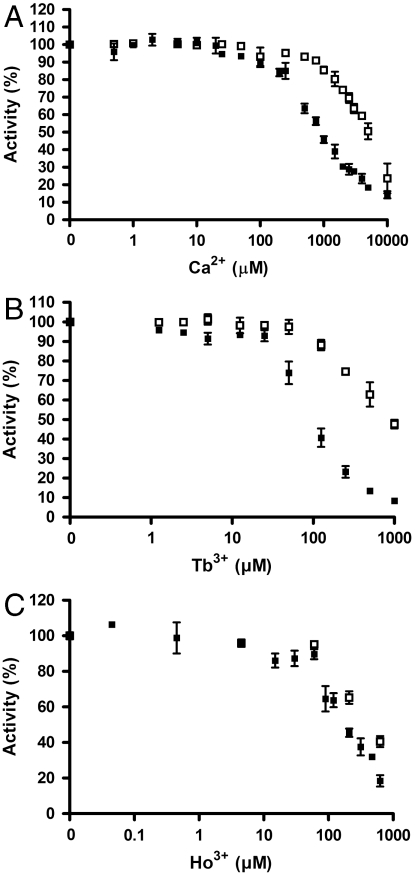
The Tb3+ and Ho3+ binding sites identified at the interface of the N and P domains are lined by highly conserved amino acid residues necessary for basic P-type ATPase function. Arg456 was suggested from the crystallographic determination to be involved in Tb3+ coordination (Fig. 1E). We constructed an R456V single point mutation of the PM H+-ATPase in order to test its sensitivity toward Tb3+, Ca2+ and Ho3+ (Fig. 3 A–C). The modified protein was expressed in yeast and affinity purified before biochemical characterization. The catalytic turnover of the R456V mutant was less than 5% of the wild-type level (Table 1) concomitant with a reduced level of EP accumulation at steady state (Fig. S3). Removal of the positive charge at position 456 in the protein did indeed increase the Ki for Tb3+ and Ca2+ significantly (Fig. 3 A and B and Fig. S4). In contrast, the Ki for Ho3+ was not significantly shifted (Fig. 3C and Fig. S4), in line with the Tb3+ and Ho3+ ions binding to separate sites at the interface of the N and P domains. The apparent affinity for ATP was significantly reduced for the R456V mutant (Fig. S5A), and the pH optimum appeared to be shifted toward more acidic values (Fig. S5B) in line with a requirement of an increased proton concentration to substitute for the removal of the positive guanidinium group of the arginine residue.
Fig. 3.
The R456V mutant PM H+-ATPase displays decreased affinity for Ca2+ and Tb3+ ions. ATP hydrolytic activity of the wild type and the R456V mutant was measured with varying concentrations of: (A) Ca2+ ions, (B) Tb3+ ions, and (C) Ho3+ ions. The activity without the addition of ions were set to 100% in A, B, and C. ▪, wild type; □, R456V. All experiments are represented as ± S.D.
The Ki for Ca2+ Is Independent of an Intact Regulatory Autoinhibitory Domain.
Ca2+ was found to have an inhibitory effect on ATP hydrolytic activity and EP accumulation of the purified PM H+-ATPase (Ki > 500 μM) (Fig. 2C and Fig. S2). Previous studies have found a profound direct inhibitory effect of Ca2+ (Ki ∼ 1 μM) on the Vicia faba plant PM H+-ATPase, indicating a potential important physiological function of the calcium concentration on PM H+-ATPase activity (13). PM H+-ATPases are subject to autoinhibition by terminal regulatory domains (17–19). The wild-type protein construct used in this study is devoid of the C-terminal part of its regulatory domain (6), and Ca2+ binding to the PM H+-ATPase could thus potentially be dependent on an intact autoinhibitory domain. However, no difference in the effect of Ca2+ on the full-length and the C-terminal truncated PM H+-ATPase could be detected (Fig. 2C). The fact that Ca2+ does not exert any significant effect on the PM H+-ATPase at concentrations below 100 μM implies that direct inhibition of the PM H+-ATPase by Ca2+ is unlikely to be physiologically relevant. Likewise, no significant differences between the full-length and the C-terminal truncated PM H+-ATPase regarding their sensitivities to Tb3+ and Ho3+ could be detected (Fig. 2 A and B).
