Abstract
Developmental programs are controlled by transcription factors and chromatin regulators, which maintain specific gene expression programs through epigenetic modification of the genome. These regulatory events at enhancers contribute to the specific gene expression programs that determine cell state and the potential for differentiation into new cell types. Although enhancer elements are known to be associated with certain histone modifications and transcription factors, the relationship of these modifications to gene expression and developmental state has not been clearly defined. Here we interrogate the epigenetic landscape of enhancer elements in embryonic stem cells and several adult tissues in the mouse. We find that histone H3K27ac distinguishes active enhancers from inactive/poised enhancer elements containing H3K4me1 alone. This indicates that the amount of actively used enhancers is lower than previously anticipated. Furthermore, poised enhancer networks provide clues to unrealized developmental programs. Finally, we show that enhancers are reset during nuclear reprogramming.
The genome comprises a large number of noncoding DNA regulatory elements, including silencers, insulators, and enhancer regions, that play important regulatory roles in maintaining gene expression programs. Enhancers have emerged as key cis-regulatory elements that can affect gene transcription independent of their orientation or distance. Global identification of these regions as well as their contribution to target gene expression has been challenging because enhancers can often reside thousands of base pairs away from their target of regulation (1, 2).
Individual enhancers have been identified by detailed studies of disease-related elements that reside in noncoding regions outside of model genes (3), followed on a larger scale by comparative genomics using ultra-conservation (4). These studies unequivocally established specific cis-regulatory elements as important regulators of development and disease. Recently efforts to globally identify enhancers have focused on the histone mark H3K4me1 to locate many cell type-specific enhancer sites (5–9). Analysis of human primary and tumor cell lines has shown that enhancer regions identified by virtue of H3K4me1 enrichment, as well as DNaseI hypersensitivity, display a cell type-specific distribution across the genome (5, 10). Another study found that the histone acetyltransferase p300 is enriched at enhancers in fetal mouse tissues and found similar tissue-specific marking of these elements. Notably, many of these enhancers were shown to contribute to transcriptional activity of proximal genes in a tissue-specific manner (11).
Although most H3K4me1 marked enhancer regions displayed activity when tested in reporter assays, a significant percentage was not active (5, 11). One hypothesis is that H3K4me1 can mark active enhancers as well as those in a poised or predetermined state. Indeed, several lines of evidence support the existence of poised enhancers. Lineage-specific enhancers that become activated upon differentiation are protected from DNA methylation in the parental ES cells, where the enhancers are not yet active (12). Furthermore, large sets of inactive developmental genes in hematopoietic stem cells are associated with H3K4me1-enriched distal enhancers (13). Finally distal H3K4me1-enriched Stat1 binding sites become selectively activated upon INF-γ treatment of HeLa cells, suggesting that these regions maintain the potential to become active (5). This raises the intriguing possibility that enhancers contain information about the current and future developmental potential of a cell, as well as its ability to respond to external cues.
Here, we identify close to 135,000 candidate distal enhancer regions in five different cell types. In a candidate approach to identify factors that could potentially discriminate between the various possible enhancer states, we establish H3K27ac as an important enhancer mark that can distinguish between active and poised enhancer elements. This mark can be deposited by both p300 and CREB binding protein (CBP) (14) and is associated with active promoters in mammalian cells (15). Using this modification to distinguish poised from active enhancer networks allows us to make predictions on the developmental potency of the cell, as well as it current state. Furthermore, it suggests that the active global enhancer network of a cell is smaller than previously anticipated.
Results
H3K27ac Distinguishes Active from Inactive Enhancers.
We first identified cell type-specific enhancers in murine ES cells using distal H3K4me1 histone marks identified by ChIP-Seq (Fig. 1A, Table S1, and SI Materials and Methods) using similar criteria as shown previously (5–9, 16). We identified 25,036 distal enhancer elements that are roughly equally distributed between introns and intergenic regions (Fig. S1A). To test the robustness of these data we compared our results with a previously published H3K4me1 data set in 129/C57BL6 F1 ES cells (17) and found that they displayed a remarkably similar distribution pattern (Pearson correlation, 0.76; Fig. S1 B and C). Second ES cells from an inbred genetic background (C57BL6/N) yielded very similar results (Pearson correlation, 0.77), indicating that cell type-specific enhancers pattern the genome with high fidelity with little effect from genetic background.
Fig. 1.
