Abstract
Here, we present estimates of heritability and selection on network traits in a single population, allowing us to address the evolutionary potential of social behavior and the poorly understood link between sociality and fitness. To evolve, sociality must have some heritable basis, yet the heritability of social relationships is largely unknown. Recent advances in both social network analyses and quantitative genetics allow us to quantify attributes of social relationships and estimate their heritability in free-living populations. Our analyses addressed a variety of measures (in-degree, out-degree, attractiveness, expansiveness, embeddedness, and betweenness), and we hypothesized that traits reflecting relationships controlled by an individual (i.e., those that the individual initiated or were directly involved in) would be more heritable than those based largely on the behavior of conspecifics. Identifying patterns of heritability and selection among related traits may provide insight into which types of relationships are important in animal societies. As expected, we found that variation in indirect measures was largely explained by nongenetic variation. Yet, surprisingly, traits capturing initiated interactions do not possess significant additive genetic variation, whereas measures of received interactions are heritable. Measures describing initiated aggression and position in an agonistic network are under selection (0.3 < |S| < 0.4), although advantageous trait values are not inherited by offspring. It appears that agonistic relationships positively influence fitness and seemingly costly or harmful ties may, in fact, be beneficial. Our study highlights the importance of studying agonistic as well as affiliative relationships to understand fully the connections between sociality and fitness.
Keywords: animal model, animal social networks, yellow-bellied marmots
Behavioral ecologists have long viewed sociality and social relationships as adaptive traits shaped by evolution (1, 2). However, if we are to study the evolution of sociality and social relationships, there must be heritable variation in traits describing individual social behavior. Numerous studies have identified heritable variation in animal dispositions (3, 4), morphological characteristics, and behavioral traits (5) that may affect how individuals interact with conspecifics, yet the role of genetics in social interactions themselves is poorly understood. If traits affecting social interactions are heritable, we may expect measures of social relationships to be explained somewhat by additive genetic factors.
There has been a recent upsurge in using animal social networks as tools for studying the ecology, evolution, and adaptive significance of sociality (6–8). Networks are based on interactions between individuals, and a variety of measures have been developed to quantify how connected individuals are with others in the group (9). Although studies of nonhuman species have explored the development of social networks (10) as well as the causes (11–13) and consequences (14–16) of network structure and individual position, no study has addressed the heritability of social network traits. If networks are to be useful tools for studying the evolution and maintenance of sociality, there must be heritable variation in network parameters.
To our knowledge, the only previous quantitative genetic study of social network traits was conducted in humans (17); that study reported sizeable heritabilities for the number of times an individual was named as a friend by others (in-degree, h2 = 0.46), the likelihood that friends of the individual were connected to each other (transitivity or clustering coefficient, h2 = 0.47), and the proportion of connections between individuals in the network that pass through the individual (betweenness, h2 = 0.29), but it but found no evidence for additive genetic variation in how many friends a person named (out-degree). The authors proposed an attract and induce model of network formation in which additive genetic variation in initiated and received behaviors was required to generate heritabilities similar to real-life estimates. This approach refuted previous models of human social network formation based on a single interaction type, suggesting that networks cannot evolve from initiated or received interactions alone (18).
We wished to understand the genetic architecture of social relationships further as well as the poorly understood link between sociality and fitness (2), in a population of free-living nonhuman mammals. Furthermore, we wished to identify patterns among similar traits in an attempt to discover which interaction types are important in animal social networks. Previous studies of yellow-bellied marmot (Marmota flaviventris) social networks have addressed the robustness of network estimates (7), the correlations between network position and dispersal (15), and the roles of age and kinship in structuring networks (10); however, the genetic basis and fitness consequences of network traits are unknown.