Calcium Does Not Regulate Activity from the Extracellular Ho3+ Binding Site.
The identification of a Ho3+ ion-binding site at the extracellular mouth of the protein could potentially signify an extracellular regulatory role of Ca2+ on the PM H+-ATPase. Single point mutations of the acidic residues Asp87, Asp92, Asp95, Asp272, and Asp275, all located at or near the identified extracellular Ho3+ ion-binding site, were therefore constructed and tested for their ability to complement a conditional yeast null PM H+-ATPase mutant in the presence of various concentrations of Ca2+ (Fig. 4A). Depending on the Ca2+ concentration in the growth medium, a modulatory extracellular binding site for calcium on the plant PM H+-ATPase would be expected to influence the ability of plant PM H+-ATPase to complement the endogenous fungal PM H+-ATPase. No significant effect of calcium on the growth of yeast cells expressing the wild-type plant PM H+-ATPase or any of the mutated versions were however found (Fig. 4A). Moreover, when purified to homogeneity, none of the mutant proteins displayed any significant changes in their sensitivity toward added Ca2+ ions compared to the wild-type PM H+-ATPase (Fig. 4B). This suggests that the observed inhibitory effect of Ca2+ on the PM H+-ATPase is mediated through binding to the identified Tb3+/Ca2+ site located in the cytoplasmic domain.
Fig. 4.
Mutation of the extracellular Ho3+ binding site does not change the affinity for Ca2+. (A) Yeast complementation assay was performed for the wild type and D87A, D92A, D95A, D272A, and D275A single point mutants of the extracellular Ho3+ binding site. In the yeast strain RS-72, the endogenous PM H+-ATPase, Pma1p, has been placed under the control of a galactose promoter, whereas the introduced, plasmid-borne plant PM H+-ATPases are under the control of the constitutive PMA1 promoter (51). Yeast growth on glucose is therefore dependent on a functional plant PM H+-ATPase. Transformed yeast cells, RS-72, were spotted on either galactose-containing media (Gal) or glucose-containing media (Glu) supplemented with the indicated concentrations of Ca2+ ions. Growth was recorded after three days. (B) ATP hydrolytic activity of the wild type and mutants of the extracellular Ho3+ binding site was measured with varying concentrations of Ca2+ ions. ▪, wild type; ◊, D87A; □, D92A; Δ, D95A; ○, D272A; ×, D275A. The activity without any addition of Ca2+ ions was set to 100%, and all measurements are represented as ± S.D.
The Extracellular Ho3+ Binding Site Might Represent a Proton Exit Site.
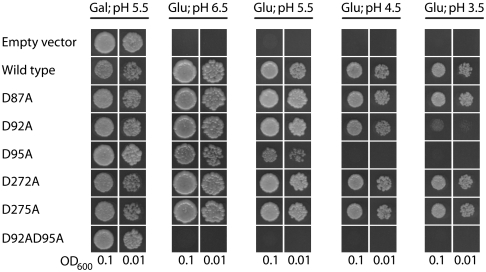
In several proton transporting pumps a series of internal proton transfer reactions take place between charged residues along a proton transport pathway (20, 21). In order to test if the acidic cation binding pocket occupied by Ho3+ could be part of the proton transport pathway of the PM H+-ATPase, mutants of the extracellular Ho3+ binding site were tested for their ability to complement the conditional yeast null PM H+-ATPase mutant described previously at different proton concentrations. As shown in Fig. 5, the D87A, D272A, and D275A mutants were able to support yeast growth to the same extent as the wild type. In contrast, the D92A and the D95A mutants displayed a decreased ability to support yeast growth at more acidic pH values (pH 4.5-3.5), indicative of the mutants being unable to pump protons against a high proton gradient. A double mutant of these two residues, D92A,D95A, was not able to complement the yeast null PM H+-ATPase mutant irrespective of the proton concentration (Fig. 5).
Fig. 5.
Mutation of the extracellular Ho3+ binding site gives a yeast phenotype that displays sensitivity toward low pH. Yeast complementation test was performed essentially as described in Fig. 4. Thus, yeast expressing single point mutants of the extracellular Ho3+ site were spotted on either galactose-containing media (Gal) at pH 5.5 or glucose-containing media (Glu) with different pH values (pH 6.5–3.5) in two different concentrations (OD600 of 0.1 and 0.01).
