Abstract
Parallel sequence and structure alignment tools have become ubiquitous and invaluable at all levels in the study of biological systems. We demonstrate the application and utility of this same parallel search paradigm to the process of protein structure determination, benefitting from the large and growing corpus of known structures. Such searches were previously computationally intractable. Through the method of Wide Search Molecular Replacement, developed here, they can be completed in a few hours with the aide of national-scale federated cyberinfrastructure. By dramatically expanding the range of models considered for structure determination, we show that small (less than 12% structural coverage) and low sequence identity (less than 20% identity) template structures can be identified through multidimensional template scoring metrics and used for structure determination. Many new macromolecular complexes can benefit significantly from such a technique due to the lack of known homologous protein folds or sequences. We demonstrate the effectiveness of the method by determining the structure of a full-length p97 homologue from Trichoplusia ni. Example cases with the MHC/T-cell receptor complex and the EmoB protein provide systematic estimates of minimum sequence identity, structure coverage, and structural similarity required for this method to succeed. We describe how this structure-search approach and other novel computationally intensive workflows are made tractable through integration with the US national computational cyberinfrastructure, allowing, for example, rapid processing of the entire Structural Classification of Proteins protein fragment database.
Keywords: p97 ATPase, likelihood functions, scoring methods, grid computing
Can access to vast quantities of computational power be leveraged to advance the study of biological systems in previously unexplored ways? Whereas many domains have driven demand for computational power and novel computational techniques in the process of scientific investigation, there remain areas where the opportunities provided by the most advanced computational infrastructures and tools have not been fully explored. The last decade has quietly seen the development of significant national and international federated cyberinfrastructures, established primarily to support the half dozen globally distributed particle physics collaborations. In the same way this community established the World Wide Web as a simple, standards-based system for information sharing, the particle physics community has also facilitated sharing of data and computing through development of what is known as “grid computing.” An area within the field of macromolecular structural biology that can leverage grid computing is harnessing the large and growing set of known protein structures to accelerate protein structure determination. The question of how to benefit from known structures was posed even as the earliest protein structures emerged, following observation of the similarity of the hemoglobin subunits to each other and to the structure of myoglobin.
The method now known as molecular replacement (MR) was first proposed for macromolecular crystallography by Rossmann and Blow (1), based on ideas developed by Hoppe in the context of small molecule crystallography (2). This was in response to the observation of evident family resemblances among different proteins and to the realization that it would be necessary to determine the structure of a particular protein in multiple states and with multiple ligands. The MR approach bootstraps the process of X-ray crystallographic phase determination by placing a known protein structure template in an orientation and position that aligns with that of the unknown protein. MR has now become the most commonly used method in protein structure determination by X-ray crystallography. It accounts for roughly half of all structures recorded in the Protein Data Bank (PDB) (3), which currently contains almost 70,000 depositions. In traditional MR, a suitable template model is selected based on sequence similarity. Other similar methods in structural biology rely on small databases of short protein fragments [e.g., the “lego” feature in O (4), and molecular fragment replacement in NMR (5)], or homologous structures [e.g., low-resolution refinement in crystallography (6)]. The selection of a suitable candidate template model remains a primary limiting factor in all of these methods. Although several approaches have been proposed for automating the selection of MR template models, either based on sequence information (7–9), or adapting MR algorithms to run in parallel on a specialized cluster (10), none have attempted molecular replacement searches using a complete, PDB-derived database of all available macromolecular domains, or considered the new insights provided by examining the aggregated results from large template model sets. Improved template selection would be expected to accelerate the structure determination process, minimize bias, and extend the range of suitable template models to proteins with negligible sequence identity.
In this paper, we ask three questions. First, can we compare results from independent molecular replacement runs and use these results to discriminate and rank solutions, thereby justifying the use of large template model databases? Second, can we develop improved criteria for recognizing correct solutions, in order ultimately to improve the convergence and speed of MR and further automatic structure determination? Third, can existing applications be scaled, deployed, and executed in a grid computing environment to enable new avenues of investigation, rather than merely faster computation? To answer the first question, we evaluated three diverse structure determination scenarios: (i) optimal selection in cases with several template model candidates; e.g., an MHC–TCR complex with 5,000 potential peptide binding or Ig domains that could be used as a template models in the MR search; (ii) structural homolog searches in cases for which sequence-based searches fail to identify usable MR template models; and (iii) “blind” cases in which the sequence of the crystallized sample is unknown. By adapting the widely used Phaser (11) MR application to the format of grid computing, we demonstrate the power of this unique wide search molecular replacement (WS-MR) approach, which can be used to search up to 100,000 domains in a few hours and to provide the range of results necessary to answer the questions posed above. WS-MR successfully identifies the closest structural homologues from a large family of candidates and does so more reliably than traditional, sequence-based approaches. The approach is also successful in identification of domains with marginal sequence identity or coverage. We use the WS-MR method to determine a structure of the full-length insect homologue of p97, a mammalian AAA+ ATPase (12–14) that was crystallized as a contaminant and reveals a previously unobserved D1 ADP-free conformation. Based on the extensive collection of results from the completed cases we demonstrate that incorporating multivariate scoring metrics [e.g., Phaser’s log likelihood gain (LLG) and translation function Z-score (TFZ)], or classification and clustering [e.g., Structural Classification of Proteins (SCOP) class and domain size], significantly improves discrimination to identify the best solutions. The computations for WS-MR were performed using the federated computing environment of the Open Science Grid (OSG) (15), illustrating how the national distributed cyberinfrastructure can be effectively used to develop and support unique computational workflows in research areas outside of physics.