We used social networks constructed from affiliative and agonistic interactions separately to estimate the heritability and fitness consequences of social network traits in marmots. Networks consisted of nodes (in this case, individual marmots) connected by binary links (i.e., 1/0 indicating the presence or absence of interactions between individuals). In our study, out-degree is the number of social partners toward which an individual initiates interactions, whereas in-degree represents the number of social partners from which an individual receives interactions. Expansiveness and attractiveness are similar to out-degree and in-degree but control for network density and reciprocity; these measures reflect an individual's tendency to initiate and receive interactions relative to the tendencies of other individuals in the network. We included two additional measures capturing more complex information about network position and integration: betweenness (reflecting an individual's importance as a connection point) and embeddedness (a measure of social cohesion) (19). These measures are partially based on the relationships among an individual's connections, and such links are known as indirect interactions (7). We categorized network measures in terms of their directionality (initiated vs. received), type (direct vs. indirect), and nature (affiliative vs. agonistic) and predicted that similar traits would have similar selective pressures, and therefore comparable heritabilities and selection differentials. We hypothesized that social interactions controlled by an individual (i.e., those that are initiated or directly involve the focal animal) would be more heritable than those determined by the behavior of conspecifics, because such traits would be less determined by environmental variation. Although this line of thought [specifically hypothesis (i) as stated below] contradicts results from the previously discussed human network study, we believed that individual control would be important for explaining additive genetic variation in marmot social networks. We therefore predicted that (i) measures capturing an animal's tendency to initiate (i.e., out-degree, expansiveness) interactions are more heritable than those capturing an animal's tendency to receive (i.e., in-degree, attractiveness) interactions and (ii) measures based on direct interactions (i.e., out-degree, expansiveness, in-degree, attractiveness) are more heritable than those based on indirect (or a mixture of direct and indirect) interactions (i.e., betweenness, embeddedness).
We tested these hypotheses by estimating the heritabilities of six social network statistics based on affiliative and agonistic interactions (for a total of 12 measures, Table 1) to understand the role of each trait category in marmot networks. Furthermore, we quantified the magnitude of selection for all traits and examined correlations (genetic and phenotypic) that may explain behavioral patterns. Here, we decompose social behavior and address the heritability and fitness consequences of its individual parts.
Table 1.
Variance estimates and selection differentials for agonistic and affiliative social network measures
| Proportion of variance |
LRS |
Longevity |
|||||||||
| Trait | Description | Mean (SD) | N1 | Additive genetic | Permanent environment | Social group | N2 | S | C | S | C |
| Agonistic | |||||||||||
| Out-degree | Direct, initiated | 2.030 (2.922) | 367 (152) | 0.017 (0.050) | 0.257 (0.074) | 0.078 (0.040) | 256 (118) | 0.401 (0.082) | −0.003 (0.001) | 0.159 (0.069) | −0.003 (0.001) |
| In-degree | Direct, received | 1.256 (1.393) | 367 (152) | 0.112 (0.068) | 0.033 (0.061) | 0.161 (0.049) | 256 (118) | 0.07 (0.066) | −0.003 (0.003) | −0.006 (0.054) | 0 (0.002) |
| Expansiveness | Direct, initiated | 0.253 (1.250) | 214 (102) | 0 | 0.100 (0.081) | 0.033 (0.060) | 136 (90) | 0.374 (0.091) | −0.006 (0.006) | 0.189 (0.069) | 0.002 (0.005) |
| Attractiveness | Direct, received | −0.154 (0.815) | 217 (107) | 0.177 (0.093) | 0 | 0 | 143 (92) | −0.243 (0.07) | −0.019 (0.009) | −0.039 (0.057) | −0.004 (0.007) |
| Betweenness | Indirect | 3.133 (6.708) | 367 (152) | 0 | 0.130 (0.053) | 0.019 (0.033) | 256 (118) | 0.322 (0.075) | −0.001 (0.001) | 0.177 (0.066) | 0 (0.001) |
| Embeddedness | Direct and indirect | 1.711 (1.446) | 367 (152) | 0.066 (0.065) | 0.155 (0.070) | 0.233 (0.057) | 256 (118) | 0.302 (0.079) | −0.001 (0.002) | 0.084 (0.063) | −0.004 (0.002) |
| Affiliative | |||||||||||
| Out-degree | Direct, initiated | 2.589 (2.830) | 367 (152) | 0.080 (0.070) | 0.240 (0.080) | 0.030 (0.030) | 256 (118) | 0.263 (0.078) | −0.002 (0.001) | 0.039 (0.065) | −0.001 (0.001) |
| In-degree | Direct, received | 2.447 (2.391) | 367 (152) | 0.106 (0.065) | 0.085 (0.069) | 0.054 (0.037) | 256 (118) | 0.191 (0.07) | −0.005 (0.001) | 0.066 (0.055) | 0 (0.001) |
| Expansiveness | Direct, initiated | −0.072 (1.263) | 268 (120) | 0 | 0.037 (0.058) | 0.153 (0.073) | 179 (102) | 0.111 (0.073) | 0 (0.005) | 0.016 (0.063) | −0.003 (0.004) |
| Attractiveness | Direct, received | −0.185 (1.184) | 268 (107) | 0.078 (0.058) | 0 | 0.067 (0.060) | 170 (100) | −0.081 (0.079) | −0.001 (0.006) | −0.039 (0.062) | 0.009 (0.004) |
| Betweenness | Indirect | 2.301 (4.726) | 367 (152) | 0 | 0.136 (0.054) | 0.082 (0.039) | 256 (118) | 0.218 (0.073) | −0.002 (0.001) | 0.039 (0.062) | 0 (0.001) |
| Embeddedness | Direct and indirect | 2.371 (2.292) | 367 (152) | 0.087 (0.067) | 0.163 (0.074) | 0.110 (0.046) | 256 (118) | −0.12 (0.064) | 0.002 (0.002) | −0.006 (0.051) | 0.003 (0.001) |
Each network trait was classified as direct (measure based solely on interactions including the individual) or indirect (measure based on interactions of others, the individual's position in the network, or attributes of the total network) and as initiated or received. Univariate animal models were used to estimate the proportion of variation (±SD) attributed to additive genetic factors (h2), permanent environment, and social group using a sample size of N1 [total number of observations (number of unique individuals)]. Directional (S) and nonlinear (C) selection differentials were estimated in bivariate animal models of sample size N2. Bivariate models included LRS and longevity as fitness proxies. All significant results (P > 0.05) are indicated in bold.