For the single and double mutants of Asp92 and Asp95, the ATPase specific activity was reduced to 45–65% of the wild-type level (Table 1). Similar Km values for ATP in all mutant enzymes were found, demonstrating that the substitutions do not abolish the ability of the enzymes to bind nucleotides and hydrolyze ATP. The pH profile of the ATP hydrolytic activity of the double mutant D92A,D95A revealed that it was largely insensitive toward the concentration of protons (Fig. S6).
The Asp92 and Asp95 mutants were reconstituted in vesicles and their ability to perform ATP-dependent proton pumping was evaluated using the ΔpH probe ACMA. The proton transport rate was severely reduced for all mutants compared to the wild type (Table 1). When a transmembrane potential was allowed to establish by the elimination of the ionophore valinomycin in the experimental setup, the capacity of the mutants to transport protons was even more inhibited relative to the wild type (Table 1).
Table 1.
Kinetic properties of PM H+-ATPase mutants
| PM H+-ATPase | Specific activity (µmol/mg/min) | pH optimum | ATP affinity, Km (µM) | Vanadate inhibition, IC50 (µM) | Proton transport, + valinomycin (%) | Proton transport, − valinomycin (%) |
| Wild type | 25.7 ± 1.8 | ∼6.8 | 45 ± 5 | ∼8 | 100 | 100 |
| R456V | 0.7 ± 0.1 | < 5.6* | 1428 ± 69 | ∼39 | n.d.† | n.d.† |
| D92A | 11.8 ± 2.9 | ∼6.6 - 6.8 | 36 ± 6 | ∼7 - 8 | 24.7 ± 10.3 | 7.3 ± 1.9 |
| D95A | 16.7 ± 0.5 | ∼6.8 | 39 ± 4 | ∼6 | 37.6 ± 4.0 | 17.3 ± 3.6 |
| D92A,D95A | 12.4 ± 1.3 | ∼6.6 - 6.8 | 35 ± 13 | ∼16 | 18.2 ± 1.0 | 5.3 ± 0.2 |
*The pH optimum was not determined within the tested range but appears to be shifted toward more acidic values (Fig. S5B).
†n.d., not determined.
In contrast to their different abilities to support yeast growth (Fig. 5), the catalytic activity and the proton transport rate of the D92A mutant and the D92A,D95A double mutant are at similar levels. Thus the ability to functionally complement the pma1 deficiency in vivo must integrate more parameters besides the ones evaluated in vitro.
Discussion
The presence of regulatory ion-binding pockets is a common feature of enzymes (22, 23). This study provides crystallographic evidence for three cation binding sites in the cytoplasmic domains of the PM H+-ATPase and an additional site in the membrane domain that is accessible from the extracellular side.
The identified Rb+/K+ ion-binding site confirms previous mutagenesis experiments (15) and corresponds to the cytoplasmic K+ site described for the Na+, K+-ATPase and for the SR Ca2+-ATPase (5, 24). The coordinating P-domain residues are highly conserved among P-type ATPases, and the structural identification of a K+ binding site in both type II (Na+/K+- and SR Ca2+-ATPases) and type III (H+) P-type ATPases hints at a fundamental function of this allosteric ion-binding site. In plant cells, the physiological concentration of K+ is usually constant at concentrations around 50 mM K+ (25). This suggests that the potassium binding site (with an apparent Km for K+ ∼1 mM) most likely is occupied by K+ in vivo (15). The K+ binding site has been suggested to be involved in the docking of the A domain during dephosphorylation of the E2P state of Na+, K+-, and SR Ca2+-ATPase (5, 24) and allows for stimulation of uncoupled dephosphorylation of the E1P state in the PM H+-ATPase (15).