Results
1. Comparison of Results from Independent Molecular Replacement Runs.
a. Selecting the Best Model from a Large Library of Homologous Structures.
We selected the MHC–TCR complex as the first system to validate the WS-MR approach. The structure contains one peptide binding domain (MHC–PBD) and six immunoglobulin (Ig) domains (Fig. 1A). There are over 5,100 candidate domains out of the 95,000 domains found in the Structural Classification of Proteins (SCOP) database (16) (Methods) that could map to parts of this structure, thus providing a useful spectrum of results to correlate the degree of structure coverage, sequence identity, and structural similarity with the quality of the initial phases. WS-MR, using the full SCOP database with the MHC–TCR reflection data [PDB code 2VLJ (17)], was used to determine whether structurally similar models rank best (in terms of various MR scoring metrics) and whether these models can be identified from incorrectly placed domains and from other structures in the database. This case is representative of using WS-MR for an unknown structure with many homologues, where it could be used to select the best model. This would be especially useful in cases where model coverage or sequence identity are low.
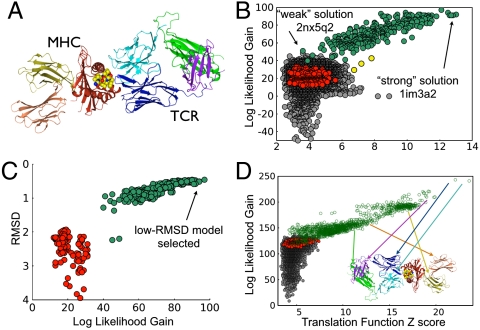
Fig. 1.
Validation of the WS-MR approach. (A) MHC–TCR complex, central peptide binding α12 domain in red (MHC–PBD), α3 domain of the MHC in orange, β2-microglobulin in blue, and four Ig domains in the TCR (yellow, cyan, green purple). Figure generated in CCP4MG (37). (B) Phaser LLG vs. TFZ from WS-MR for MHC–TCR complex. First round search, 300 correctly placed MHC–PBDs (green), 270 incorrectly placed MHC–PBDs (red), three identifiable and correctly placed Ig domains (yellow), and all other SCOP domains with MR results (gray). (C) Structure alignment RMSD vs. Phaser LLG score for MHC–PBDs. Correctly placed domains in green, incorrectly placed domains in red. Note that domains with RMSD as high as 2.2 Å were correctly placed by Phaser. Correlation coefficient of -0.9 for correctly placed domains (highly anticorrelated). (D) Phaser LLG vs. TFZ from WS-MR for MHC–TCR complex. Second round search, with MHC–PDB from first round fixed, 1,212 correctly placed Ig domains (green), 3,393 incorrectly placed Ig domains (red), all other SCOP domains with MR results (gray). Arrows indicate best result for each Ig domain.
The WS-MR search was completed in 12 hours of elapsed time (800 processor-days of computing time) utilizing a small subset of idle computers in the otherwise highly subscribed resources in OSG. This level of performance was typical of all the WS-MR iterations described here. Collected results allowed quick identification of a group of distinct, viable, MR models. Whereas several scoring functions were used to evaluate the quality of Phaser placement results (see section 2), a two-dimensional quality measure based on the LLG and the TFZ provides the best discrimination of results, producing a cluster of approximately 300 candidate domains from the search set of 95,000 (the “top cluster,” Fig. 1B). Domains in the top cluster all belong to SCOP class d.19.1.1, the MHC–PBD domain that represents 20% of the full model, and are all placed correctly by Phaser, in reference to the actual structure (SI Text). Three Ig domains are also identified in the top cluster (12% of search model), and no false positives are observed. The above results provide the boundary for the molecular replacement search to produce correct and identifiable placements for the MHC–TCR example with a model completeness between 12% (in case of the Ig domains) and 20% (for the MHC–PBD). The likelihood of obtaining the correct and identifiable placement with high quality models is very small when searching with 12% of the target (3 in 4,500 Ig domains, Fig. 1B) and dramatically increases for a search with 20% of the target (300 in 550 MHC–PBD domains, Fig. 1B).