Results
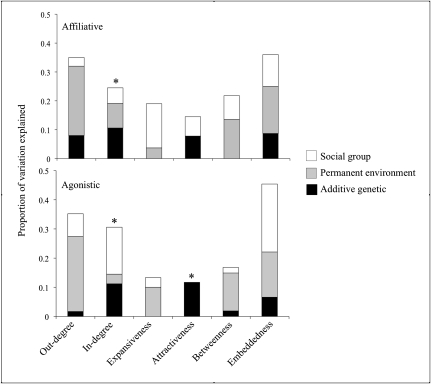
Contrary to our first hypothesis, network traits capturing initiated interactions did not possess significant additive genetic variation (Table 1), although three (agonistic in-degree and attractiveness, affiliative in-degree) of the four received measures were moderately (h2 < 0.2) heritable. These heritable received measures represent a subset of the measures based on direct interactions (3 of 8). Although the majority of network traits describing direct interactions lack significant additive genetic variance, none of the measures describing indirect interactions were found to be heritable. In our study, social network measures based on direct interactions tend to be more heritable than those based on indirect interactions. Furthermore, variation in measures of indirect interactions is largely explained by permanent environment and social group, whereas these random factors play a less consistent role among measures of direct interactions (Fig. 1).
Fig. 1.
Proportion of variation explained by social group, permanent environment, and additive genetic factors (h2) for agonistic and affiliative social network measures. All estimates are from univariate animal models. Significant (P < 0.05) heritabilities are denoted by an asterisk.
The majority of affiliative and agonistic network traits are phenotypically, although not genetically, correlated. Only agonistic in-degree and affiliative in-degree were genetically correlated [rG = 0.959 ± 0.102 SE, log-likelihood ratio test (LRT) = 3.86, P = 0.049]. There were significant phenotypic correlations between agonistic and affiliative out-degree (rP = 0.689 ± 0.029 SE, LRT = 16.43, P < 0.001), in-degree (rP = 0.570 ± 0.040 SE, LRT = 18.02, P < 0.001), betweenness (rP = 0.444 ± 0.042 SE, LRT = 7.36, P = 0.001), and embeddedness (rP = 0.648 ± 0.033 SE, LRT = 18.25, P < 0.001).
We found evidence of directional selection in the majority (8 of 12) of social network measures (Table 1). Stabilizing selection was less common (6 of 12) and was consistently lower in magnitude (all |C| < 0.01) than estimates of directional selection. Significant directional selection differentials for lifetime reproductive success (LRS) tended to be more common (LRS, 8 of 12; longevity, 3 of 12) and larger in magnitude (all |S| > 0.19) than those observed for longevity (all |S| < 0.19). Both initiated agonistic measures are under moderate selection (|S| > 0.37), although neither is heritable. Affiliative out-degree and in-degree are under some selection, although only in-degree is heritable, and consequently possesses some evolutionary potential. Individuals with high agonistic betweenness, agonistic embeddedness, and affiliative embeddedness have increased reproductive success (all |S| > 0.30); however, neither betweenness nor embeddedness is heritable in agonistic or affiliative networks.