Intrinsic lanthanide binding sites, or novel lanthanide binding sites introduced by protein engineering, serve extensive applications in spectroscopic and crystallographic studies of proteins (16, 26, 27). The identification of highly conserved P-type ATPase lanthanide binding sites implies that they can serve as a direct tool in the biophysical and structural analysis of other members of the P-type ATPase family of proteins. Although both Tb3+ and Ho3+ were found positioned near the nucleotide at the interface of the N and the P domains, the localization of these two ions was not identical. Tb3+ has previously been reported to inhibit the activity of the PM H+-ATPase from Schizosaccharomyces pompe (12) and of the SR Ca2+-ATPase (28–30). However, a binding site for Tb3+ ions in P-type ATPases has so far not been described structurally. The Tb3+ ion identified in the present study is located in close vicinity of the nucleotide and could be coordinated by Asp487 and Arg456. Asp487 is part of the highly conserved D487PPR segment of the P-type ATPase family and is located at the hinge between the N and P domains. In the Na+, K+-ATPase and the SR Ca2+-ATPase, the corresponding residues have been reported to be very sensitive to even conservative substitutions (31–34). Based on structural data from the SR Ca2+-ATPase, it was speculated that this aspartate residue has an important role in the positioning of the N domain with respect to the P domain (34). Arg456 is conserved in type II and III P-type ATPases, and the corresponding residue in the Na+, K+-ATPase has been shown to be important for high-affinity binding of both ATP and ADP (35). The presence of a Tb3+/Ca2+ ion at the identified position will most likely influence the side chains of Arg456 and Asp487, thereby interfering with ATP binding and/or phosphoryl transfer. Indeed, the R456V mutant displayed a significant reduction in the apparent affinity for ATP. For the R456V mutant, EP formation was not inhibited to the same extent by the presence of Tb3+/Ca2+ ions as the wild type, and the apparent acidic shift in the pH profile indicates that electrostatic interactions are crucial for ATP binding and/or phosphoryl transfer.
The Ho3+ ion identified adjacent to the nucleotide at the interface between the N and the P domains resides near the canonical aspartate residue Asp329 as well as Asp588. Asp588, placed in the P-type ATPase motif TGD588GVND, is well-conserved within the haloacid dehalogenase superfamily, and for several members of this family the aspartate residue has been demonstrated to be required for Mg2+ coordination (36). Likewise, by use of mutational analysis and peptide binding studies, the corresponding residues in the SR Ca2+-ATPase and the Na+, K+-ATPase have been shown to coordinate the Mg2+ ion during phosphoryl transfer (32, 34, 37, 38). Thus, correct positioning of this aspartate is of crucial importance for phosphorylation of Asp329, and Ho3+ must therefore be assumed to have an inhibitory effect on the PM H+-ATPase through a distortion of the γ-phosphate group away from the conserved Asp329. Ho3+ was indeed found to inhibit EP formation in both the wild-type protein and in the R456V mutant.
In this study, structural evidence for Ho3+ binding to an extracellular negatively charged binding pocket on the PM H+-ATPase was obtained. This area has been suggested to function as a proton acceptor/donor relay cluster during proton release from the intramembranous proton binding site (6). Substitution of either Asp92 or Asp95 with alanine residues led to loss of yeast complementation at high proton concentrations. This indicates that these mutants cannot sustain proton transport against a high proton electrochemical gradient. Further, the double substituted mutant (D92A,D95A) was completely unable to support yeast growth. The ability of the mutants to transport protons was evaluated with and without a potential difference across the vesicle membrane, and for all the mutants tested, the proton transport rate was found to be significantly inhibited by a transmembrane potential. These results support the proposal that Asp92 and Asp95 line the proton exit pathway of the PM H+-ATPase. Several of the amino acids found to coordinate Ho3+ at the extracellular mouth of the pump are conserved in both plant and fungal PM H+-ATPases, and in the Saccharomyces cerevisiae PM H+-ATPase Pma1p, the corresponding aspartate residues are important for pump activity (39).
The mechanism of P-type ATPases is usually discussed in terms of the E1–E2 model developed from the Post–Albers scheme for the Na+, K+-ATPase (40, 41). The model proposes that P-type pumps can exist in one of two distinct forms, E1 and E2. In its original form, the model further proposes alternating exposure of the same ion-binding sites toward the intracellular and extracellular medium in the E1 and E2 conformations, respectively. While our results are compatible with the alternating-site model, the model does not predict additional sites, such as an exit site as shown in this work.