WS-MR not only discriminates correctly placed models but, in this case, also orders them by the similarity of the structure and the target molecule (Fig. 1C). For the correctly placed MHC–PBD models, LLG/TFZ is highly correlated to RMSD between the model and the reference structure. For example, the lowest RMSD model also scores the highest on the LLG/TFZ scale. In comparison, selecting models based exclusively on sequence identity results in a wide range of LLG/TFZ values, even for the subset with identities > 90% (Fig. S1B). In this test case LLG-based selection provides superior distinction of correct solutions compared to sequence similarity and would therefore provide an advantage for MR model selection.
As expected, placement of the best first domain identified by WS-MR (an MHC–PBD) facilitated completion of structure determination. Repeating WS-MR with the MHC–PBD domain fixed placed over 1,000 Ig domains in the top cluster result from the second WS-MR iteration, and further analysis confirmed that all six MHC–TCR Ig domains are found in this set (Fig. 1D). Here the LLG scores correlate strongly with the structural similarity (see linear fit lines in Fig. 2A). Whereas the search for the first MHC–TCR fragment required a minimum 60% sequence identity to obtain identifiable solutions, in the secondary search individual Ig domains with as little as 11.6% sequence identity produced identifiable results (see Section 1b), a noteworthy success of the partially phased MR approach with 20% placed, a 12% search fragment and 68% of the structure still missing.
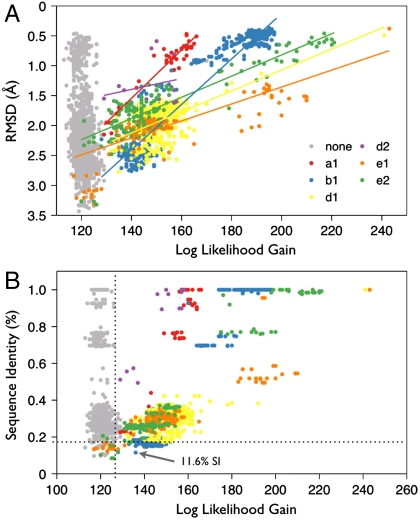
Fig. 2.
Second round search for Ig domains with best first round MHC–PBD template fixed. Colored points correspond to correctly placed Ig domains, Whereas gray points indicate incorrectly placed domains. Domains with sequence identity as low as 11.6% are clearly identified by LLG and TFZ scores and correctly placed. Note that both sequence identity and RMSD (to actual structure) are poor indicators of successful MR placement. (A) RMSD vs. LLG of Ig domains in second round WS-MR search. Data points for matching Ig domains are fitted with linear regression. Domains with lower RMSD to the target score higher. (B) Sequence Identity vs. LLG of Ig domains in second round WS-MR search.
b. Identifying Good MR Models with Marginal Sequence Identity.
Perhaps the most intriguing opportunity for the WS-MR structure determination technique is the application of a blind search in cases where traditional MR techniques fail, and before attempting further experimental phasing methods. Blind WS-MR, where no template filtering is applied and the full template database is searched, can reveal structures that would not otherwise be identified by sequence alignment algorithms [which generally provide poor results when the best sequence-based homologues have an identity of less than 30% (9)]. Such searches make no a priori assumptions about the target structure and can utilize large databases of PDB-derived models. The infrastructure described in section 3 makes this approach feasible, and the trend of decreasing cost per unit of processing power is such that in the next few years such a workflow could be executed solely by the internal computational resources of a single laboratory.
In a limited number of completed searches we observe that models with borderline sequence identity (between 10–20%) can work well. For example, in the MHC–TCR example described above, in the secondary search with the MHC–PBD placed, the majority of Ig domains with sequence identity below 20% failed to be correctly placed, but the placement of 244 domains was correct (gray vs. colored dots in Fig. 2). All but 17 of the correctly placed domains could be readily identified based on LLG and TFZ scores, indicating a false negative rate for this set (sequence identity below 20%) of 7%, and a clear LLG cut-off of 130, above which 100% of the results were correct, including domains with sequence identity as low as 11.6%.
Further tests of WS-MR were carried out on structures that had previously been determined by experimental phasing methods. A search with data for EmoB (18) (PDB code: 2VZF) was performed with the SCOP database, and returned a clear cluster of 14 solutions (Fig. 3A). All 14 models that belong to SCOP flavoprotein classes c.23.5.4 and c.23.5.8 are positioned properly, while all remaining 182 flavoprotein domains in the bottom cluster, except for 4, are incorrectly placed. The 14 correctly placed and identifiable models have sequence identities of 13% to 21%, and RMSD between 2.2 and 2.7 Å relative to the reference structure (Fig. S2 A and B). The top solution can be used to rapidly refine the structure. In contrast, four iterations of a PSI-BLAST search identified 313 candidates, of which two were in the group of 14 identified by the WS-MR approach as suitable MR templates. These two were conformationally similar structures with mutually identical sequence, but a sequence identity to the target structure of only 17%. PSI-BLAST failed to identify any of the other twelve structures, despite having sequence identities in the same range (13–21%). Whereas in this particular case sequence-based approaches should converge on a correct solution, the unpredictability of successful molecular replacement results combined with the difficulty of selecting models by sequence-based searches explain why viable MR models may be missed in other similar cases.