Discussion
We hypothesized that social network measures capturing interactions controlled by an individual (direct and initiated) would be more heritable than measures based on indirect and received interactions. This rationale was confirmed in our second prediction (ii); measures of indirect interactions, which depend on the behavior of other group members, were not heritable. Interestingly, our first prediction (i) was strongly refuted; the majority of received (and both received agonistic) measures were heritable, whereas none of the initiated measures were explained by additive genetic variation. These results clearly refute the attract and induce model of human network formation (17), which requires additive genetic variation in measures of initiated and received interactions to explain real-life heritability estimates. Clearly, more studies addressing the quantitative genetics of social networks are needed to understand trends across species. In our study, patterns among agonistic traits may provide insight into the role of aggressive interactions in marmot societies.
Sociality should evolve when the benefits of group living (e.g., protection from predators, mating opportunities) exceed the costs [e.g., resource competition, exposure to disease (2, 20)]. However, group living inevitably leads to competition over limited resources (e.g., mates, habitat, food), and variation in aggressive behavior may therefore influence fitness (when more competitive individuals gain access to resources). Assuming that additive genetic variation is present, agonistic behavior should evolve and spread via natural selection. In yellow-bellied marmots, traits describing individual initiated agonistic relationships are not heritable. These traits are, however, under strong selection (longevity, S > 0.15; LRS, S > 0.37), and we expect such fitness-related traits to be rapidly fixed in the population (21). Thus, the lack of additive genetic variation in initiated aggression may suggest a crucial role for agonistic behavior in marmot societies. In fact, dominant individuals obtain increased access to high-quality burrows and foraging sites (22) as well as reproductive opportunities (23); furthermore, female marmots classified as aggressive produce more offspring than those classified as having a social or submissive-avoider behavioral phenotype (24). It appears that within-group competition is strong in marmot societies; initiated agonistic behavior therefore has profound fitness consequences, and additive genetic variation was likely depleted by past selection.
Interestingly, we found stronger overall selection on LRS than longevity for traits describing initiated agonistic interactions. Marmots initiating agonistic interactions with more social partners have increased longevity, but individual survival is largely unaffected by received agonistic interactions (as measured by in-degree and attractiveness) or affiliative relationships. These results may differ in more cooperative species or in those that form coalitions, because affiliative interactions may have a profound impact on survival in such societies. Future studies could address the fitness consequences (longevity vs. LRS) of social interactions in other types of animal societies to clarify this issue.
Although we may expect genetic variation in received aggression to influence fitness similarly and be depleted by selection, agonistic attractiveness (h2 = 0.177) and in-degree (h2 = 0.112) are heritable. Furthermore, individuals receiving more agonistic interactions (relative to others in the network) have decreased LRS (S = −0.243), and agonistic attractiveness may therefore evolve. Heritability in received aggression traits may be linked to additive genetic variation in dominance rank (25, 26) or personality (3, 4), with animals of low rank or certain dispositions easily victimized. We must also note the substantial (R2 = 0.959) genetic correlation between agonistic and affiliative in-degree. The magnitude of this genetic correlation is surprising because we expect opposing selection pressures for agonistic and affiliative traits. However, we found no evidence for fitness consequences associated with agonistic in-degree, and change in this trait will likely result from directional selection on affiliative in-degree.
Although we report only one significant genetic correlation, we found a tendency for agonistic and affiliative network measures (specifically in-degree, out-degree, betweenness, and embeddedness) to be correlated at the phenotypic level. In other words, individuals involved in aggressive interactions with many individuals also socialize affiliatively with numerous conspecifics. Furthermore, marmots that are well connected and integrated in affiliative networks are also well connected and integrated in agonistic networks. Although affiliative and agonistic betweenness and embeddedness are not genetically correlated, certain individuals play more crucial roles in their group than others and this tendency is phenotypically expressed across network types.
In addition, we found embeddedness and betweenness (for both affiliative and agonistic networks) to covary with LRS and longevity, although these measures are not heritable and unlikely to evolve. It has long been assumed that, for social animals, interactions with conspecifics somehow influence reproductive success; however, the links between social relationships and fitness are still largely unresolved (2). In savanna baboons (Papio cynocephalus), infants of socially integrated female baboons (as measured by a composite index reflecting the proportion of time spent engaged in social behaviors) are more likely to survive (27), and it appears that social centrality and integration influence marmot fitness as well. Although anthropologists have long acknowledged an important role for social relationships in primates (28), rodents and nonprimate mammals have received less attention (cf. 29). Our results suggest that social integration and network position may have reproductive consequences in a wider range of species, including those with rudimentary sociality (30).