Recently, a low affinity Ca2+ binding site was suggested in the luminal Ca2+ exit pathway of the SR Ca2+-ATPase (42). This site involves Glu90 of the SR Ca2+-ATPase, which is homologous to Asp95 in the PM H+-ATPase, part of the Ho3+ cation binding site identified in this work. It therefore seems a possibility that all P-type pumps could have a cation leaving site. In the proton transporting bacteriorhodopsin, two acidic residues at the extracellular side of the protein form a proton release group (43–45) suggesting that a leaving site for protons could be a general feature of H+ pumps. In the E1 crystal structure, the approximate distance from the Asp684 to the proton release group is approximately 15 Å. Proton transport from Asp684 could entail proton release directly to the proton exit group and the extracellular side along a hydrogen bonded network of water upon deocclusion and opening of the release pathway. Additional studies are needed to address this and if the proton release group takes form of a H+ or H3O+ binding site.
The proposed leaving site for transported protons identified in this work would have been difficult to locate without the use of the positively charged Ho3+ to reveal a negative patch of residues important for proton unloading. Ho3+ is a relatively small cation (ionic radii 0.96 Å) with a high charge density and well known to form complexes with the deprotonated form of carboxylates in protein structures (46, 47). Our structural and biochemical data identify Ho3+ as a novel chemical tool for structural identification of proton binding sites.
Materials and Methods
Construction of Mutants.
The multicopy vector Yep-351 (48) containing a modified cDNA of the Arabidopsis thaliana AHA2 plasma membrane H+-ATPase isoform under the control of the PMA1 promoter (49) was used as starting point. The modified cDNA encodes the AHA2 PM H+-ATPase with a C-terminal deletion of 73 amino acid residues and the insertion of a C-terminal Met-Arg-Gly-Ser-His6 (MRGSH6) tag (aha2Δ73-6xHis) (10), in here denoted wild type. The C-terminal deletion renders the PM H+-ATPase constitutively active, while the addition of the His6-tag allows for affinity purification of the constructed mutants. Site-directed mutagenesis was performed by standard procedures using polymerase chain reaction, and all mutated sequences were verified by DNA sequencing.
Expression of PM H+-ATPases in Yeast.
The S. cerevisiae strain RS-72 (50) was transformed and cultured essentially as described previously (51). In RS-72 (MATa ade1-100 his4-519 leu2-3,112), the natural constitutive promoter of the yeast endogenous PM H+-ATPase PMA1 has been replaced by the galactose-dependent GAL1 promoter (50).
Protein Purification.
Plasma membrane enriched fractions from yeast expressing either full-length AHA2 (i.e., including the C-terminal 73 residues) or an empty vector were purified as previously described (19). The PM H+-ATPases was purified by membrane solubilization using n-dodecyl-β-D-maltoside and Ni2+-affinity chromatography according to established procedures (6).
Yeast Complementation Assay.
Yeast complementation assay was performed on solid growth media (pH 5.5) as previously described (19), except that various concentration of Ca2+ (0–20 mM) was included.
Crystallographic Identification of Cation Binding Sites.
For the Ho3+ and Tb3+ bound form, protein crystals were grown as described previously (6) and then soaked with 10 mM HoCl3 or 5 mM TbCl3. For the Rb+ bound form, the protein was dialyzed by 50 mM RbCl, 50 mM MES pH 6.5, 10% sucrose and 1xCMC C12E8. 5 mM Mg-AMPPCP was added to the dialyzed protein, which was then centrifuged (70.000 rpm 10 min) and used directly at 4 °C in hanging drops by mixing equal volumes of protein and crystallization buffer [29% polyethylene glycol-400, 100 mM RbCl, 100 mM MES 6.0, 5% sucrose (w/v)]. Large, single crystals grew over a week and were dehydrated using the protocol previously established (6) and flash-frozen in liquid nitrogen. All diffraction data were collected at the beamline X06SA at the Swiss Light Source and were processed and merged using XDS and analyzed using programs from the CCP4 package.
Enzyme Activity Measurements.
ATPase activity was determined using either plasma membrane enriched fractions or purified protein diluted in a reactivation buffer as described (10). Varying concentrations of CaCl2 (0–10 mM), TbCl3 (0–1 mM), or HoCl3 (0–630 μM) were included as indicated. Formation of the phosphorylated intermediate was performed as described (10).
Supplementary Material
Acknowledgments.