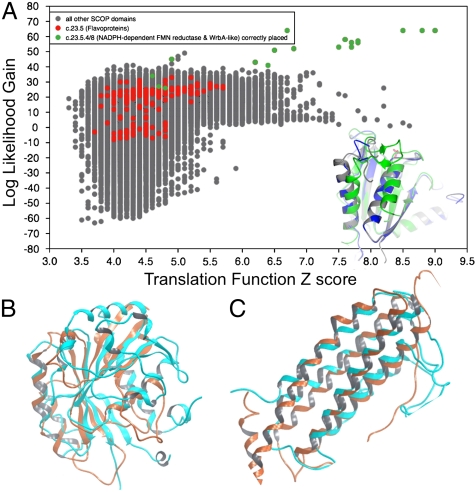
Fig. 3.
WS-MR with distant homologues. (A) Phaser LLG vs. TFZ for EmoB protein. 12 distinct flavoprotein (SCOP SCCS class c.23.5.4/8) MR templates identified and correctly placed (green), and a further 200 (red) from the same SCOP class incorrectly placed. All other SCOP domains in gray. Foreground: actual oxidoreductase structure in gray, Phenix Autobuild structure in blue, and top WS-MR result in green. Using density modification and automated build procedures, with final R-factor/R-free statistics that are comparable to the deposited structure (20.0%/23.6% for the deposited EmoB vs. 22.1%/25.5% for the WS-MR model). (B) Distant homologue search for structure 3CIO returns structurally similar results. (C) Distant homologue search for structure 3CEX returns structurally similar results.
We have also recorded several cases, when performing full SCOP WS-MR searches, where the identified solutions share significant structural characteristics with the target but are too divergent to produce the correct placement. For example WS-MR with experimental data for the kinase domain of Escherichia coli tyrosine kinase ETK (PDB code: 3CIO) retrieves no strong results, but after closer inspection of the LLG/TFZ profile, we selected two solutions with relatively high TFZ score (> 6), and LLG scores separated from other results. One of those peaks corresponds to SCOP model 1Z0Fa1—a Rab GTPase (Fig. 3B). The two structures have 12.5% pairwise sequence identity, a misleading metric given that the two proteins can only be superposed in a sequence independent manner (Fig. S2C). In another case, a structure of a four helix protein recently deposited by the Midwest Center for Structural Genomics (PDB code: 3CEX) can be superposed on a SCOP domain from ferritin (1IESa_) (Fig. 3C). The superposition of the four helical elements is sequence independent (Fig. S2D). Clustalw fails to provide sufficient insight, and produces a relatively high pairwise identity of 16.6%, but this does not correspond to the actual sequence identity for the aligned structures.
c. An Example of a Blind Search Without Prior Sequence Information: Structure of ADP-Free p97 Homolog.
We have also tested our method on five cases provided for evaluation by colleagues in response to our solicitation for recalcitrant datasets—those that resisted molecular replacement efforts with the most obvious models. For each submitted dataset there was a concern that the crystallized sample was a contaminant rather than the target protein, as the identity of proteins could not be confirmed experimentally due to the limited sample availability. In some cases dissolved crystals had characteristics consistent with the target protein (e.g., migration on SDS-PAGE or mass spectrometry profile). For each dataset we performed WS-MR with the full SCOP database. Four datasets were immediately confirmed as contaminants. The most striking was a homolog from Trichplusia ni (order Lepidoptera, Hi-5 cells) of a mammalian p97, a hexameric AAA+ ATPase, which is characterized by poorly diffracting crystals (6) and multiple nucleotide binding states (19). The T.ni protein remains unsequenced, but we expect it to be very similar to the sequenced Bombyx mori transitional endoplasmic reticulum ATPase TER94 (also order Lepidoptera, accession codes: BAE54254 and NP_001037003), which in turn is 83% identical to the full-length Mus musculus p97. WS-MR clearly identified nine domains in a distinct high scoring cluster (Fig. 4A). The overall architecture of T.ni p97 closely resembles the structure of M.musculus p97, and the space group matches the 1R7R structure. Inspection of fo-fc electron density maps suggests, however, that in contrast to other p97 crystal structures (1E32; 1YQ0; 1YQ1; 1YPW) (Fig. 4B), the T.ni p97 is nucleotide-free in the D1 binding pocket (Fig. 4C). Although spectroscopic analysis of the protein sample will be required to confirm that indeed all of the symmetry-related molecules in T.ni p97 are ADP-free, the unexpected results of WS-MR in this case reveals another potentially valuable utility of the method. Other contaminants retrieved by WS-MR include carbonic anhydraze (1I6Oa_), inorganic phosphatase (1MJWb_), and pyruvate kinase (1AQFg2). In each case, WS-MR provided a quick, conclusive answer to problems that could not be readily addressed using standard biochemical tools.