Furthermore, we present evidence for selection on social group position in agonistic networks. By constructing affiliative and agonistic networks separately, we were able to tease apart fitness consequences associated with each interaction type. This approach led us to a unique view of agonistic relationships, namely, that they are somehow beneficial. Previous studies linking social bonds and reproductive success have primarily focused on affiliative interactions (2), because such relationships have more immediate and obvious benefits [e.g., reduced stress (31, 32), enhanced immune competence (33)]. However, we found that the extent to which a marmot is integrated and centrally positioned in its agonistic network positively covaries with LRS and longevity. It is possible that a central and integrated position in the social group (which presumably results in more affiliative and agonistic partners and interactions) produces more benefits than the costs associated with the aggressive interactions themselves. Additionally, aggressive interactions in marmot societies may function to structure dominance hierarchies or maintain social order rather than seriously injuring or affecting recipients of aggressive interactions. We encourage future studies of adaptive social relationships to consider all interaction types, because seemingly costly or harmful ties may, in fact, benefit individual fitness.
Materials and Methods
Marmots were studied under research protocol ARC 2001-191-01 as well as permits issued by the Colorado Division of Wildlife. The research protocol was approved by the University of California Los Angeles Animal Care Committee on May 13, 2002, and was renewed annually. All subjects were members of a natural population living in and around the Rocky Mountain Biological Laboratory (RMBL; 38° 57′ North, 106° 59′ West), Gunnison County, CO. This population has been well studied since 1962 (34), and individuals remaining in the study area are live-trapped, individually marked (35), and closely monitored during their active season.
Marmots were observed several times a week over the 6-y period (total hours watched: 2003 = 698.48 h, 2004 = 783.76 h, 2005 = 775.04 h, 2006 = 847.21 h, 2007 = 1019.60 h, 2008 = 720.61 h) during times of peak activity (0700–1000 h and 1600–1900 h) from a distance that did not obviously affect the animals’ behavior (20–150 m depending on habitat features). Social interactions between identified individuals were recorded using a detailed ethogram (15). Interactions were classified as affiliative (i.e., allogrooming, forage together within 1 m, greet, sit <1 m apart, play, sniff anogenital region) or agonistic (i.e., aggression, displacement) for analyses, with a small number of ambiguous interactions excluded from the dataset. Rare interactions between individuals who did not frequently associate were retained in the dataset. Although some studies of fission-fusion societies filter social relationships based on the number of observations (36), we believed rare interactions accurately reflected real relationships in marmot networks, which are geographically delimited and stable within a year. Eliminating weak relationships would potentially bias networks and discount the significance of infrequent social interactions.
Social networks were constructed in UCINET (37), and network measures were calculated using UCINET and the graphing software igraph (38) in the program R (version 2.10.1; R Development Core Team). We defined social networks as four geographically distinct colonies and determined yearly network membership from observations and trapping. Only individuals seen in the colony more than five times that year were included in network analysis (i.e., transient animals and their interactions were left out). Furthermore, we defined social networks annually because colony membership changes from year to year. Networks incorporated interactions between yearling and adult marmots; however, quantitative genetic analyses focused on adults (i.e., animals >2 y old), because the consistency of behavioral types is known to increase with age (39).
In total, network statistics were calculated for 24 social group-years over a 6-y period (2003–2008). Each social group represented a discrete network, and 12 measures were calculated for each member of the social group. Out-degree, in-degree, expansiveness, and attractiveness were based on directed networks (i.e., interactions have an initiator and recipient), whereas betweenness and embeddedness were calculated based on symmetrical networks (i.e., interactions are present or absent, with no directionality). Detailed information on these measures and how they are related to more traditional methods of quantifying social behavior (i.e., number and rate of interactions) is available in SI Text and elsewhere [degree and betweenness (9, 40, 41), expansiveness and attractiveness (42, 43), and embeddedness (15, 19)].
For all quantitative genetic analyses, we used a previously published (44) pedigree based on microsatellite genotypes and likelihood parentage assignment methods. We focused genetic analyses on a subset of individuals included in the pedigree for whom social network measures were available (sample sizes provided in Table 1). Heritabilities, genetic correlations, and fitness consequences were estimated using restricted maximum likelihood animal models (45, 46) in the program ASReml (47).