We thank the staff of X06SA at the Swiss Light Source for assistance with synchrotron data collection.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1010416107/-/DCSupplemental.
References
- 1.Møller JV, Juul B, le Maire M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biochim Biophys Acta. 1996;1286:1–51. doi: 10.1016/0304-4157(95)00017-8. [DOI] [PubMed] [Google Scholar]
- 2.Morth JP, et al. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- 3.Shinoda T, Ogawa H, Cornelius F, Toyoshima C. Crystal structure of the sodium-potassium pump at 24 Å resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- 4.Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- 5.Sørensen TL, Møller JV, Nissen P. Phosphoryl transfer and calcium ion occlusion in the calcium pump. Science. 2004;304:1672–1675. doi: 10.1126/science.1099366. [DOI] [PubMed] [Google Scholar]
- 6.Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- 7.Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- 8.Olesen C, Sørensen TL, Nielsen RC, Møller JV, Nissen P. Dephosphorylation of the calcium pump coupled to counterion occlusion. Science. 2004;306:2251–2255. doi: 10.1126/science.1106289. [DOI] [PubMed] [Google Scholar]
- 9.Olesen C, et al. The structural basis of calcium transport by the calcium pump. Nature. 2007;450:1036–1042. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- 10.Buch-Pedersen MJ, Venema K, Serrano R, Palmgren MG. Abolishment of proton pumping and accumulation in the E1P conformational state of a plant plasma membrane H+-ATPase by substitution of a conserved aspartyl residue in transmembrane segment 6. J Biol Chem. 2000;275:39167–39173. doi: 10.1074/jbc.M007537200. [DOI] [PubMed] [Google Scholar]
- 11.Kühlbrandt W, Zeelen J, Dietrich J. Structure, mechanism, and regulation of the Neurospora plasma membrane H+-ATPase. Science. 2002;297:1692–1696. doi: 10.1126/science.1072574. [DOI] [PubMed] [Google Scholar]
- 12.Ronjat M, Lacapere JJ, Dufour JP, Dupont Y. Study of the nucleotide binding site of the yeast Schizosaccharomyces pombe plasma membrane H+-ATPase using formycin triphosphate-terbium complex. J Biol Chem. 1987;262:3146–3153. [PubMed] [Google Scholar]
- 13.Kinoshita T, Nishimura M, Shimazaki K. Cytosolic Concentration of Ca2+ Regulates the plasma membrane H+-ATPase in guard cells of fava bean. Plant Cell. 1995;7:1333–1342. doi: 10.1105/tpc.7.8.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Briskin DP, Gawienowski MC. Role of the plasma membrane H+-ATPase in K+ transport. Plant Physiol. 1996;111:1199–1207. doi: 10.1104/pp.111.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buch-Pedersen MJ, Rudashevskaya EL, Berner TS, Venema K, Palmgren MG. Potassium as an intrinsic uncoupler of the plasma membrane H+-ATPase. J Biol Chem. 2006;281:38285–38292. doi: 10.1074/jbc.M604781200. [DOI] [PubMed] [Google Scholar]
- 16.Pidcock E, Moore GR. Structural characteristics of protein binding sites for calcium and lanthanide ions. J Biol Inorg Chem. 2001;6:479–489. doi: 10.1007/s007750100214. [DOI] [PubMed] [Google Scholar]
- 17.Portillo F, de Larrinoa IF, Serrano R. Deletion analysis of yeast plasma membrane H+-ATPase and identification of a regulatory domain at the carboxyl-terminus. FEBS Lett. 1989;247:381–385. doi: 10.1016/0014-5793(89)81375-4. [DOI] [PubMed] [Google Scholar]
- 18.Palmgren MG, Sommarin M, Serrano R, Larsson C. Identification of an autoinhibitory domain in the C-terminal region of the plant plasma membrane H+-ATPase. J Biol Chem. 1991;266:20470–20475. [PubMed] [Google Scholar]