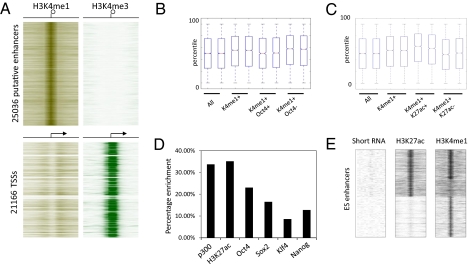
H3K27ac is a good candidate to distinguish active from inactive enhancer elements. (A) Heatmap of 25,036 putative enhancers based on H3K4me1 enrichment and H3K4me3 depletion in ES cells. Eight-kilobase pairs around the center of the enhancer region are displayed. (B) Gene expression of microarray data generated in duplicates in ES cells. Box-plots show all genes (All) and genes found associated with enhancers (H3K4me1+) either enriched (+) or unenriched (−) for Oct4 or (C) H3K27ac. Solid bars of boxes display the 25–75% of ranked genes with the mean indicated as an intersection. (D) Graph showing the percentage of enhancers identified in A that are called bound at P = 10−8 by the indicated factors or chromatin marks. (E) Heatmap showing a composite of distal H3K27ac- and H3K4me1-enriched regions ordered by presence and absence of these two histone marks. Left: Occurrence of short RNA reads based on this distribution within a −8 kb to +8 kb window (SI Materials and Methods).
Proximal gene expression was previously shown to be preferentially affected by enhancer elements (11), and enhancers seem to cluster near the genes they regulate (5). Similarly, the overall presence of the enhancers identified in this study correlated well with increased gene activity of proximal genes when we compared these ChIP data with microarray expression data (Fig. 1 B and C; P = 4.4E-16). Consistent with its role as a master regulator of ES cell state, motif search analysis identified the Oct4 (POU) binding domain at ES cell enhancers (P = 1.0E-250), suggesting a role for this transcription factor in enhancer regulation. Indeed, this transcription factor was previously seen enriched at promoter distal elements similar to other ES cell regulators, such as Sox2 and Nanog (18). We decided to test a number of enhancer-associated transcription factors and chromatin modifications in ES cells as potential candidates to distinguish between active and inactive enhancer elements. We generated data sets for p300 and H3K27ac and adapted datasets for Oct4, Sox2, Klf4, p300, and Nanog from previous work (18, 19) and found that a significant amount of the putative ES cell enhancers identified here were enriched for the factors tested (Fig. 1D and Fig. S2). For instance, the histone acetyltransferase p300 has been used as a reliable marker to identify distal enhancers (10, 11, 16) and in agreement with this could be found associated with a significant portion of H3K4me1-enriched regions (Fig. 1D). Notably, genes closest to H3K4me1-positive p300-bound enhancers are not preferentially more active compared with genes associated with H3K4me1-positive enhancers that do not contain p300 (P < 1.0), suggesting that similar to H3K4me1, p300 can be used to identify both active and less active enhancer types (Fig. S2). Similarly, proximal genes to Nanog-bound (P = 0.46) and Klf4-bound enhancers (P < 1.0) were not significantly different from average enhancers. Interestingly Oct4- and Sox2-bound enhancers correlated with less active genes (Fig. 1B and Fig. S2; P = 0.008 and P = 0.002), suggesting these factors might be involved in keeping enhancers in an inactive state until differentiation ensues. Because this idea is similar to what has been shown for polycomb group proteins at poised promoter regions that combine multiple different factors that in differentiated cells behave in a mutually exclusive manner (20, 21), we asked whether Oct4- and Sox2-positive enhancers were enriched for the polycomb-associated mark H3K27me3 (Table S1) but could find no significant overrepresentation of H3K27me3 at Oct4-enriched (P = 0.19) or Sox2-enriched (P = 0.88) enhancer elements. Furthermore, only 1.2% of identified enhancer elements were enriched for H3K27me3. In contrast to Oct4 and Sox2, H3K27ac-modified enhancer regions correlated well with enhanced proximal gene activity (P = 1.0E-5). Importantly we also found that enhancer proximal genes that lack H3K27ac enrichment displayed lower expression levels compared with the average enhancer proximal gene (Fig. 1C; P = 4.06E-7). These data indicate that H3K27ac is a good candidate to discriminate between active and poised enhancer states. In support of this, we found that Oct4 (P = 1.8E-8) and Sox2 (P = 1.7E-8) are selectively enriched at H3K27ac-negative enhancers.