For each social network measure, we created separate univariate models, including fixed effects when significant, of social group size (for all traits except agonistic attractiveness, affiliative attractiveness, and affiliative expansiveness) and gender (for, agonistic out-degree expansiveness, and betweenness as well as affiliative out-degree, expansiveness, attractiveness, and betweenness); random effects were added to the model in an additive stepwise manner and evaluated with LRTs (df = 1). We examined random effects of permanent environment, maternal and paternal environment, paternal and maternal genetics, and social group (which accounts for year and location), with environmental effects retained in the model even when nonsignificant (48). Final models included random effects of social group, permanent environment, and individual identity (i.e., additive genetic) because these were found to explain significant variation in social network traits (Table 1). Because the phenotype of a given individual is invariably affected by other members of the network, social group was also retained as a random effect in all animal model analyses to control for nonindependence and the clustering of network measures. We estimated the proportion of variance explained by each random factor by dividing the variance from each random term by the total phenotypic variance (conditional on fixed effects). The identification and inclusion of fixed and random effects are therefore extremely important for accurately calculating heritability, because the estimate is dependent on the amount of additive genetic variance as well as variance explained by fixed effects (49). Heritability estimates are also especially vulnerable to the possibility that phenotypic similarities among relatives reflect shared environments rather than additive genetic variation (48). We have therefore tested and accounted for such common environment effects (i.e., social group, maternal and paternal environment) according to the method of Kruuk and Hadfield (48) and are confident that our heritability estimates truly represent genetic effects.
For each network statistic, we used bivariate models (with fixed and random effects from univariate models) to estimate the genetic and phenotypic covariance between agonistic and affiliative measures. The significance of each covariance estimate was assessed using the LRT, in which a model with the covariance term was compared with a model with the covariance term constrained to 0. Correlations were calculated by dividing the phenotypic [COVP(XY)] and genetic [COVA(XY)] covariances between the measured traits by the product of the single-trait SDs (VPX and VPY) for phenotypic and genetic variance [rP = COVP(XY)/√VPXVPY and rA = COVA(XY)/√VPXVPY].
We used measures of LRS and longevity to assess the fitness consequences of each social network trait. LRS and longevity were calculated only for deceased individuals so as to avoid the problem of censored data. LRS was calculated directly from the pedigree, and longevity estimates were based on observation and trapping records; marmots at RMBL are extremely well trapped and observed, and individuals not sighted within a given year were therefore assumed to be dead (44).
We estimated linear (S) and nonlinear (C) standardized selection differentials for each measure using standard methods (50). S was estimated as the covariance between the measured trait and relative individual fitness. C was estimated as the covariance between relative individual fitness and the orthogonal quadratic estimate of the measured trait and represents the strength of stabilizing or disruptive selection independent of the effects of directional selection (50). Covariances were estimated in bivariate models, including the two fitness proxies as well as the measured trait (51). All bivariate models included a random effect of permanent environment as well as fixed effects from univariate models. Furthermore, both fitness proxies were corrected for gender to account for differential mortality and reproductive strategies among female and male marmots (52, 53). Covariance significance was assessed using the LRT statistic as described above.
Supplementary Material
Acknowledgments
We thank our many marmoteers for collecting data over the years, especially Lucretia Olson for help with genetic analyses. We also thank Jennifer Smith, Ferenc Jordán, and two astute reviewers for comments on previous drafts. This work was supported by the University of California Los Angeles Academic Senate and Division of Life Sciences, The National Geographic Society, and the National Science Foundation (Grant NSF-IDBR-0754247 to D.T.B. and Grant DBI 0242960, 0731346 to the Rocky Mountain Biological Laboratory).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009882107/-/DCSupplemental.