- 19.Ekberg K, Palmgren MG, Veierskov B, Buch-Pedersen MJ. A novel mechanism of P-type ATPase autoinhibition involving both termini of the protein. J Biol Chem. 2010;285:7344–7350. doi: 10.1074/jbc.M109.096123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lanyi JK. Proton transfers in the bacteriorhodopsin photocycle. Biochim Biophys Acta. 2006;1757:1012–1018. doi: 10.1016/j.bbabio.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Wikström M, Verkhovsky MI. Mechanism and energetics of proton translocation by the respiratory heme-copper oxidases. Biochim Biophys Acta. 2007;1767:1200–1214. doi: 10.1016/j.bbabio.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 22.Di Cera E. A structural perspective on enzymes activated by monovalent cations. J Biol Chem. 2006;281:1305–1308. doi: 10.1074/jbc.R500023200. [DOI] [PubMed] [Google Scholar]
- 23.Page MJ, Di Cera E. Role of Na+ and K+ in enzyme function. Physiol Rev. 2006;86:1049–1092. doi: 10.1152/physrev.00008.2006. [DOI] [PubMed] [Google Scholar]
- 24.Schack VR, et al. Identification and function of a cytoplasmic K+ site of the Na+, K+ -ATPase. J Biol Chem. 2008;283:27982–27990. doi: 10.1074/jbc.M803506200. [DOI] [PubMed] [Google Scholar]
- 25.Leigh RA. Potassium homeostasis and membrane transport. J Plant Nutr Soil Sc. 2001;164:193–198. [Google Scholar]
- 26.Vazquez-Ibar JL, Weinglass AB, Kaback HR. Engineering a terbium-binding site into an integral membrane protein for luminescence energy transfer. Proc Natl Acad Sci USA. 2002;99:3487–3492. doi: 10.1073/pnas.052703599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cha A, Snyder GE, Selvin PR, Bezanilla F. Atomic scale movement of the voltage-sensing region in a potassium channel measured via spectroscopy. Nature. 1999;402:809–813. doi: 10.1038/45552. [DOI] [PubMed] [Google Scholar]
- 28.Highsmith SR, Head MR. Tb3+ binding to Ca2+ and Mg2+ binding sites on sarcoplasmic reticulum ATPase. J Biol Chem. 1983;258:6858–6862. [PubMed] [Google Scholar]
- 29.Squier TC, Bigelow DJ, Fernandez-Belda FJ, deMeis L, Inesi G. Calcium and lanthanide binding in the sarcoplasmic reticulum ATPase. J Biol Chem. 1990;265:13713–13720. [PubMed] [Google Scholar]
- 30.Henao F, Orlowski S, Merah Z, Champeil P. The metal sites on sarcoplasmic reticulum membranes that bind lanthanide ions with the highest affinity are not the ATPase Ca2+ transport sites. J Biol Chem. 1992;267:10302–10312. [PubMed] [Google Scholar]
- 31.Farley RA, et al. Site-directed mutagenesis of the sodium pump: analysis of mutations to amino acids in the proposed nucleotide binding site by stable oxygen isotope exchange. Biochemistry. 1997;36:941–951. doi: 10.1021/bi962153y. [DOI] [PubMed] [Google Scholar]
- 32.Clarke DM, Loo TW, MacLennan DH. Functional consequences of alterations to amino acids located in the nucleotide binding domain of the Ca2+-ATPase of sarcoplasmic reticulum. J Biol Chem. 1990;265:22223–22227. [PubMed] [Google Scholar]
- 33.Inesi G, Ma H, Lewis D, Xu C. Ca2+ occlusion and gating function of Glu309 in the ADP-fluoroaluminate analog of the Ca2+-ATPase phosphoenzyme intermediate. J Biol Chem. 2004;279:31629–31637. doi: 10.1074/jbc.M403211200. [DOI] [PubMed] [Google Scholar]
- 34.Ma H, Lewis D, Xu C, Inesi G, Toyoshima C. Functional and structural roles of critical amino acids within the “N”, “P”, and “A” domains of the Ca2+ ATPase (SERCA) headpiece. Biochemistry. 2005;44:8090–8100. doi: 10.1021/bi050332m. [DOI] [PubMed] [Google Scholar]
- 35.Jacobsen MD, Pedersen PA, Jorgensen PL. Importance of Na,K-ATPase residue α1-Arg544 in the segment Arg544-Asp567 for high-affinity binding of ATP, ADP, or MgATP. Biochemistry. 2002;41:1451–1456. doi: 10.1021/bi015891h. [DOI] [PubMed] [Google Scholar]