Fig. 4.
WS-MR discovery of the insect analogue of mammalian AAA+ p97 structure from crystallized T. ni protein contaminant. (A) LLG vs. TFZ for p97 WS-MR search. Green points correspond to domains from known structure of mouse p97 protein (SCOP SCCS class c.37.1.20), showing 9 domains that form a distinct cluster, and 2 that are “buried.” Red points correspond to all other SCOP domains that produced MR results (45,700 in total). (B) Fo - Fc difference map calculated using p97 coordinates (PDB accession code 3CF2), with ADP molecule omitted and contoured at 3 sigma level shows clear density for the ADP. (C) Fo - Fc difference map calculated using the refined T. ni model with a side chain of His 385 omitted and contoured at 3 sigma level shows a clear density for Histidine side chain, and no density for the ADP.
2. Improved Criteria for Recognizing Correct Solutions.
By collecting a large number of data points in many dimensions for several different target structures, we are able to consider techniques beyond the traditional TFZ score to identify viable MR models. We find that Phaser LLG and TFZ scores, in particular, combine to provide good discrimination of templates when strong MR models exist. When combined with LLG, TFZ scores as low as 3.5 are associated with positive results in the correctly placed top cluster. High TFZ (greater than 7) indicates a good MR solution, but our findings show that a low TFZ can, in some cases, also represent a usable MR solution. It is already well known that the LLG scores for different template models are comparable for the same set of reflection data, and this feature is used by Phaser when presented simultaneously with multiple candidate models. WS-MR greatly expands the number and efficiency of intermodel comparisons by LLG that are possible, and thus, we hypothesized, would improve the process of identifying good MR models.
We can further augment the sensitivity of the scoring function by incorporating additional dimensions, such as rotation function Z-score (Fig. S3A), domain length (Fig. S4), or domain class clustering. Other measures such as R-factor improvement or contrast as provided by Molrep (SI Text and Fig. S3B) (20) are less suitable for cross-model comparison. For example we carried out Phenix refinement protocols for several single domain MR solutions to the MHC–TCR example. Only the best solution has an R-factor that falls below 50.0, and for other cases R-free does not improve, most likely because of the limited convergence of refinement with partial model information.
3. Efficiency and Reliability of Molecular Replacement Computations Executed on Grids.
All computations in this project were carried out on “opportunistic” resources of OSG. This required accessing 20–30 computing centers that participate in the OSG federation and have allowed our scientific domain (structural biology) to utilize the otherwise idle computing resources of their clusters. To benefit fully from this national cyberinfrastructure, we established a software and hardware environment that can manage and support both general and specialized types of grid computations. Unlike a desktop or cluster computing environment, where the configuration of the system is fixed and well known, grid computing introduces complexities that require new approaches rather than simple reconfiguration of existing programs. The dynamic nature of grids with a high level of unpredictable faults, federation, geographic distribution, and system heterogeneity present significant challenges. We have therefore developed unique strategies for the synchronization and flow of data and applications at four grid levels: “static” (constantly available), “workflow” (a related set of computations), “grid job” (a single instance of grid resource utilization), and “atomic job” (the smallest computational unit that produces a distinct result as part of the workflow, but may be too small to efficiently run as an independent grid job) (Fig. 5). By tracking application and script versioning, and by considering the permanence and relevance of data, we can reduce the obstacles presented by network congestion and multiple levels of caching to maximally localize data and computations while minimizing data movement. We have combined these efforts with fault management techniques at the workflow, grid, and atomic job level to detect unfavorable conditions for computation in advance of execution or to track failures post execution. In all cases, the grid job manager can correct the situation and retry the computations where possible. Our mechanisms for moving data and initiating executions on remote systems have relied heavily on the OSG Virtual Data Toolkit (21), Globus Toolkit (22), Condor (23), and GridSite (24), with an underlying security layer provided by X.509-based public-key cryptography and higher layer workflow scheduling decisions managed through a combination of Condor DAGMan (25) and the OSG Match Maker, or GlideinWMS (26). We have tuned these various systems to maximize correct scheduling and successful completion of computations (e.g., setting an execution timeout suitable to capture all viable results; Fig. S5). We can reach computing levels of over 50,000 CPU hours in a single day, and concurrent execution in excess of 7,000 grid jobs at dozens of computing centers. We have created a Web portal, which acts as the hub and clearing house for these computations. It enables secure access to create, run, analyze, visualize, and share workflows and data.