Because genes can be associated with multiple enhancers that are either H3K4me1+, H3K27ac+, or a combination of both, we analyzed the relationship between H2K27ac and H3K4me1 and proximal gene activity according to a simple hierarchy to filter out the effects of single enhancer regions on the expression of neighboring genes (SI Materials and Methods). We found that genes associated with distal H3K4me1 regions that are H3K27ac negative and also lack any other neighboring H3K27ac-positive enhancer region express at a much lower level than neighboring genes of average enhancers (P = 1.93E-13; Fig. S3A). This suggests that H3K27ac enrichment is a deterministic feature of active enhancers. In support of this, genes that are associated with distal H3K27ac-enriched regions that are negative for H3K4me1 showed overall similar expression levels as proximal genes of enhancers enriched for both H3K27ac and H3K4me1 (P = 0.94). Furthermore, these data suggest that in addition to H3K4me1, distal H3K27ac sites that do not show significant H3K4me1 enrichment should also be considered as potential enhancer elements.
It has previously been shown that enhancers that associate with promoters and support transcription of neighboring genes produce small RNA products that depend on the presence of the proximal promoter (8). We asked whether H3K27ac-enriched enhancers selectively expressed these short RNAs using a small RNA-Seq data set previously published (22) and found that these short RNAs are indeed selectively transcribed from H3K27ac-positive over H3K27ac-negative enhancer elements (Fig. 1E; P = 1E-154) regardless of H3K4me1 enrichment. This again supports the notion that H3K27ac is a deterministic factor of active enhancer elements.
H3K27ac Marking of Active Enhancers Is a General Phenomenon.
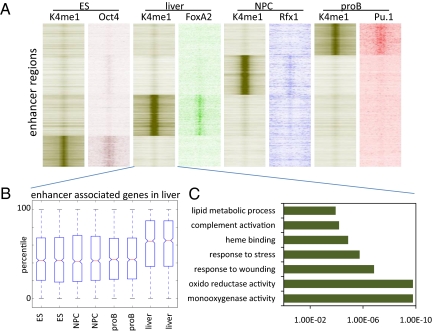
We next generated chromatin state maps for H3K4me1 and H3K4me3 and H3K27ac enrichment in proB cells, neural progenitor cells (NPCs), and adult liver to further test the relationship between this modification and active enhancers (Table S1). We identify a total of 94,437 different putative enhancer regions on the basis of distal H3K4me1 enrichment (Fig. 2A and Fig. S1). H3K4me1 and H3K4me3 datasets in NPCs were adapted from previous work (17). The genomic distribution of these sites was highly cell type specific (Pearson correlation, between 0.01 and 0.19), and genes that were associated with distal H3K4me1 in a single cell type displayed enhanced transcriptional activity in that specific cell type [Fig. 2B (e.g., liver) and Fig. S4]. Furthermore, these particular genes correlated well with tissue-specific function [Fig. 2C (e.g., liver)]. Motif search analysis on conserved DNA sequence at these cell type-specific enhancers identified DNA binding motifs for the known enhancer regulators Foxa2 (P = 5.3E-18) in liver and PU.1 (P = 5.2E-06) in proB cells (7, 23) (Fig. S5). Examination of ChIP-Seq data (7, 24) confirmed that both Foxa2 and PU.1 occupy these enhancer regions in the corresponding adult tissues (P = 7.53E-147; Fig. 2A and Fig. S5), providing support for the robustness of these putative enhancer data sets. In neural progenitors, we determined that a cis-regulatory element called the x-box that is recognized by the transcription factor Rfx (25) was highly enriched at enhancers (P = 4.5E-125). Interestingly, Drosophila RFX is required for sensory neuron differentiation (26). Because mammalian Rfx proteins have not been previously linked to global enhancer patterning, we performed ChIP-Seq using antibodies specific for Rfx1 in neural progenitors (Table S1) and found that Rfx1 is selectively enriched at neural precursor enhancers (P = 8.9E-139). This suggests that, similar to Foxa2 in liver and PU.1 in proB cells, Rfx family members are involved in enhancer patterning during neural development.
Fig. 2.