References
- 1.Alexander RD. The evolution of social behavior. Annu Rev Ecol Syst. 1974;5:25–38. [Google Scholar]
- 2.Silk JB. The adaptive value of sociality in mammalian groups. Philos Trans R Soc Lond B Biol Sci. 2007;362:539–559. doi: 10.1098/rstb.2006.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drent PJ, van Oers K, van Noordwijk AJ. Realized heritability of personalities in the great tit (Parus major) Proc Biol Sci. 2003;270:45–51. doi: 10.1098/rspb.2002.2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sinn DL, Apiolaza LA, Moltschaniwskyj NA. Heritability and fitness-related consequences of squid personality traits. J Evol Biol. 2006;19:1437–1447. doi: 10.1111/j.1420-9101.2006.01136.x. [DOI] [PubMed] [Google Scholar]
- 5.Mousseau TA, Roff DA. Natural selection and the heritability of fitness components. Heredity. 1987;59:181–197. doi: 10.1038/hdy.1987.113. [DOI] [PubMed] [Google Scholar]
- 6.Croft DP, James R, Krause J. Exploring Animal Social Networks. Princeton: Princeton Univ Press; 2008. [Google Scholar]
- 7.Wey TW, Blumstein DT, Shen W, Jordon F. Social network analysis of animal behaviour: A promising tool for the study of sociality. Anim Behav. 2008;75:333–344. [Google Scholar]
- 8.Sih A, Hanser SF, McHugh KA. Social network theory: New insights and issues for behavioral ecologists. Behav Ecol Sociobiol. 2009;63:975–988. [Google Scholar]
- 9.Wasserman S, Faust K. Social Network Analysis: Methods and Applications. New York: Cambridge Univ Press; 1994. [Google Scholar]
- 10.Wey TW, Blumstein DT. Social cohesion in yellow-bellied marmots is established through age and kin structuring. Anim Behav. 2010;79:1343–1352. [Google Scholar]
- 11.Lusseau D, Newman MEJ. Identifying the role that animals play in their social networks. Proc Biol Sci. 2004;271(Suppl 6):S477–S481. doi: 10.1098/rsbl.2004.0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pike TW, Samanta M, Lindström J, Royle NJ. Behavioural phenotype affects social interactions in an animal network. Proc Biol Sci. 2008;275:2515–2520. doi: 10.1098/rspb.2008.0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Madden JR, Drewe JA, Pearce GP, Clutton-Brock TH. The social network structure of a wild meerkat population: 2. Intragroup interactions. Behav Ecol Sociobiol. 2009;64:81–95. [Google Scholar]
- 14.McDonald DB. Predicting fate from early connectivity in a social network. Proc Natl Acad Sci USA. 2007;104:10910–10914. doi: 10.1073/pnas.0701159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Blumstein DT, Wey TW, Tang K. A test of the social cohesion hypothesis: Interactive female marmots remain at home. Proc Biol Sci. 2009;276:3007–3012. doi: 10.1098/rspb.2009.0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Voelkl B, Kasper C. Social structure of primate interaction networks facilitates the emergence of cooperation. Biol Lett. 2009;5:462–464. doi: 10.1098/rsbl.2009.0204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fowler JH, Dawes CT, Christakis NA. Model of genetic variation in human social networks. Proc Natl Acad Sci USA. 2009;106:1720–1724. doi: 10.1073/pnas.0806746106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jackson MO. Genetic influences on social network characteristics. Proc Natl Acad Sci USA. 2009;106:1687–1688. doi: 10.1073/pnas.0813169106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moody J, White DR. Social cohesion and embeddedness. Am Sociol Rev. 2003;68:103–127. [Google Scholar]
- 20.Krause J, Ruxton GD. Living in Groups. Oxford: Oxford Univ Press; 2002. [Google Scholar]
- 21.Kruuk LEB, et al. Heritability of fitness in a wild mammal population. Proc Natl Acad Sci USA. 2000;97:698–703. doi: 10.1073/pnas.97.2.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frase BA, Armitage KB. Foraging patterns of yellow-bellied marmots: Role of kinship and individual variability. Behav Ecol Sociobiol. 1984;16:1–10. [Google Scholar]
- 23.Armitage KB. Individuality, social behavior, and reproductive success in yellow-bellied marmots. Ecology. 1986;67:1186–1193. [Google Scholar]
- 24.Svendsen GE. Behavioral and environmental factors in the spatial distribution and population dynamics of a yellow-bellied marmot population. Ecology. 1974;55:760–771. [Google Scholar]
- 25.Moore AJ. The inheritance of social dominance, mating behavior and attractiveness to mates in male Nauphoeta cinerea. Anim Behav. 1990;39:388–397. [Google Scholar]
- 26.Nol E, Cheng K, Nichols C. Heritability and phenotypic correlations of behaviour and dominance rank of Japanese quail. Anim Behav. 1996;52:813–820. [Google Scholar]