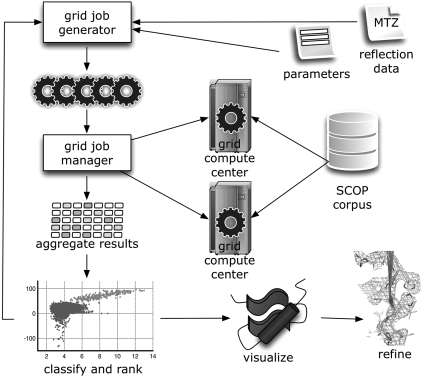
Fig. 5.
Process workflow, illustrating the inputs (search parameters and reflection data), the key grid components, and the division of computational jobs with slices of the SCOP database. Results are aggregated, classified and ranked, and then manually analyzed for further refinement or iterative model building if appropriate.
Discussion
We have demonstrated that WS-MR is able to discriminate strong molecular replacement template models with marginal sequence identity and coverage, identifying top candidates for subsequent density modification, model building, and refinement steps. In rare cases templates comprising as little as 6% of the scattering matter (27), or having sequence identity below 20% (28), have been shown to produce correct MR placement results. Validating or utilizing templates with such characteristics is typically difficult. A routine evaluation of all marginal fragments typically requires several cycles of model building and refinement, can be time consuming, and it is not always clear if the results are correct. Wide search comparison of several domains based on multidimensional scoring metrics greatly accelerates the validation process. Our results suggest that the limit of sequence identity for successful WS-MR search is low enough to allow our method to extend to models that would otherwise be missed by methods that are based on sequence alignment for template selection (29). Both remote homologues and structural analogs (30) can be detected by WS-MR, with specific examples where models with an identity of 11.6% and an RMSD under 3 Å can be correctly placed and distinguished from negative results. We also show that low completeness with structure coverage of as little as 12% can be sufficient for good WS-MR template models, however in these cases high sequence identity and structural similarity for the covered area are required.
By using an approach in which no a priori knowledge or primary sequence information is required for search model selection, we have expanded the probability of success for difficult molecular replacement problems in X-ray crystal structure determination. Utilizing this system is straightforward, as the only required input is the reflection data. Additionally, initial search constraints (e.g., sequence, predicted secondary structure profile, molecular weight, oligomerization state) can be provided to optimize the search, or previously placed domains in the case of subsequent domain searches for multidomain structures. The output of WS-MR provides both graphical and tabular summary representations of the results, allowing rapid identification of the best candidate MR template models. The user would then attempt to validate a few top scoring solutions using standard approaches, such as packing analysis or interpretation of density modified difference maps. If a particular solution looked plausible, a search for missing components of a given structure, or a manual or automatic rebuilding process could be attempted. To encourage rapid convergence to the best MR models (if they exist), the WS-MR strategy can proceed iteratively, starting with the most promising models based on the specified constraints, for example using the top 100 sequence-similar models, and include a small control set that is widely representative of known domains (for contrasting expected negative results). If no promising models are returned from the initial constrained search, subsequent iterations can relax the selection criteria to associated domain classes, thus expanding the number of search models, eventually considering all known domains. Although it is not possible to predict whether a less-than-exhaustive WS-MR search is necessary (if obvious models existed, conventional MR would suffice), this iterative approach will avoid an exhaustive search if promising models are discovered from the constrained search set. The WS-MR method is accessible and applicable to many crystallographic projects, as it allows the search of arbitrary structure databases, constructed dynamically from selection criteria or from preexisting sets. The WS-MR approach becomes increasingly powerful as more structures are determined and made publicly available.
A benefit of the large result sets produced by WS-MR is the ability to evaluate algorithmic improvements that should result in better scoring and discrimination of search models, in particular a reduction of false negatives. Our work on several WS-MR test cases has provided unique insights leading to improved scoring and model discrimination strategies. By using multiple scoring metrics (such as LLG and TFZ) from the high quality maximum likelihood algorithm in Phaser, it is possible to distinguish correct solutions by cluster identification. In the case of weak (but still valid) MR templates, we have shown that effective model discrimination is significantly aided by these additional metrics. Fig. S4 illustrates how the additional consideration of model size allows for the clear identification of several correctly placed Ig domain models for the MHC–TCR case that were not identifiable from only the LLG and TFZ data. LLG led to the selection of several correctly placed models in the EmoB case (Fig. 3A). Classification (e.g., SCOP class) or MR placement clustering (similar domains placed in the same orientation and location) can also provide a mechanism to identify groups of viable MR models. One important observation for the results of exhaustive WS-MR is that small domains can lead to anomalously high TFZ scores (greater than 10), due either to insufficient statistics or the ability of very small fragments to match accurately to some region of a large unknown structure. Nevertheless, these anomalous results also benefit from the addition of LLG scoring, as they consistently have LLG scores below 20 and can therefore be easily identified and discounted.