Distal putative enhancers (n = 94,437) identified according to distal H3K4me1 enrichment in different tissue types reveals cell-specific distributions. (A) Heatmap displaying k-means clustering of ChIP-Seq datasets for H3K4me1 and the indicated transcription factors. Clusters are generated according to H3K4me1 enrichment at the enhancer regions. The different transcription factor data sets are arranged to match the order of enhancers found by clustering H3K4me1. Four-kilobase pairs around the enhancer peaks are displayed. (B) Gene expression of microarray data generated in duplicates from the different tissues as displayed (x axis). Box-plots show genes found specifically associated with enhancers in liver. The different boxes display how these genes behave in the different cell types tested as shown along the axis. Solid bars of boxes display the 25–75% of ranked genes, with the mean indicated as an intersection. (C) GO analysis displaying gene functions for genes specifically associated with enhancer activity in adult liver according to H3K4me1 enrichment but not associated with distal H3K4me1 enriched elements in the other cell types tested.
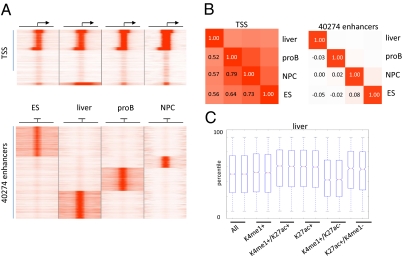
We next analyzed the chromatin state maps for H3K27ac enrichment (Fig. 3A and Table S1), identifying a total of 40,274 enriched promoter distal regions according to the same criteria used to identify distal H3K4me1 regions. Similar to H3K4me1-enriched regions, cell-specific patterning was observed for the H3K27ac mark at identified enhancer sites (Fig. 3B; correlation between −0.05 and 0.08). These data show that distal H3K27ac enrichment is at least as diverse between cell types as distal H3K4me1 enrichment. Similarly to H3K4me1, the majority of distal H3K27ac enrichment was found within introns and intergenic regions at a roughly equal distribution. In agreement with our findings in ES cells, we find that the activity of proximal genes correlates positively with distal H3K27ac enrichment in all adult tissues [Fig. 3C (e.g., liver) and Fig. S4] and that H3K4me1-enriched distal regions that are void of H3K27ac do not correlate with enhanced gene activity. Thus, the finding that H3K27ac distinguishes active from inactive enhancers seems to be a general phenomenon. We were able to identify known active enhancers in a tissue-specific manner, such as for example the Eμ enhancer of the Ig heavy chain locus in proB cells (27), as well as the 3′ enhancer in the same region by using H3K27ac as a marker (Fig. S6). Notably two of the transcription factors identified by motif analysis in the adult cell types tested also correlate well with more active enhancers being PU.1 in proB cells (P < 3.67E-4) and Rfx1 in NPCs (P < 6.0E-4) (Fig. S5). Enhancer elements negative for these factors were, however, not consistently different from average enhancers. This indicates that select transcription factors mark specific subsets of active enhancers.
Fig. 3.
Enhanced proximal gene activity of distal H3K4me1-enriched regions is a function of H3K27ac. (A) Heatmap of H3K27ac enrichment at known TS sites (Upper) and 40,274 distal enhancers according to H3K27ac enrichment and H3K4me3 absence (Lower) in the four indicated cell/tissue types showing tissue-specific distribution. Four-kilobase pairs around the enhancer peaks are displayed. (B) Correlation analysis for the H3K27ac-enriched regions shown in A. Color intensities are a measure of correlation (Pearson), which is also indicated by numbers. Left: Correlation for H3K27ac at known transcriptional start sites. Right: Correlation for the 40,274 distal H3K27ac-enriched regions found in the four cell types combined. (C) Gene expression of microarray data generated in duplicate from adult liver. Box-plots show all genes (All) and genes found specifically associated with enhancers in liver either enriched (+) or unenriched (−) for H3K4me1 or H3K27ac. A hierarchical model corrects for multiple enhancers being associated with single genes (SI Materials and Methods). Solid bars of boxes display the 25–75% of ranked genes, with the mean indicated as an intersection.
Global Enhancer Patterns Are Reset During Nuclear Reprogramming.