- 27.Silk JB, Alberts SC, Altmann J. Social bonds of female baboons enhance infant survival. Science. 2003;302:1231–1234. doi: 10.1126/science.1088580. [DOI] [PubMed] [Google Scholar]
- 28.Cheney DL, Seyfarth RM, Smuts B. Social relationships and social cognition in nonhuman primates. Science. 1986;234:1361–1366. doi: 10.1126/science.3538419. [DOI] [PubMed] [Google Scholar]
- 29.Cameron EZ, Setsaas TH, Linklater WL. Social bonds between unrelated females increase reproductive success in feral horses. Proc Natl Acad Sci USA. 2009;106:13850–13853. doi: 10.1073/pnas.0900639106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson LE, Blumstein DT. Applying the coalitionary-traits metric: Sociality without cooperation in male yellow-bellied marmots. Behav Ecol. 2010;21:788–793. [Google Scholar]
- 31.Hennessy MB, Zate R, Maken DS. Social buffering of the cortisol response of adult female guinea pigs. Physiol Behav. 2008;93:883–888. doi: 10.1016/j.physbeh.2007.12.005. [DOI] [PubMed] [Google Scholar]
- 32.Wittig RM, et al. Focused grooming networks and stress alleviation in wild female baboons. Horm Behav. 2008;54:170–177. doi: 10.1016/j.yhbeh.2008.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaplan JR, et al. The relationship of agonistic and affiliative behavior patterns to cellular-mediated immune function among cynomolgus monkeys (Macaca fascicularis) living in unstable social groups. Am J Primatol. 1991;25:157–173. doi: 10.1002/ajp.1350250303. [DOI] [PubMed] [Google Scholar]
- 34.Ozgul A, et al. Phenotypic mechanisms underlying a demographic response to climate change. Nature. 2010;466:482–485. [Google Scholar]
- 35.Armitage KB. Yellow-Bellied Marmot. Boca Raton, FL: CRC Press; 1982. [Google Scholar]
- 36.Rubenstein DI, Sundaresan S, Fischoff I, Saltz D. In: Exploration into the Biological Resources of Mongolia. Stubbe A, Kaczensky P, Wesche K, Samjaa R, Stubbe M, editors. Halle, Germany: Martin Luther University Halle-Wittenberg; 2007. [Google Scholar]
- 37.Borgatti SP, Everett MG, Freeman LC. Cambridge, MA: Analytic Technologies, Harvard University; 2006. UCINET for Windows: Software for Social Network Analysis. [Google Scholar]
- 38.Csardi G, Nepusz T. The igraph software package for complex social network research. 2006 (InterJournal, Complex Systems p. 1695) [Google Scholar]
- 39.Barash D. Marmots: Social Behavior and Ecology. Stanford, CA: Stanford Univ Press; 1989. [Google Scholar]
- 40.Freeman LC. Centrality in social networks: Conceptual clarification. Soc Networks. 1979;1:215–239. [Google Scholar]
- 41.Friedkin NE. Theoretical foundations for centrality measures. Am J Sociol. 1991;96:1478–1504. [Google Scholar]
- 42.Skvoretz J, Faust K. Logit models for affiliation networks. Sociol Methodol. 1999;29:253–280. [Google Scholar]
- 43.Robins G, Pattison P, Kalish Y, Lusher D. An introduction to exponential random graph (p*) models for social networks. Soc Networks. 2007;29:173–191. [Google Scholar]
- 44.Blumstein DT, Lea AJ, Olson LE, Martin JGA. Heritability of anti-predatory traits: Vigilance and locomotor performance in marmots. J Evol Biol. 2010;23:879–887. doi: 10.1111/j.1420-9101.2010.01967.x. [DOI] [PubMed] [Google Scholar]
- 45.Kruuk LEB. Estimating genetic parameters in natural populations using the “animal model.”. Philos Trans R Soc Lond B Biol Sci. 2004;359:873–890. doi: 10.1098/rstb.2003.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson AJ, et al. An ecologist's guide to the animal model. J Anim Ecol. 2010;79:13–26. doi: 10.1111/j.1365-2656.2009.01639.x. [DOI] [PubMed] [Google Scholar]
- 47.Gilmour AR, Gogel BJ, Cullis BR, Thompson R. ASReml User Guide Release 2.0. Hemel Hempstead, United Kingdom: VSN International; 2004. [Google Scholar]
- 48.Kruuk LEB, Hadfield JD. How to separate genetic and environmental causes of similarity between relatives. J Evol Biol. 2007;20:1890–1903. doi: 10.1111/j.1420-9101.2007.01377.x. [DOI] [PubMed] [Google Scholar]
- 49.Wilson AJ. Why h2 does not always equal V A/V P? J Evol Biol. 2008;21:647–650. doi: 10.1111/j.1420-9101.2008.01500.x. [DOI] [PubMed] [Google Scholar]
- 50.Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 51.Hadfield JD. Estimating evolutionary parameters when viability selection is operating. Proc Biol Sci. 2008;275:723–734. doi: 10.1098/rspb.2007.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Downhower JF, Armitage KB. The yellow-bellied marmot and the evolution of polygamy. Am Nat. 1971;105:355–370. [Google Scholar]
- 53.Armitage KB. Reproductive strategies of yellow-bellied marmots: Energy conservation and differences between the sexes. J Mammal. 1998;79:385–393. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.