- 36.Ridder IS, Dijkstra BW. Identification of the Mg2+-binding site in the P-type ATPase and phosphatase members of the HAD (haloacid dehalogenase) superfamily by structural similarity to the response regulator protein CheY. Biochem J. 1999;339(Pt 2):223–226. [PMC free article] [PubMed] [Google Scholar]
- 37.Girardet JL, Bally I, Arlaud G, Dupont Y. Localization of a putative magnesium-binding site within the cytoplasmic domain of the sarcoplasmic reticulum Ca2+-ATPase. Eur J Biochem. 1993;217:225–231. doi: 10.1111/j.1432-1033.1993.tb18237.x. [DOI] [PubMed] [Google Scholar]
- 38.Pedersen PA, Jorgensen JR, Jorgensen PL. Importance of conserved α-subunit segment 709GDGVND for Mg2+ binding, phosphorylation, and energy transduction in Na, K-ATPase. J Biol Chem. 2000;275:37588–37595. doi: 10.1074/jbc.M005610200. [DOI] [PubMed] [Google Scholar]
- 39.Seto-Young D, Na S, Monk BC, Haber JE, Perlin DS. Mutational analysis of the first extracellular loop region of the H+-ATPase from Saccharomyces cerevisiae. J Biol Chem. 1994;269:23988–23995. [PubMed] [Google Scholar]
- 40.Albers RW. Biochemical aspects of active transport. Annu Rev Biochem. 1967;36:727–756. doi: 10.1146/annurev.bi.36.070167.003455. [DOI] [PubMed] [Google Scholar]
- 41.Post RL, Hegyvary C, Kume S. Activation by adenosine triphosphate in the phosphorylation kinetics of sodium and potassium ion transport adenosine triphosphatase. J Biol Chem. 1972;247:6530–6540. [PubMed] [Google Scholar]
- 42.Clausen JD, Andersen JP. Glutamate 90 at the luminal ion gate of sarcoplasmic reticulum Ca2+-ATPase is critical for Ca2+ binding on both sides of the membrane. J Biol Chem. 2010;285:20780–20792. doi: 10.1074/jbc.M110.116459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown LS, et al. Glutamic acid 204 is the terminal proton release group at the extracellular surface of bacteriorhodopsin. J Biol Chem. 1995;270:27122–27126. doi: 10.1074/jbc.270.45.27122. [DOI] [PubMed] [Google Scholar]
- 44.Dioumaev AK, et al. Existence of a proton transfer chain in bacteriorhodopsin: Participation of Glu-194 in the release of protons to the extracellular surface. Biochemistry. 1998;37:2496–2506. doi: 10.1021/bi971842m. [DOI] [PubMed] [Google Scholar]
- 45.Phatak P, Ghosh N, Yu H, Cui Q, Elstner M. Amino acids with an intermolecular proton bond as proton storage site in bacteriorhodopsin. Proc Natl Acad Sci USA. 2008;105:19672–19677. doi: 10.1073/pnas.0810712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nieboer E. The lanthanide ions as structural probes in biological and model systems. Struct Bond. 1975;22:1–47. [Google Scholar]
- 47.Martin RB, Richardson FS. Lanthanides as probes for calcium in biological systems. Q Rev Biophys. 1979;12:181–209. doi: 10.1017/s0033583500002754. [DOI] [PubMed] [Google Scholar]
- 48.Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- 49.Villalba JM, Palmgren MG, Berberian GE, Ferguson C, Serrano R. Functional expression of plant plasma membrane H+-ATPase in yeast endoplasmic reticulum. J Biol Chem. 1992;267:12341–12349. [PubMed] [Google Scholar]
- 50.Cid A, Perona R, Serrano R. Replacement of the promoter of the yeast plasma membrane ATPase gene by a galactose-dependent promoter and its physiological consequences. Curr Genet. 1987;12:105–110. doi: 10.1007/BF00434664. [DOI] [PubMed] [Google Scholar]
- 51.Regenberg B, Villalba JM, Lanfermeijer FC, Palmgren MG. C-terminal deletion analysis of plant plasma membrane H+-ATPase: Yeast as a model system for solute transport across the plant plasma membrane. Plant Cell. 1995;7:1655–1666. doi: 10.1105/tpc.7.10.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.