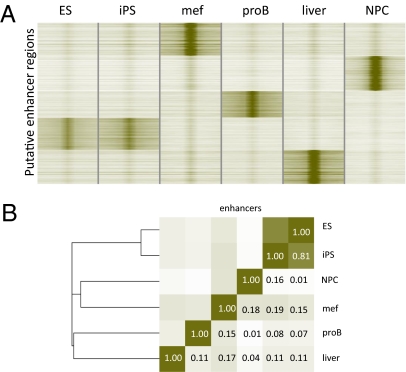
Previous studies have shown that the bivalent chromatin conformation comprising H3K27me3 and H3K4me3 marks are reset in induced pluripotent stem (iPS) cells (28). To test whether enhancer elements are also reset we generated chromatin state maps for H3K4me1 in fibroblasts and fibroblast-derived iPS cells (Table S1). We used the iPS cell line that was previously shown to express all markers of pluripotency, give rise to fully developed embryos by tetraploid complementation, and contribute to the germ line (29). We found that enhancer distribution was indeed reset to the ES cell configuration in iPS cells (Fig. 4 A and B; correlation, 0.81) relative to the parental fibroblasts (correlation, 0.19). This result again supports the notion that proper reestablishment of the histone marks that pattern enhancer networks is important for changes in cell fate determination.
Fig. 4.
Global enhancer networks are reset during nuclear reprogramming. (A) Heatmap of 118,935 distal enhancers based on H3K4me1 enrichment (green) and H3K4me3 depletion in the six indicated cell/tissue types showing tissue-specific enhancer distribution. Four-kilobase pairs around the center of the enhancer region are displayed. (B) Correlation analysis for the H3K4me1-enriched regions shown in A across the whole set of enhancer regions. Color intensities are a measure of correlation (Pearson), which is also indicated by numbers.
Distal H3K27ac Enrichment Is a Good Predictor of Current Developmental State.
Because there seem to be two classes of enhancers, active (H3K27ac+) and poised (H3K27ac-), this raises the intriguing possibility that enhancers contain information about the current and future developmental potential of a cell, as well as its ability to respond to external cues. To test this hypothesis, we determined the functions associated with enhancer proximal genes using Gene Ontology (GO) analysis. We analyzed distal enhancers according to H3K4me1 and H3K27ac enrichment separately and found similar enriched annotations associated with the adult developmental state of the different cell types, although with higher confidence for H3K27ac than found for H3K4me1. We also uncovered additional functions that could not be classified by H3K4me1 association alone, such as developmental processes (P = 4.5E-4) and embryonic development (P = 7.1E-3) in ES cells. This suggests that using proximal genes to H3K27ac-positive enhancers can more accurately predict developmental state than H3K4me1 alone. Interestingly, we also found developmental functions that were more specific for genes proximal to the H3K4me1 mark, such as activation of plasma proteins in response to acute inflammatory response in liver (Fig. S7), which is a function of the liver in response to an external stimulus. Typically, we would expect such a function to be supported by enhancers that are poised for activation under normal conditions until an inflammatory stimulus is given. This idea is consistent with the presence of the distal H3K4me1 enhancer mark but the absence of enhanced proximal gene expression associated with these marks (Fig. 3D) and thus supports the idea that these enhancers exist in a poised state.
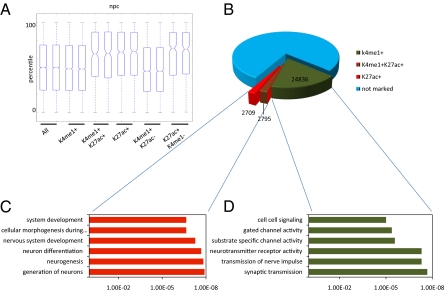
In neural progenitors the diversity between GO functions found for H3K4me1 proximal genes and H3K27ac proximal genes was most pronounced (Fig. 5). We found that GO annotations for genes could be faithfully split in two groups: functions associated with enhancers that are H3K4me1 positive and H3K27ac negative and functions associated with enhancers that are H3K27ac positive (Fig. 5B). These two groups displayed two very distinct sets of GO functions (Fig. 5 C and D) with essentially no overlap. Functions related to an adult neuronal state, such as synaptic transmission and neurotransmitter receptor activity, were found while interrogating genes associated with silent H3K4me1-based enhancers (Fig. 5D), whereas the more active H3K27ac-associated genes generated GO functions that are more typical of a multipotent stem cell, such as anatomical structure development and nervous system development (Fig. 5C). Given the fact that neural progenitors function as precursors of the neural lineages that can give rise to many adult cell types in the brain, such as adult neurons, oligodendrocytes, and astrocytes (30), it fits that the genes proximal to active H3K27ac-positive enhancers reflect such a progenitor state. Conversely, genes proximal to H3K27ac-negative enhancer regions reflect the future more committed state of these cells, which again supports the notion that these enhancers exist in a predetermined state.