Without existing infrastructure, a transition to grid computing requires a significant time investment and presents numerous unexpected hurdles. The challenge in accessing and deploying applications into a grid environment can be simplified for the end user by the development of web-based portals, an approach that has proved successful for many other grid environments [e.g., TeraGrid Science Gateways (31)]. The SBGrid Science Portal (http://www.sbgrid.org) that we have developed will make the WS-MR technique described here widely available to the entire community. Using OSG to perform WS-MR for the cases described here, we typically accessed 2,000–5,000 computing cores concurrently, thus completing what would otherwise have required several years of computing within one day. Access to the national cyberinfrastructure makes it possible for any individual research group to develop novel computational workflows that take advantage of large federated resources, in particular idle cycles that would otherwise be wasted. Computers in a typical scientific computing cluster spend around half their time lightly utilized (less than 10% load), but even then they typically consume more than 80% of the maximum power consumption at full load (32). This presents a tremendous computational opportunity with relatively minor cost overhead. With a transition to a new resource access and scheduling mechanism, using GlideinWMS (26), we have been able to execute up to 7,000 concurrent computations using this pool of otherwise idle computers, well above of what is currently available to a typical research group.
Arguably more important than the WS-MR technique itself are the opportunities to reuse the framework that has been developed for large scale data processing and computation. We have started work on problems in NMR, electron microscopy and in other areas of X-ray crystallography that use this foundational infrastructure and the capacity provided by OSG. Any scientific application that can run without active user interaction can be deployed into a grid environment with a suitable workflow management protocol for data staging, results aggregation, and analysis. We have shown that it is not necessary to redesign applications and algorithms to benefit from these advances. Existing applications can be used in new ways with statistical and data visualization techniques applied to aggregate and filter orders of magnitude higher data volumes than the application designers intended, leading to new challenges for interpretation and discovery.
Methods
The SCOP domains utilized for WS-MR were taken from the November 2007 (1.73) release (16, 33). Molecular replacement computations were performed with Phaser (version 2.1.4), and Molrep (version 10.2.3). We used a modified version of TM-Align (34) to perform structural alignment and combination of TM-Align and Reforigin (CCP4, version 6.1.2) (35) to calculate placement quality and placement correctness. Scheduling of jobs to OSG sites was managed through a combination of Condor DAGMan (25) and the OSG Match Maker. Density modification and model building of the MHC–TCR and EmoB models were performed in Phenix Autobuild (36) starting with Phaser Sigma(A)-type weighted fourier maps (FWT/PHWT) (37) and amplitudes with standard deviations from the Protein Data Bank structure factor files. Detailed protocols are described in SI Text.
Supplementary Material
Acknowledgments.
We thank Peter Doherty for grid computing support, Mats Rynge for Matchmaker customizations, Steve Timm for operational assistance, and Stephen C. Harrison and Yunsun Nam for discussion and critical review of the manuscript. The work was supported by National Science Foundation Grant 0639193 (P.S.), and National Institutes of Health Grant P01 GM062580 (to Stephen C. Harrison). This research was done using resources provided by the Open Science Grid, which is supported by the National Science Foundation and the US Department of Energy’s Office of Science.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1012095107/-/DCSupplemental.
References
- 1.Rossmann MG, Blow DM. The cetection of sub-units within the crystallographic asymmetric unit. Acta Crystallogr. 1962;D15:24–32. [Google Scholar]
- 2.Hoppe W. Faltmolekülmethode. Acta Crystallogr. 1957;10:750–751. [Google Scholar]
- 3.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for the building of protein models in electron density maps and the location of errors in these models. Acta Crystallogr. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 5.Delaglio F, Kontaxis G, Bax A. Protein structure determination using molecular fragment replacement and NMR dipolar couplings. J Am Chem Soc. 2000;122:2142–2143. [Google Scholar]
- 6.Schroeder GF, Levitt M, Brunger AT. Super-resolution biomolecular crystallography with low-resolution data. Nature. 2010;464:1218–1223. doi: 10.1038/nature08892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keegan RM, Winn MD. MrBUMP: An automated pipeline for molecular replacement. Acta Crystallogr. 2008;D64:119–124. doi: 10.1107/S0907444907037195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Long F, Vagin AA, Young P, Murshudov GN. BALBES: A molecular-replacement pipeline. Acta Crystallogr. 2008;D64:125–132. doi: 10.1107/S0907444907050172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwarzenbacher R, Godzik A, Jaroszewski L. The JCSG MR pipeline: optimized alignments, multiple models and parallel searches. Acta Crystallogr. 2008;D64:133–140. doi: 10.1107/S0907444907050111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schmidberger JW, et al. High-throughput protein structure determination using grid computing. Proceedings of the IEEE International Parallel and Distributed Processing Symposium. 2009:1–8. [Google Scholar]