Fig. 5.
Poised and active enhancers can discriminate between current and future developmental states. (A) Gene expression of microarray data generated in duplicate from neural progenitors. Box-plots show all genes (All) and genes found specifically associated with enhancers in neural progenitors either enriched (+) or unenriched (−) for H3K4me1 or H3K27ac. A hierarchical model corrects for multiple enhancers being associated with single genes (SI Materials and Methods). Solid bars of boxes display the 25–75% of ranked genes, with the mean as an intersection. (B) Pie chart showing the distribution of H3K4me1 marked (green) and H3K27ac marked (red) enhancers in neural progenitors as indicated. In blue are unmarked enhancers. Total enhancers are based on combining distal H3K4me1- and H3K27ac-enriched regions totaling 136,397 regions enriched. Lines indicate the section of enhancers used to derive the genes for GO analysis in C and D. (C and D) GO analysis displaying gene functions for genes specifically associated with enhancer activity in neural progenitors split according to either H3K4me1 enrichment but absence of H3K27ac enrichment (C, green bars) or H3K27ac enrichment (D, red bars). Displayed is a representation of GO functions found.
Select Poised Enhancers Are Activated During Differentiation to Support the New Developmental State.
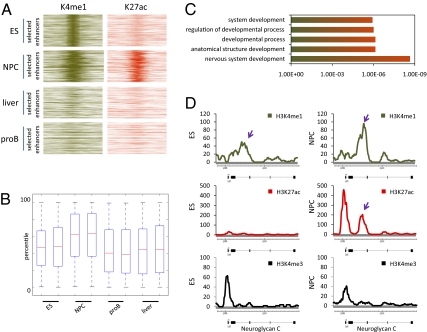
To test whether poised enhancers that are activated during lineage commitment would be indicative of a new developmental state, we used data from ES cells and NPCs, which can be directly derived from ES cells in culture (30). On the basis of our model, we predict that ES cells would contain poised enhancers that become active (H3K27ac+) in neural progenitors (Fig. 7). Proximal genes to these enhancers should become selectively activated and reflect the new developmental state of the neural progenitors. We were indeed able to identify poised enhancers in ES cells that became specifically activated (H3K27ac+) in neural progenitors but not in liver or proB cells (Fig. 6A). In agreement with these data we found that proximal genes to these enhancers were selectively activated in neural progenitors but not in any of the other cell types (Fig. 6B). Finally, we performed GO analysis on these genes and found functions such as nervous system development and differentiation indicating that they reflect the newly acquired cellular function (Fig. 6C). Neuroglycan-C (Fig. 6D) and Neurophilin-2 (Fig. S8) are two such genes that harbor poised enhancers in ES cells that become active in neural progenitors. These data support the notion that enhancers play a deterministic role in cellular differentiation by activating poised enhancers when a cell switches developmental state and that H3K27ac can be used to discriminate between the two enhancer states.
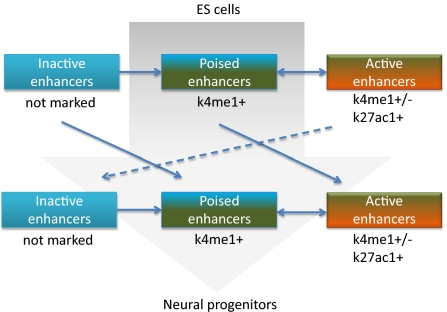
Fig. 7.
Model for regulation of poised enhancers during differentiation to a new developmental state. While the cell is locked in a differentiation state it can activate poised enhancers in response to external stimuli. The repertoire of poised enhancers determines which responses are available to the cell.
Fig. 6.
Poised enhancers become activated during differentiation to activate key genes that drive the new developmental state. (A) Heatmap of 361 enhancers that are H3K4me1 enriched (Left, green) but negative for H3K27ac in ES cells and gain H3K27ac enrichment in neural progenitors (Right, orange) for the different cell types displayed. Four-kilobase pairs around the enhancer region are displayed. (B) Gene expression of microarray data generated in duplicate in the different cell types indicated (x axis) for the genes proximal to the 361 enhancers that become H3K27ac positive during differentiation from an ES cell to a neural progenitor. Box-plots show expression of these genes in the indicated cell types. Solid bars of boxes display the 25–75% of ranked genes, with an intersection as the mean. (C) GO analysis displaying gene functions for the genes in B that are specifically associated with enhancers that are poised in ES cells (H3K4me1+/H3K27ac-) and become active (H3K27ac+) in neural progenitors. (D) Gene track for H3K4me1 (green, Top), H3K27ac (red, Middle), and H3K4me3 (black, Bottom) for a 20,792-bp large region containing the Neuroglycan C gene. Tracks are shown in ES cells (Left) and NPCs (Right). Arrows indicate a poised enhancer that gains the H3K27ac mark. y axis shows number of reads.
Discussion
Here we have identified a composite set of ≈135,000 putative enhancer elements in embryonic stem cells and several adult cell types using both distal H3K4me1- and distal H3K27ac-enriched regions for global enhancer discovery. We show that the genomic location of marked enhancer regions is highly diverse between the different cell types tested, and proximal genes to these regions are not only more active but also reflect the associated developmental state of the cell. Additionally, we show that enhancers are faithfully redistributed to an ES cell-like pattern during reprogramming of fibroblasts into iPS cells. These results offer further support for the importance of global enhancer networks in establishing cellular identity.
H3K27ac is one of the modifications that were shown to be present at a subset of enhancers (16), yet its relevance was unclear. We establish the H3K27ac modification as an important mark for enhancer identification that can distinguish between active and inactive enhancer regions according to the expression of proximal genes and preferential eRNA production. Because distal H3K27ac-enriched regions are less abundant than distal H3K4me1-enriched regions, these data suggest that the amount of enhancers actively used by a cell is significantly less than was previously postulated, on the basis of H3K4me1.
The fact that distal enhancers without H3K27ac do not correlate with more active proximal gene expression as compared with average genes suggests these elements exist in a poised state ready to become active after external cues. Data that support the existence of poised enhancers have been previously presented (5, 12, 13). By using H3K27ac we can now discriminate between the two states on a global scale, thus producing more general information on current and possible future developmental states (Fig. 7).
Some transcription factors bound to enhancers, such as Rfx1 in neural precursors, also correlated with enhanced gene activity, whereas others correlated with less active proximal gene activity. Proximal genes to transcription factor-negative enhancers were in all cases not significantly different from average enhancers, suggesting that these transcription factors bind subsets of active or inactive enhancers. This suggests that enhancers can be subdivided into multiple different functional classes involved at different stages of enhancer activation and could be used to identify poised enhancer networks that respond to more specific signals. Interestingly, Oct4 and Sox2 both associated with less active enhancer elements in ES cells, suggesting that they might be involved in keeping developmental regulators in a poised state, similar to polycomb group proteins at promoters.
Finally, genome-wide association studies have identified disease-associated polymorphisms that reside outside of coding regions (31). Global enhancer identification across different tissues could identify subsets of these polymorphisms as enhancer elements, thus providing valuable insight into how some of these regions cause disease in a tissue-specific manner.
Materials and Methods
Full details can be found in SI Materials and Methods. Briefly, cells or tissues were chemically cross-linked and sonicated. After clearance, IPs were performed overnight using previously established antibodies (Abcam: H3K4me3 ab8580, H3K4me1 ab8895, H3K27ac ab4729, and H3 ab1791; Santa Cruz Biotechnology: Rfx1 sc-10652X and p300 sc-585). Samples were run using the GA2X genome sequencer (SCS v2.6, pipeline 1.5). Raw data are available online at the Gene Expression Omnibus ascension viewer series (accession nos. GSE24164, GSE24165, and GSE23907).
Supplementary Material
Acknowledgments
We thank Alla Leshinski, Charlie Whittaker, and Dick Cook at the Massachusetts Institute of Technology Koch Institute for running the Solexa pipeline; Jeong-Ah Kwon and Jennifer Love at the Genome Technology Core for help with the microarrays; Thijn Brummelkamp for comments and suggestions; and Frank Soldner at the Whitehead Institute for Biomedical Research in Cambridge for EDTA. This work was supported by National Institutes of Health Grants 5-RO1-CA87869, 5-RO1-HDO45022, and 5-R37-CA084198 (to R.J.) and HG002668 (to R.A.Y.); by US Public Health Service Grant PO1-CA42063 from the National Cancer Institute (to T.K. and P.A.S.); and partially by Cancer Center Support (core) Grant P30-CA14051 from the National Cancer Institute (to T.K. and P.A.S.). A.W.C. was supported by a Croucher scholarship.
Footnotes
The authors declare no conflict of interest.