- 11.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLaBarre B, Brunger AT. Complete structure of p97/valosin-containing protein reveals communication between nucleotide domains. Nat Struct Biol. 2003;10:856–863. doi: 10.1038/nsb972. [DOI] [PubMed] [Google Scholar]
- 13.Huyton T, et al. The crystal structure of murine p97/VCP at 3.6A. J Struct Biol. 2003;144:337–348. doi: 10.1016/j.jsb.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 14.Davies JM, Brunger AT, Weis WI. Improved structures of full-length p97, an AAA ATPase: Implications for mechanisms of nucleotide-dependent conformational change. Structure. 2008;16:715–726. doi: 10.1016/j.str.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Pordes R, et al. The Open Science Grid. J Phys Conf Ser. 2007;78:012057. [Google Scholar]
- 16.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: A structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 17.Ishizuka J, et al. The structural dynamics and energetics of an immunodominant T cell receptor are programmed by its Vbeta domain. Immunity. 2008;28:171–182. doi: 10.1016/j.immuni.2007.12.018. [DOI] [PubMed] [Google Scholar]
- 18.Nissen MS, et al. Crystal structures of NADH:FMN oxidoreductase (EmoB) at different stages of catalysis. J Biol Chem. 2008;283:28710–28720. doi: 10.1074/jbc.M804535200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Briggs LC, et al. Analysis of nucleotide binding to P97 reveals the properties of a tandem AAA hexameric ATPase. J Biol Chem. 2008;283:13745–13752. doi: 10.1074/jbc.M709632200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vagin A, Teplyakov A. Molecular replacement with MOLREP. Acta Crystallogr. 2010;D66:22–25. doi: 10.1107/S0907444909042589. [DOI] [PubMed] [Google Scholar]
- 21.Roy A. Building and testing a production quality grid software distribution for the Open Science Grid. J Phys Conf Ser. 2009;180 [Google Scholar]
- 22.Foster I. Lecture Notes in Computer Science. Vol. 3779. Berlin: Springer; 2005. Globus toolkit version 4: Software for service-oriented systems; p. 2. [Google Scholar]
- 23.Thain D, Tannenbaum T, Livny M. Grid Computing: Making the Global Infrastructure a Reality. London: Wiley; 2003. Condor and the grid; pp. 299–335. [Google Scholar]
- 24.McNab A. Grid-based access control and user management for Unix enviroments, filesystems, Web sites and virtual organisations; Computing in High Energy Physics; 2003. [Google Scholar]
- 25.Couvares P, Kosar T, Roy A, Weber J, Wegner K. Workflow in Condor. In: Taylor I, Deelman E, Gannon D, Shields M, editors. Workflows for e-Science. New York: Springer; 2007. pp. 357–375. [Google Scholar]
- 26.Sfiligoi I. Making science in the Grid world: Using glideins to maximize scientific output. 2007 Nuclear Science Symposium Conference Record (IEEE) 2007:1107–1109. [Google Scholar]
- 27.Bernstein BE, Hol WG. Probing the lmits of the mlecular rplacement mthod: The case of Trypanosoma brucei phosphoglycerate kinase. Acta Crystallogr. 1997;D53:756–754. doi: 10.1107/S0907444997010214. [DOI] [PubMed] [Google Scholar]
- 28.Jones DT. Evaluating the potential of using fold-recognition models for molecular replacement. Acta Crystallogr. 2001;D57:1428–1434. doi: 10.1107/s0907444901013403. [DOI] [PubMed] [Google Scholar]
- 29.Peterson M, et al. Evolutionary constraints on structural similarity in orthologs and paralogs. Protein Sci. 2009;18:1306–1305. doi: 10.1002/pro.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng H, Kim BH, Grishin NV. Discrimination between distant homologs and structural analogs: Lessons from manually constructed, reliable data sets. J Mol Biol. 2008;377:1265–1278. doi: 10.1016/j.jmb.2007.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wilkins-Diehr N, Gannon D, Klimeck G, Oster S, Pamidighantam S. TeraGrid science gateways and their impact on science. IEEE Computer. 2008;41:32–41. [Google Scholar]
- 32.David M, Brian TG, Thomas FW. PowerNap: Eliminating server idle power; Proceedings of the 14th International Conference on Architectural Support for Programming Languages and Operating Systems; Washington, DC: ACM; 2009. [Google Scholar]
- 33.Andreeva A, et al. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Skolnick J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005;33:2302–2309. doi: 10.1093/nar/gki524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Collaborative Computational Project N. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 36.Zwart PH, et al. Automated structure solution with the PHENIX suite. Method Mol Biol. 2008;426:419–435. doi: 10.1007/978-1-60327-058-8_28. [DOI] [PubMed] [Google Scholar]
- 37.Read RJ. Improved Fourier coefficients for maps using phases from partial structures with errors. Acta Crystallogr. 1986;A42:140–149. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.