Data deposition: Raw data are available online at the Gene Expression Omnibus (GEO) ascension viewer series, www.ncbi.nlm.nih.gov/geo (accession nos. GSE24164, GSE24165, and GSE23907).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1016071107/-/DCSupplemental.
See Commentary on page 21240.
This article is a PNAS Direct Submission.
References
- 1.Heintzman ND, Ren B. Finding distal regulatory elements in the human genome. Curr Opin Genet Dev. 2009;19:541–549. doi: 10.1016/j.gde.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visel A, Rubin EM, Pennacchio LA. Genomic views of distant-acting enhancers. Nature. 2009;461:199–205. doi: 10.1038/nature08451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bulger M, Groudine M. Looping versus linking: Toward a model for long-distance gene activation. Genes Dev. 1999;13:2465–2477. doi: 10.1101/gad.13.19.2465. [DOI] [PubMed] [Google Scholar]
- 4.Boffelli D, Nobrega MA, Rubin EM. Comparative genomics at the vertebrate extremes. Nat Rev Genet. 2004;5:456–465. doi: 10.1038/nrg1350. [DOI] [PubMed] [Google Scholar]
- 5.Heintzman ND, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghisletti S, et al. Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity. 2010;32:317–328. doi: 10.1016/j.immuni.2010.02.008. [DOI] [PubMed] [Google Scholar]
- 7.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim TK, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Birney E, et al. ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children's Hospital Oakland Research Institute. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xi H, et al. Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 2007;3:e136. doi: 10.1371/journal.pgen.0030136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visel A, et al. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature. 2009;457:854–858. doi: 10.1038/nature07730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, et al. Transcriptional competence and the active marking of tissue-specific enhancers by defined transcription factors in embryonic and induced pluripotent stem cells. Genes Dev. 2009;23:2824–2838. doi: 10.1101/gad.1861209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui K, et al. Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell. 2009;4:80–93. doi: 10.1016/j.stem.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tie F, et al. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development. 2009;136:3131–3141. doi: 10.1242/dev.037127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang Z, et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet. 2008;40:897–903. doi: 10.1038/ng.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heintzman ND, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet. 2007;39:311–318. doi: 10.1038/ng1966. [DOI] [PubMed] [Google Scholar]
- 17.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marson A, et al. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell. 2008;134:521–533. doi: 10.1016/j.cell.2008.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 20.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 21.Creyghton MP, et al. H2AZ is enriched at polycomb complex target genes in ES cells and is necessary for lineage commitment. Cell. 2008;135:649–661. doi: 10.1016/j.cell.2008.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seila AC, et al. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zaret KS, et al. Pioneer factors, genetic competence, and inductive signaling: Programming liver and pancreas progenitors from the endoderm. Cold Spring Harb Symp Quant Biol. 2008;73:119–126. doi: 10.1101/sqb.2008.73.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wederell ED, et al. Global analysis of in vivo Foxa2-binding sites in mouse adult liver using massively parallel sequencing. Nucleic Acids Res. 2008;36:4549–4564. doi: 10.1093/nar/gkn382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reith W, et al. Congenital immunodeficiency with a regulatory defect in MHC class II gene expression lacks a specific HLA-DR promoter binding protein, RF-X. Cell. 1988;53:897–906. doi: 10.1016/s0092-8674(88)90389-3. [DOI] [PubMed] [Google Scholar]
- 26.Dubruille R, et al. Drosophila regulatory factor X is necessary for ciliated sensory neuron differentiation. Development. 2002;129:5487–5498. doi: 10.1242/dev.00148. [DOI] [PubMed] [Google Scholar]
- 27.Perlot T, Alt FW. Cis-regulatory elements and epigenetic changes control genomic rearrangements of the IgH locus. Adv Immunol. 2008;99:1–32. doi: 10.1016/S0065-2776(08)00601-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maherali N, et al. Directly reprogrammed fibroblasts show global epigenetic remodeling and widespread tissue contribution. Cell Stem Cell. 2007;1:55–70. doi: 10.1016/j.stem.2007.05.014. [DOI] [PubMed] [Google Scholar]
- 29.Wernig M, et al. In vitro reprogramming of fibroblasts into a pluripotent ES-cell-like state. Nature. 2007;448:318–324. doi: 10.1038/nature05944. [DOI] [PubMed] [Google Scholar]
- 30.Conti L, et al. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.